Abstract
Purpose:
To (a) create a survival risk-score using radiomic features from the tumor habitat on routine MRI to predict progression-free survival (PFS) in Glioblastoma, and (b) obtain a biological basis for these prognostic radiomic features, by studying their radio-genomic associations with molecular signaling pathways.
Experimental Design:
203 patients with pre-treatment Gd-T1w, T2w, T2w-FLAIR MRI were obtained from 3 cohorts: TCIA (n=130), Ivy-GAP (n=32), and Cleveland Clinic (n=41). Gene expression profiles of corresponding patients were obtained for TCIA cohort. For every study, following expert segmentation of tumor sub-compartments (necrotic-core, enhancing tumor, peri-tumoral edema), 936 3D-radiomic features were extracted from each sub-compartment across all MRI protocols. Using Cox regression model, radiomic risk score (RRS) was developed for every protocol to predict PFS on the training cohort (n=130) and evaluated on the hold-out cohort (n=73). Further, Gene Ontology and single-sample Gene Set Enrichment Analysis was used to identify specific molecular signaling pathway networks that were associated with RRS features.
Results:
25 radiomic features from the tumor habitat yielded the RRS. A combination of RRS with clinical (age, gender) and molecular features (MGMT, IDH status) resulted in a concordance index of 0.81 (p <0.0001) on training and 0.84 (p = 0.03) on the test set. Radiogenomic analysis revealed associations of RRS features with signaling pathways for cell differentiation, cell adhesion, and angiogenesis, that contribute to chemo-resistance in GBM.
Conclusions:
Our findings suggest that prognostic radiomic features from routine Gd-T1w MRI may also be significantly associated with key biological processes that impact response to chemo-therapy in GBM.
Keywords: Glioblastoma, Radiomics, Radiogenomics, risk-stratification, Progression Free Survival, Magnetic Resonance Imaging
Statement of Translational Relevance
Progression-free survival (PFS) in Glioblastoma (GBM) remains a surrogate end-point for patient’s response to standard or experimental treatments. However, currently there are no clinically-validated prognostic biomarkers for predicting PFS in GBM. In this work, we attempt to address this gap in knowledge by developing a radiomic risk score (RRS) using radiomic features from the tumor habitat (i.e. necrotic, enhancing tumor, edema) to predict PFS on pre-treatment MRI. Further, in an attempt to establish a biological basis for these prognostic radiomic features, we investigate their associations with corresponding gene expression data using Gene Ontology and single-sample Gene Set Enrichment Analysis. Our prognostic RRS consisted of 25 radiomic features from the tumor habitat on Gd-T1w MRI. These features were found to be significantly associated with cell differentiation, cell adhesion, and angiogenesis signaling pathways. With additional validation, such radiogenomic analysis may allow for identification of patients who may be candidates for targeted therapies.
1. INTRODUCTION
Glioblastoma (GBM) is a highly aggressive, rapidly fatal, and the most common primary malignant tumor in the brain. Current standard of care treatment for GBM is maximal safe surgical resection followed by Stupp protocol - cranial radiation therapy and concomitant chemotherapy, with temozolomide (TMZ).1 Despite this aggressive and multimodal treatment, median survival has only slightly improved to approximately 15 months, with less than 25% of patients surviving up to 2 years, and less than 10% surviving for over 5 years.2 This poor prognosis can be attributed to genetic instability and intra- and inter-tumor heterogeneity of GBM that leads to treatment resistance, progression, and tumor recurrence. 3,4
Recent investigative studies have identified several driver mutations (i.e., IDH - Isocitrate dehydrogenase), chromosomal anomalies (i.e. 1p19q chromosome deletions),5 epigenetic alterations (such as MGMT - O-6-Methylguanine-DNA Methyltransferase)6 as specific targets for personalizing anticancer therapeutics, enhancing treatment response, and improving survival. While these genomic biomarkers have shown some prognostic value in GBM, biopsies are tissue destructive, rely on a small sample of tissue that do not account for the inherent intra-tumoral heterogeneity extant in GBMs, and are usually expensive and time-consuming to perform during routine screening. With almost 40% of GBM patients diagnosed with cancer recurrence within 6 months even after aggressive treatment management, alternate treatment options such as anti-angiogenesis therapies, immunotherapy vaccinations and clinical trials could be more suited in patients that may be at high risk of tumor recurrence, to achieve prolonged progression free survival (PFS).7–9 There is hence a need to develop imaging based biomarkers that are prognostic of patient outcome to potentially assist in building comprehensive and patient-centric treatment plans.
Multiple studies suggest that tumor-associated cellular alterations are not just limited to the visible tumor enhancement of GBM, but infiltrate beyond the tumor margins into the peri-tumoral brain zone.10 It is also known that 90% of GBM tumors recur in the peri-tumoral region and also the inner necrotic compartments.11 Radiomics has provided a surrogate mechanism to non-invasively characterize the tumor by capturing sub-visual cues of morphologic diversity (e.g. roughness, image homogeneity, regularity and edges) on routine MRI scans for diseases diagnosis, patient prognosis and treatment response.12 Recently, a few research groups have demonstrated the added prognostic value in investigating radiomic features, not just from within these visible tumor margins, but also from the surrounding peri-tumoral regions (i.e. edema) as well as the features from necrotic core.13–15 This concept of “tumor habitat” involves interrogating the radiomic features from the enhancing tumor as well as the necrotic core, and peri-tumoral edema sub-compartments across different MRI protocols.
While a few recent studies have attempted to develop prognostic radiomic based models from the tumor habitat to predict progression-free and overall survival in GBM, there has been limited work on creating a reliable survival risk assessment model using radiomic features from the tumor habitat, using a large multi-institutional cohort. Further, a more critical missing link in previous work has been on identifying the molecular associations of habitat-specific radiomic features with the underlying signaling pathways that drive different biological processes, via radiogenomic analysis. Improved understanding of how the changes in biological processes at the molecular level impact changes at a radiological scale, could help drive adoption of the prognostic radiomic features in a clinical setting. Further, such cross-scale associations of radiomic-based prognostic markers with molecular processes in GBM could allow for designing patient-centric treatments based on their risk profile for poor survival.
In this work, we have two objectives. Firstly, we seek to create a prognostic radiomic risk score (RRS) using radiomic features from different sub-compartments of the tumor habitat on routine multi-parametric MRI protocols (i.e. Gd-T1w, T2w, T2w-FLAIR), to risk-stratify GBM patients based on their PFS. The RRS model will be trained using a multi-institutional training cohort of n=130 patients via least absolute shrinkage and selection operator (LASSO) Cox regression model. We will then evaluate the ability of this RRS to stratify an independent test cohort (n=73) into two groups of ‘low-risk’ and ‘high-risk’ of poor survival, based on the fixed median cut-off value of the RRS. In our second objective, in an attempt to provide a biological basis for the radiomic features driving the RRS, we will seek to identify radiogenomic correlations between the most prognostic radiomic features from different sub-compartments of the tumor habitat with molecular signaling pathway networks in GBM using Gene Ontology (GO) and single-sample Gene set enrichment analysis (ssGSEA).16–18 Figure 1B illustrates an overview of our framework.
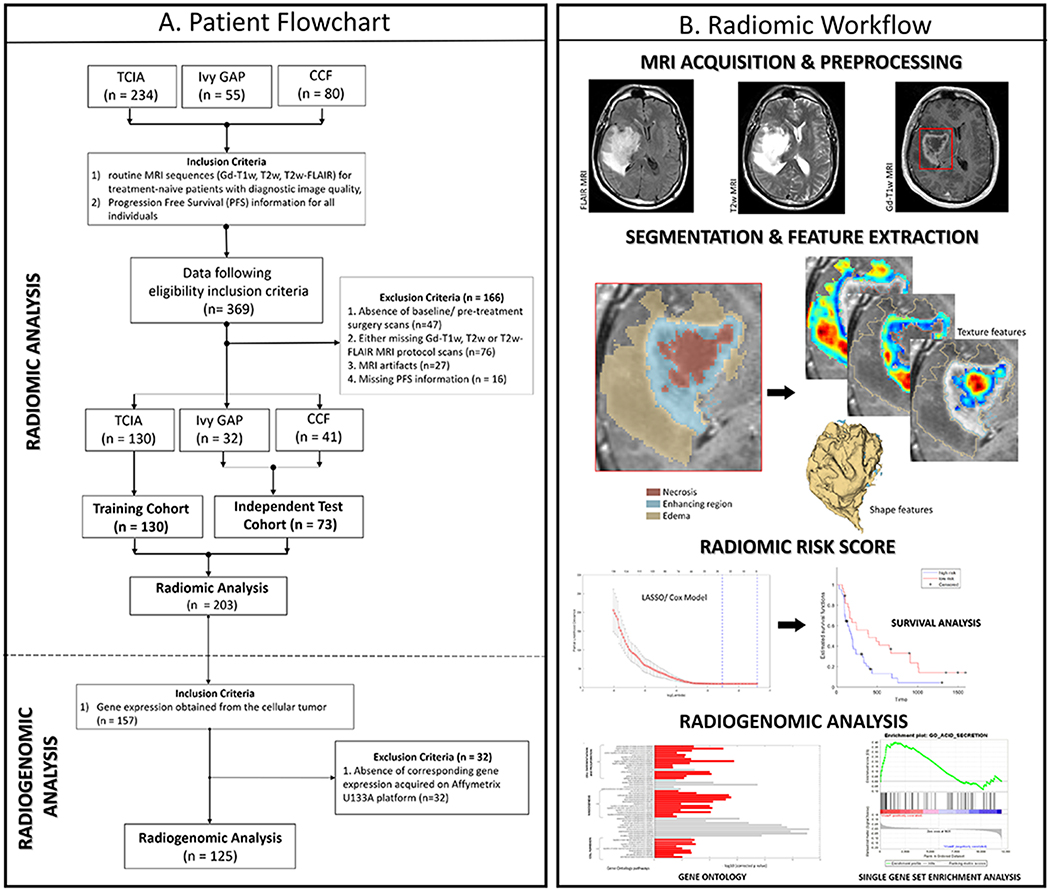
Figure 1:
(A) Flow diagram of patient enrolment, eligibility and exclusion criteria of the dataset (B) Radiomic Workflow
2. MATERIALS AND METHODS
2.1. Study Population
Our retrospective cohort consisted of treatment-naive multi-parametric MRI scans from three cohorts. Two of the three datasets were curated from publicly available datasets - The Cancer Imaging Archive (TCIA)19,20 and The Ivy Glioblastoma Atlas Project (Ivy GAP).21,22 TCIA is an open archive of cancer-specific medical images and associated clinical metadata established by the collaboration between the National Cancer Institute (NCI) and participating institutions in the United States. Similarly, Ivy-GAP is freely accessible online data resource that aims to identify important morphologic hallmarks of GBM.
The third dataset was curated from a Health Insurance Portability and Accountability Act (HIPAA) compliant and institution review board (IRB) approved participating institution – Cleveland Clinic Foundation (CCF), where the need for an informed consent from all patients was waived. Between December 1st, 2011 and May 1st, 2018, radiology image archives of this participating institution were searched to identify 80 GBM patients as confirmed via histopathology. Further information on the images acquired can be found in Supplementary Section 1.
The retrospectively collected 369 GBM cases from three sites were further triaged using inclusion criteria that involved the availability of 1) routine MRI sequences (Gd-T1w, T2w, T2w-FLAIR) for treatment-naive patients with diagnostic image quality and 2) PFS information for all individuals. A total of 166 patients with the following criteria were excluded: (i) absence of baseline/ pre-treatment surgery scans (n=47), (ii) missing either Gd-T1w, T2w or T2w-FLAIR MRI protocol scans (n=76), (iii) presence of MRI artifacts (n=27) and (iv) missing PFS information (n = 16). Following the patient exclusion criteria, 203 patients were enrolled for the radiomic analysis. The patient enrolment flowchart is shown in Figure 1A.
Of the 203 GBM patients that followed the inclusion/exclusion criteria for radiomic analysis, 130 patients from TCIA cohort comprised our training cohort and the remaining 73 cases were used as independent test cohort. Further, from the 203 GBM cases, 157 tumors were selected for radiogenomic analysis after applying an additional inclusion criterion that required RNA sequencing data obtained from cellular tumor. A total of 32 GBM tumors were further excluded from the study due to the absence of corresponding gene expression data acquired on Affymetrix U133A platform. Finally, a total of 125 GBM tumors were investigated for the radiogenomic analysis.
Multi-site data distribution has been reported in Table 1A. Our data selection for training and testing was performed in a randomized fashion, while ensuring that the distribution of patient demographics such as clinical factors (patient’s age at diagnosis, gender), and molecular characteristics (MGMT promoter methylation status, IDH status) were balanced between the training and test sets (2:1 ratio with n = 130 in the training and n = 73 in the independent test set). The clinical characteristics and treatment follow-up of the patients within the training and test cohort have been summarized in Table 1B. Within the training cohort, the extent of resection (gross total resection [GTR] or near total resection [NTR] or sub-total resection [STR]) was not known in 106 cases. From the test cohort (n=73), 40 GBM cases had undergone GTR, 12 GBM cases underwent NTR, and 12 GBM tumors underwent STR. 5 cases within the test cohort underwent laser ablation.
Table 1:
(A) MRI Data acquisition sites for the Training cohort – TCIA. (B) Patient demographics for the training and test cohort.
| A. DATA DISTRIBUTION | ||||||
|---|---|---|---|---|---|---|
| Training Cohort (n = 130) | Independent Test Cohort (n= 73) | |||||
| Site | MR Imaging | Genomics | Site | MR Imaging | Genomics | |
| TCIA | MD Anderson | 24 | 22 | Ivy GAP | 32 | 29 |
| Henry Ford Hospital | 46 | 46 | CCF | 41 | 0 | |
| UCSF | 23 | 23 | ||||
| Duke | 9 | 9 | ||||
| Emory | 9 | 9 | ||||
| Case Western Reserve | 10 | 10 | ||||
| Thomas Jefferson | 9 | 7 | ||||
| Total cases | 130 | 125 | Total cases | 73 | 29 | |
| B. PATIENT DEMOGRAPHICS | ||||||
| Clinical Parameter | Training Cohort | Test Cohort | p-value | |||
| Age (Mean, Range in years) | 57.9 (19 – 84.8) | 58.2 (26 – 77) | 0.7597 | |||
| Gender (n, %) | Female | 47 (36%) | 31 (42%) | 0.4524 | ||
| Male | 83 (64%) | 42 (58%) | ||||
| Mean Progression Free Survival (PFS, in months) | 9.5 | 10.8 | 0.3710 | |||
| Therapy class (n, %) | RT + TMZ | 98 (75%) | 73 (100%) | |||
| RT | 32 (25%) | 0 (0%) | ||||
| TMZ | 0 (0%) | 0 (0%) | ||||
| Extent of resection [ EOR ¤ ] (n, %) | Tumor resected † | 106 (81.5%) | 3 (4%) | |||
| Gross total resection (GTR) | N/A | 40 (54.7%) | ||||
| Near total resection (NTR) | N/A | 12 (16.4%) | ||||
| Sub-total resection * (STR) | 16 (12.3%) | 12 (16.4%) | ||||
| Biopsy | 8 (6%) | 1 (1.3%) | ||||
| Laser Ablation | 0 (0%) | 5 (6.8%) | ||||
| MGMT Status (n, %) | Methylated | 39 (30%) | 32 (44%) | 0.398 | ||
| Unmethylated | 32 (25%) | 37 (51%) | ||||
| Information Unavailable | 59 (45%) | 4 (5%) | ||||
| IDH Status (n, %) | Wild type | 102 (79%) | 56 (77%) | 0.1484 | ||
| Mutated | 6 (4%) | 8 (10%) | ||||
| Information Unavailable | 22 (17%) | 9 (13%) | ||||
% - percentage values have been rounded off by one decimal. P values were computed by using Student t-test for continuous variable and Fisher exact test for categorical data. Abbreviations: MGMT - O-6-Methylguanine-DNA Methyltransferase, IDH - Isocitrate dehydrogenase. EOR ¤ : Gross total resection = No evidence of enhancing tumor in post-surgery MR imaging scans, Near total resection = > 90% tumor resection, Sub-total resection = 51-90% tumor resection .
Extent of tumor resection (GTR/NTR/STR) unknown for most of the TCIA cases.
Sub-total resection status information was available for the training cohort (TCGA-GBM) only in the cases curated from Henry Ford Hospital.
No statistically significant difference was found between the training and test cohorts with respect to the age (p = 0.75), gender (p = 0.45), and PFS (p = 0.37).
2.2. Post-acquisition pre-processing and Segmentation
Pre-processed TCIA cases along with annotations on every 2D slice were obtained from Bakas et al.,23,24 Cases obtained from Ivy GAP and CCF were pre-processed using the same pipeline as the TCIA cohort.25 For every study, MR images were reoriented to the LPS (left-posterior-superior) coordinate system. All MRI protocols were then co-registered to the T1w anatomical template of SRI24 Multi-Channel Normal Adult Human Brain Atlas via affine registration and resampled to 1mm3 voxel resolution.26 Subsequent skull-stripping was implemented on all cases.25 N4-bias correction algorithm27 was not implemented in our pre-processing pipeline as it known to obliterate the T2w-FLAIR signal.23 However, a low level image processing smoothing filter, Smallest Univalue Segment Assimilating Nucleus (SUSAN) was used to reduce high frequency intensity variations of noise.28 Lastly, we corrected the MRI protocols for intensity non-standardness, that refers to the issue of MR image “intensity drift” across different imaging acquisitions. The intensity non-standardness results in MR image intensities lacking tissue-specific numeric meaning within the same MRI protocol, for the same body region, or for images of the same patient obtained on the same scanner. Therefore, intensity standardization was implemented in MATLAB R2014b (Mathworks, Natick, MA) using the method presented in Madabhushi et al., 29
Every GBM tumor was defined into 3 sub-compartments of the tumor habitat: (1) tumor necrosis, (2) enhancing region of the tumor, and (3) peri-tumoral edema. Gd-T1w MR images were used to delineate hypo-intense signals to capture the necrotic core of GBM tumor, usually located in the central region of the tumor. Similarly, enhancing region of GBM is represented as hyper-intense regions on Gd-T1w MR images when compared pre-contrast T1w. When pre-contrast T1w scans were not available, T2w images and T2w-FLAIR scans were used to evaluate the presence of blood.30 T2w and T2w-FLAIR scans were also used to identify edema and necrotic core.
The independent test cohort, that consisted of Ivy GAP and CCF cases, was manually annotated using 3D Slicer software.31 Every 2-D slice of each MRI scan with visible tumor was manually annotated by the expert readers. A total of three experts were asked to perform the manual annotations on a total of 73 test studies. The senior-most expert (expert 1, V.H, >10-years of experience in neuroradiology) independently annotated the studies obtained from IVY Gap, while expert 2 (V.S) with 7 years of experience in neuroradiology supervised expert 3 (K.B, with 3 years of radiology experience), to manually annotate the CCF cases from the test set. In rare cases with disagreement across the two readers (expert 2 and expert 3), the senior-most radiologist (V.H, expert 1) was consulted to obtain the final segmentations.
2.3. 3D Texture feature extraction from multi-parametric MRI
A total of 936 3D texture based radiomic and 19 shape-based features were extracted individually from every sub-compartment (edema, necrosis, enhancing tumor) on every MRI protocol. The feature set for every study on each MRI protocol included 19 shape-based features, 52 Haralick features (capturing tumor heterogeneity), 501 Laws energy (captures presence of spots, edges, waves, and ripples in an image), 383 Gabor wavelet (capturing structural detail at different orientations and scales) based features on a per-voxel basis. Statistics of median, standard deviation, skewness and kurtosis were then calculated from the feature responses of all voxels within the region of interest. Therefore, a total of 2850 features extracted for every study on every MRI protocol (Gd-T1w, T2w, T2w-FLAIR) across all tumor sub-compartments. List of the extracted features is summarized in Supplementary Sheet 1. All feature values were normalized (mean of 0 and standard deviation of 1). Feature extraction and statistic calculations were performed using in-house software implemented in MATLAB R2014b platform (MathWorks, Natick, MA, USA).
List of extracted shape-based features have been provided in Supplementary Section 2, Table 1. Detailed description of the set of features employed in this work and its possible relationship to the pathophysiology of GBM is provided in Supplementary Section 2, Table 2. Details of implementation and parameters of every 3D texture feature extracted has been provided in Supplementary Section 2, Table 3. The feature extraction pipeline has been made publicly available at https://github.com/ccipd/BrIC_Lab.
2.4. Statistical Analysis
2.4.1. Radiomic risk score (RRS)
The experimental design was setup to evaluate the prognostic ability of radiomic features from every MRI protocol (Gd-T1w, T2w, T2w-FLAIR). Within the training set, univariable cox regression was used as the feature pruning method, where all features were investigated to find the most statistically significant prognostic subset radiomic features with a p-value less than 0.05. After feature pruning, three least absolute shrinkage and selection operator (LASSO) Cox regression models were setup in 5-fold cross validation with 1000 iterations, to further select the most prognostic radiomic features within each of the MRI protocols based on their frequency of occurrence. LASSO L1 regularization technique iteratively shrinks the feature coefficient estimates towards zero, and chooses an optimal tuning parameter lambda (λ) that increases in a cross-validation setup until features with only non-zero coefficients are selected. Therefore, in cases with very large number of features, a LASSO model can help both shrink and find the sparse model that involves a small subset of the features. Thus, features picked by the LASSO models within the training cohort, and then pooled in a linear combination and multiplied with their respective coefficients to construct a RRS:
where n is the number of features selected by LASSO for a given MRI protocol, β is the weighted coefficient of the selected feature and τ is the selected MRI protocol based radiomic feature. Finally, the constructed RRS was then applied in the test set for validation.
2.4.2. Survival Analysis
For every MRI protocol, each patient within the training and test set was stratified into high-risk and low-risk group based on the median value of the RRS as a fixed cut-off. The prognostic ability of the RRS was evaluated by implementing the Kaplan Meier (KM) survival analysis and cox regression. The KM survival curves were used to compare survival times across high-risk and low-risk groups, within the training and testing cohorts for: (a) Gd-T1w MRI protocol, (b) T2w MRI protocol and (c) T2w-FLAIR MRI protocol. The horizontal axis on the KM survival curve represents the time and the vertical axis shows the probability of survival. At any given point on the survival curve, the probability that a patient in each group to remain alive at that time is presented. Optimal classifier predictions would show maximum separation between the survival curves.13
Further, Hazard ratios (HR) were used to quantify the effect of individual feature on survival. Features yielding negative regression coefficients (i.e. low feature values correlated with long term survival) in our cox model produce a HR between 0 and 1; features yielding positive regression coefficients (i.e. low feature values correlated with short term survival) produce a HR between 1 and infinity. We also computed Concordance indices (C indices or C statistic) for each of our univariable and multivariable analysis experiments in R. C indices is the fraction of all pairs of subjects whose predicted survival times are correctly ordered (i.e. concordant with actual survival times). C indices = 1 indicates that the model has perfect predictive accuracy, and C indices = 0.5 indicates that the model is no better than random chance.
2.5. Radio-genomic analysis
Within the training cohort of TCIA patients, pre-processed gene expression data using Robust Multichip Average (RMA) algorithm32 was available for a total of 125 patients (Table 1A) and hence was used for our radio-genomic analysis. This TCGA GBM gene expression data (Level3 - Affymetrix HT HG U133A) is publicly available for download from the Broad Institute (http://gdac.broadinstitute.org/runs/stddata__2016_01_28).33 Further information regarding the acquisition and expression profiling has been published by TCGA network.34 This transcriptomic data, consisting of 12,042 annotated genes, was used to investigate the possible underlying biological processes of the RRS. The most differentially expressing genes (DEGs) of the RRS were selected using the Wilcoxon rank sum test. Benjamini and Hochberg method was used to adjust the p-values and control for the false discovery rate (FDR) in multiple testing.35 Statistically significant DEGs (p-value < 0.03) were then used to identify distinct Gene Ontology (GO) based biological processes.16,17 GO highlights the most overrepresented genes and finds the systematic linkages between those genes and biological processes.
To further gain insight into the GO based biological processes and their association with the individual radiomic features from different sub-compartments that were used to create the Gd-T1w RRS, we performed single-sample Gene-set enrichment analysis (ssGSEA).18,36 For a predefined set of genes, ssGSEA captures the significantly enriched or depleted biological processes and calculates an enrichment score for every patient in the cohort. These predefined set of genes for the GO based biological processes were acquired from the Molecular Signatures Database (MSigDB v6.2) platform (http://software.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=BP).37 Within the training cohort, ssGSEA score was calculated for 125 GBM patients, and then a Spearman rank correlation coefficient (ρ) matrix was constructed to identify the contribution of the 25 individual Gd-T1w radiomic features (that were used to create the RRS) and their association with the GO biological processes.
All the statistical analysis was performed on MATLAB R2014b platform (MathWorks, Natick, MA, USA). P values <0.05 were considered significant. The Cox’s proportional hazard regression models were built using the ‘coxphfit’ command in MATLAB.
3. RESULTS
3.1. Radiomic risk score created using radiomic features from the tumor habitat on Gd-T1w MRI is prognostic of Progression Free Survival
Within the training cohort, 212 prognostic radiomic features were obtained from 2850 Gd-T1w radiomic features after feature pruning from univariable cox regression. For the Gd-T1w protocol, the LASSO model selected 25 radiomic features with lambda value of 0.088 (see Figure 2A). Details of the features selected and their coefficients have been listed in Figure 2B. The complete RSS formulation can be found in Supplementary Section 3. Of the 25 Gd-T1w radiomic features, 8 were picked from the peri-tumoral edema region, 9 were selected from the enhancing region of the tumor, and the remaining 8 were picked from the necrotic core of GBM. The Gd-T1w RRS for every patient within the training and testing cohort has been provided in Supplementary Sheet 2.
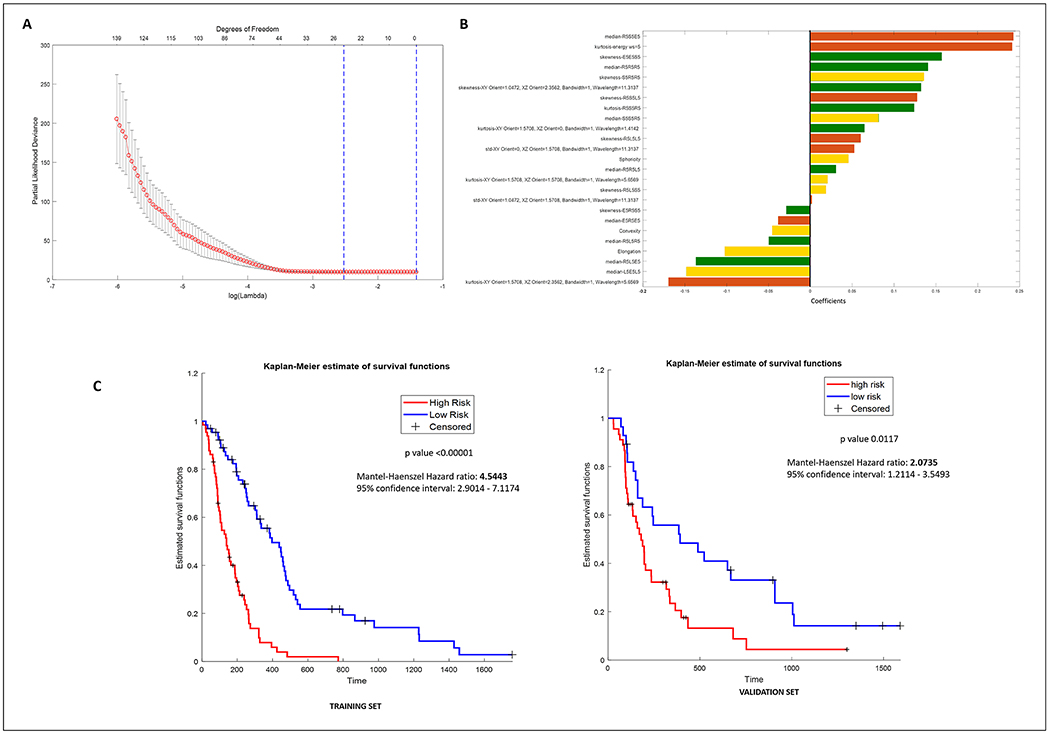
Figure 2:
(A) Radiomic feature selection using the least absolute shrinkage and selection operator (Lasso) cox regression model. Tuning penalization parameter lambda using 5-fold cross validation and minimum criterion in lasso model. The partial likelihood deviance was plotted versus log lambda. Log Lamda = −2.4308, with lambda = 0.0880 was chosen. (B) Forest plot of the beta coefficients/weights of the 25 radiomic features selected in the radiomic risk score. Brown, green, and yellow represent features obtained from the necrotic core, enhancing region and edema of the tumor habitat respectively, from Gd-T1w MRI. (C) Kaplan-Meier curves for patients stratified into low-risk and high-risk groups according to the radiomic risk score (cutoff = −0.1044) in the training cohort and independent validation set respectively. X-axis represents the progression free survival days in days, and Y-axis represents the estimated survival function.
In a multivariable Cox analysis, the RRS built using the 25 Gd-T1w radiomic features resulted in statistically significant KM curves (log-rank test, p < 0.00001, HR = 4.5, Figure 2C Left). The Gd-T1w RRS was also found to be a prognostic indicator of PFS in the independent test cohort (log-rank test, p value = 0.0117, HR = 2.0735, Figure 2C Right). Other clinical parameters (such as age, gender) between the low-risk and high-risk groups based on Gd-T1w RRS median cut-off were also investigated (Supplementary Section 4, Table S3).
Similarly, 321 and 496 prognostic features were obtained from 2850 T2w and T2w-FLAIR MRI based radiomic features respectively using the univariable Cox regression method. Then, a total of 25 T2w and 12 T2w-FLAIR radiomic features were picked by the LASSO models respectively. Similar to Gd-T1w RRS, statistically significant KM survival curves were reported with 25 T2w (log-rank test, p-value < 0.00001, HR = 28.6736) and 12 T2w-FLAIR (log-rank test, p-value = 0.0013, HR = 4.8434) MRI radiomic features respectively. However, the RRS created using radiomic features from the tumor habitat of T2w and T2w-FLAIR studies did not yield statistically significant differences in the “high risk” and “low risk” patient populations upon validation (log-rank test, T2w p-value = 0.49, T2w-FLAIR p-value = 0.25).
3.2. Added clinical utility of Gd-T1w radiomic risk score to existing clinical parameters
We further assessed the added clinical utility of the RRS to existing clinical parameters (age, gender), molecular features (MGMT, IDH mutation status) and extent of resection (EOR, on test set only) to evaluate improvement in prediction of PFS in GBM patients. In a multivariable setting, we found that the Gd-T1w RRS resulted in a training concordance-index (C-index) of 0.76 (95% Confidence Interval [CI]: 0.67 – 0.85) and predicted PFS on the independent test cohort with a C-index of 0.75 (95% CI: 0.68 – 0.84, p-value = 0.012). Interestingly, combining clinical (age, gender),molecular features (MGMT, IDH mutation status) and EOR with the Gd-T1w RRS, improved the prediction of PFS within the training (C-index = 0.81, 95% CI: 0.71 – 0.90, p-value = <0.0001) and test set (C-index = 0.84, 95% CI: 0.67 – 0.95, p-value = 0.03) respectively, as compared to validation on test set using either clinical (Age: C-index = 0.55, 95% CI: 0.46 – 0.64, Gender: C-index = 0.52, 95% CI: 0.46 – 0.64) or molecular features (MGMT status: C-index = 0.65, 95% CI: 0.57 – 0.72, IDH status: C-index = 0.55, 95% CI: 0.49 – 0.60) or EOR (C-index = 0.56, 95% CI: 0.44 – 0.67) or Gd-T1w RRS (C-index = 0.75, 95% CI: 0.68 – 0.84) alone. Table 2 lists the HR and concordance indices for the clinical parameters (age, gender), molecular features (MGMT status, IDH status), EOR and combined radiomic features for predicting PFS.
Table 2:
Cox regression analysis within the training and test cohorts. Hazard ratios, concordance using clinical and radiomic features and statistical significance (via p-value) obtained from different sub-compartments (edema, necrosis, enhancing tumor) of GBM tumor habitat on Gd-T1w MRI. Cox regression analysis was implemented in a univariable setting for clinical and molecular features, and EOR (test set only). Multivariable cox regression model was implemented for the radiomic risk score as well as the combined features. Abbreviations: 95% CI – 95% Confidence Interval, C-Index – Concordance Index, MGMT - O-6-Methylguanine-DNA Methyltransferase, IDH - Isocitrate dehydrogenase, EOR – Extent of resection.
| Cox Regression Analysis | ||||||||
|---|---|---|---|---|---|---|---|---|
| Feature | Training Cohort | Independent Test Cohort | ||||||
| Hazard Ratio (95% CI) | C- Index (95% CI) | p-value | Hazard Ratio (95% CI) | C-Index (95% CI) | p-value | |||
| Univariable Analysis | Clinical features | Age | 1.017 (0.99 – 1.032) |
0.54 (0.46 - 0.63) |
0.145 | 0.9789 (0.9441 – 0.9937) |
0.55 ( 0.46 - 0.64) |
0.015* |
| Gender | 1.467 (0.8481 - 2.539) |
0.53 (0.45 – 0.60) |
0.17 | 0.7263 (0.4126 - 1.278) |
0.52 (0.46 – 0.59) |
0.268 | ||
| Molecular features | MGMT Status | 0.694 ( 0.4061 - 1.187) |
0.52 (0.48 – 0.64) |
0.182 | 0.2646 (0.1406 - 0.4982) |
0.65 (0.57 - 0.72) |
< 0.0001* | |
| IDH Status | 0.9349 (0.2868 - 3.048 ) |
0.51 (0.46 – 0.55) |
0.724 | 0.2471 (0.0755 - 0.808) |
0.55 (0.49 – 0.60) |
0.021* | ||
| Extent of resection (EOR) | - | - | - | 0.8075 (0.60 – 2.53) |
0.56 (0.44 – 0.67) |
0.5562 | ||
| Multivariable Analysis | Radiomic risk score | 4.544 (2.901 - 7.117) |
0.76 (0.67 – 0.85) |
< 0.0001* | 2.073 (1.211 - 3.549) |
0.75 (0.68 – 0.84) |
0.012* | |
| Clinical features + Molecular features + EOR (test set only) + Radiomic risk score | - |
0.81 (0.71 – 0.90) |
< 0.0001* | - |
0.84 (0.67 – 0.95) |
0.03* | ||
3.3. Gene Ontology identifies biological processes associated with Gd-T1w radiomic risk score
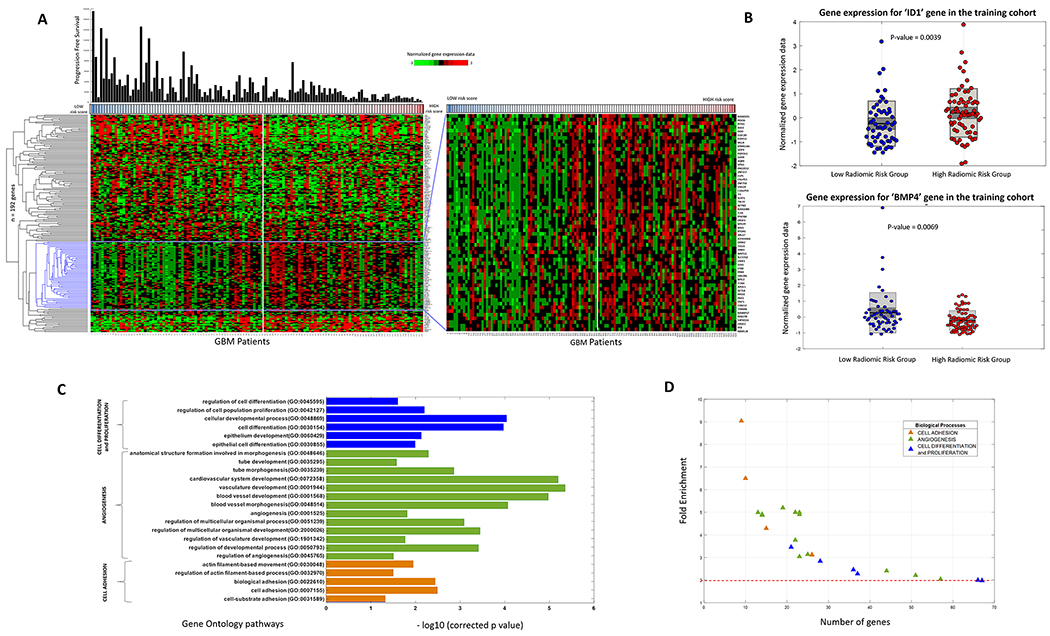
Empirical analysis of the 12,042 annotated genes (obtained from 125 patients of the training cohort) using the Wilcoxon rank sum test resulted in a total of 192 DEGs that had an p-value of less than 0.03 (with an FDR of 3%). These 192 DEGs were identified to be differentially expressed between the ‘high-risk’ and ‘low-risk’ groups of the Gd-T1w RRS, and were further used for GO analysis. Figure 3A shows the supervised hierarchical clustering of these 192 DEGs using the correlation distance metric. Figure 3B shows the boxplot of two representative DEGs ‘ID1’ (Inhibitor of differentiation/DNA binding) and ‘BMP4’ (Bone Morphogenetic Protein 4). Complete list of the 192 DEGs can be found in Supplementary Sheet 3.
Figure 3:
(A) Hierarchical clustering of the differentially expressing genes (n=192, p<0.03, false discovery rate = 3%) for radiomic risk score within the training cohort of 125 GBM patients. The genes were clustered based on the ‘correlation’ distance metric. (B) (left) Box-plot of the ID1 gene expression within the training cohort for the low-risk and high-risk radiomic groups. (right) Box-plot of the BMP4 gene expression within the training cohort for the low-risk and high-risk radiomic groups. High BMP4 expression is known to be associated with better prognosis. In consensus, we report that high BMP4 expression was found in the low radiomic risk group. (C) Gene Ontology analysis revealed several biological processes that were associated with the radiomic risk score. Fisher’s exact text (with Bonferroni correction for multiple testing of 5%), related the radiomic risk score of progression free survival with biological processes of cell differentiation, proliferation, angiogenesis and cell adhesion. (D) 2-D scatter plot to show the number of genes involved in each biological process.
Radiogenomic analysis of the training cohort using GO, revealed that the Gd-T1w RRS was associated with 57 biological processes that have been listed in Figure 3C. Complete list of all the biological processes has been provided in Supplementary Sheet 4. It was observed that 24 biological processes were implicated in cell adhesion, cell proliferation, differentiation and angiogenesis. A directed acyclic graph investigating the inter-relationships between these 57 biological processes have been provided in Supplementary Section 5.38 In Figure 3D, the fold enrichment change and number of genes involved in the various biological processes have been listed. It can be seen that biological processes of cell adhesion, that involved over 100 genes, had the highest fold enrichment change. Most of the GO terms involved in cell differentiation, proliferation, and angiogenesis had less than 100 overrepresented genes, but their fold enrichment change was greater than 2.
3.4. Gene-set enrichment analysis provides insights into the relationship of individual tumor habitat specific radiomic features with biological processes
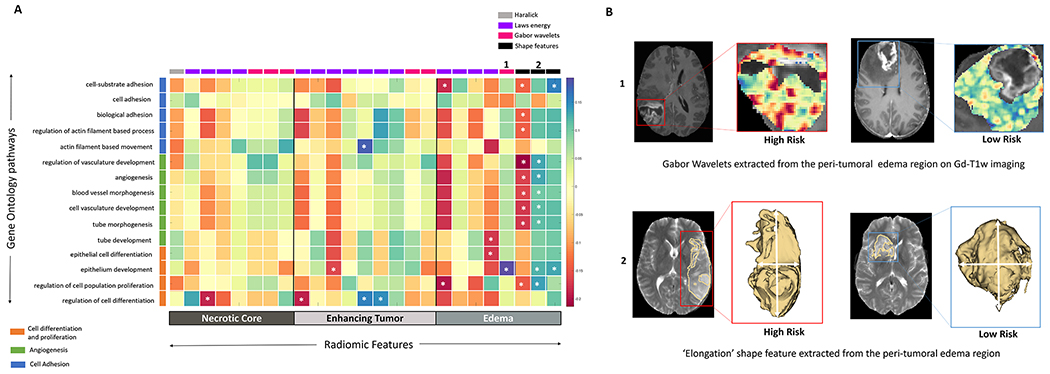
Predefined gene set annotations for 15 GO based biological processes (associated with cell differentiation, cell adhesion and angiogenesis) were available from MSigDB, and thus used to calculate the ssGSEA scores. Supplementary Sheet 5 has the complete list of pre-defined set of genes that were used to describe these 15 GO based biological processes. ssGSEA scores helped understand the associations of these biological processes with the individual 25 radiomic features that were obtained from different tumor sub-compartments and used to create the Gd-T1w RRS. As may be observed in Figure 4, statistically significant correlations (p<0.05) were found between the shape features (i.e., sphericity, elongation and convexity) of the peri-tumoral edema region and biological processes of cell proliferation, angiogenesis and cell adhesion. Additionally, Laws energy and Gabor wavelet texture features from within the peri-tumoral edema region of the GBM tumor habitat, were also found to be correlated with cell proliferation (ρLaws Energy = − 0.186, 95% CI: −0.350 - −0.011) and angiogenesis (ρGabor = 0.193, 95% CI: 0.018 - 0.356) respectively.
Figure 4:
(A) Spearman rank correlation coefficient matrix of the single-sample Gene set enrichment analysis scores and the radiomic features extracted from the GBM tumor habitat (necrotic core, enhancing tumor and peri-tumoral edema). ‘*’ signifies a statistically significant (p<0.05) relationship (B) Top row – Gabor wavelet based radiomic features extracted from the peri-tumoral edema region of the GBM tumor habitat was found to be positively correlated with the cell differentiation and proliferation biological process. Bottom row – Shape feature ‘Elongation’ of the peri-tumoral edema was found to be positively correlated with angiogenesis and cell proliferation biological processes.
4. DISCUSSION
Pre-treatment estimation of progression free survival (PFS) can help predict therapeutic efficacy of conventional first-line treatment. Additionally, recent randomized clinical trials have reported that PFS can be used as a surrogate endpoint that correlates well with overall survival (OS).39,40 Currently, there are no clinically validated imaging based markers that are prognostic of PFS in primary Glioblastoma (GBM).41
In this work, we had two objectives. In our first objective, we created and independently evaluated a radiomic risk score (RRS) using radiomic features from the tumor habitat on pre-treatment MR imaging to stratify GBM patients based on their PFS on n=203 studies from a multi-institutional cohort. In our second objective, we reported significant associations of the prognostic radiomic features that contributed to RRS with specific molecular signaling pathways, via radiogenomic analysis.
In our first objective, the survival risk score created using radiomic features from Gd-T1w MRI was found to be statistically significantly different across the ‘low risk’ and ‘high risk’ groups, both on training (p<0.001, n=130) as well as the hold-out test set (p=0.03, n=73). In consensus with recent studies42–44, we further discovered that integrating the risk score from Gd-T1w MRI with clinical parameters (age, gender) and molecular features (MGMT status, IDH status) improved the performance of the prognostic model. Our prognostic RRS consisted of a total of 25 radiomic features including textural features belonging to Laws energy and Gabor wavelet families, and as shape features from the peri-tumoral edema region, on Gd-T1w MRI. Laws energy radiomic features capture wavy, ripple, and spot-like patterns which may be reflective of increased peri-tumoral neovascularization and atypical blood vessels. Our findings also corroborate with our own previous work, where we identified Laws energy features from the edematous and enhancing region on T2w-FLAIR and Gd-T1w MRI to be predictive of OS in GBM.13,43 Similarly, Gabor wavelet features capture changes in structural orientation by quantifying prominent MRI intensity changes across multiple directional gradients in a given region of interest (ROI).45 These findings are in line with multiple studies that have demonstrated the efficacy of Gabor wavelets in predicting tumor malignancy46, disease aggressiveness47, and treatment response48 on imaging for various cancers.
In our second objective, we identified significant radiogenomic correlations (p<0.05) between our 25 prognostic Gd-T1w radiomic features from different sub-compartments of the tumor habitat with molecular signaling pathways in GBM using Gene Ontology (GO) and single-sample Gene set enrichment analysis (ssGSEA). Specifically, using the corresponding gene expression data available for the training set (n=125), we identified a total of 192 differentially expressing genes (DEGs) across the ‘low risk’ and ‘high risk’ groups obtained from the Gd-T1w RRS. Prognostic value of some of these genes has been demonstrated within various cancers. For example, ID1 is a known to be over-expressed in breast, and GBM cancers.49,50 In consensus, we found that ID1 had an adjusted p-value of 0.0039. Similarly, high expression of BMP4 has been known to be significantly associated with better prognosis in GBM.51 Our analysis revealed that the ‘low risk’ group was associated with statistically significant BMP4 expression when compared to the ‘high risk’ group, within the training set (p-value = 0.0069). TP53 (tumor protein p-53) inducible proteins such as TP53I11 (tumor protein p53 inducible protein 11) and TP73-AS1 (TP73 antisense RNA 1) were also found to be statistically significant between the ‘low-risk’ and ‘high-risk’ radiomic risk groups with p-values of 0.005 and 0.007 respectively. CD34, a transmembrane phosphoglycoprotein, known to be a prognostic marker with potential to be a therapeutic target, was differentially expressed with an adjusted p-value of 0.017.52
Using the 192 DEGs, GO analysis further revealed that cell adhesion, cell differentiation, proliferation and angiogenesis were the key biological processes that were significantly different across the high-risk and low-risk groups (as defined via our RRS). Biological processes surrounding cell adhesion, cell differentiation, proliferation, and angiogenesis are key contributors in driving malignant tumor behavior, support resistance to chemo-radiation therapy, and contribute to poor PFS in primary GBM.53,54 Previous work by Liu et al., has similarly shown via radiogenomic analysis that biological processes such as immune response, programmed cell death, cell proliferation and vasculature development dictate PFS in low grade gliomas.42
Finally, we demonstrated that multiple biological processes that are associated with poor PFS were also significantly correlated (p < 0.05) with radiomic features extracted from the peri-tumoral edema region of GBM. For instance, we found that sphericity (a measure of 3D compactness) of the edematous region, was negatively correlated with most of the biological processes involved in cell adhesion (ρavg = − 0.140, 95% CI: −0.308 - 0.036), and angiogenesis (ρavg = − 0.178, 95% CI: −0.343 - −0.002). This suggests that a more infiltrative pattern of tumor growth as reflected on peri-tumoral edema may be associated with poor prognosis in GBM patients. Further, Ismail et al., have previously reported that lower values of elongation of the edematous region (ratio between major and minor axes of the tumor) are indicative of tumor progression in conventional post-treatment primary GBM.55 Along similar lines, we found association of lower elongation values of edema with angiogenesis (ρavg = 0.118, 95% CI: −0.287 - 0.058) and cell differentiation (ρavg = 0.128, 95% CI: −0.048 - 0.299), which suggests that better prognosis may be linked with low RRS in GBM.
This study did have some limitations. The validation was done retrospectively and radiogenomic analysis was only possible on 1 out of the 3 cohorts (due to unavailability of the corresponding gene expression data in the Cleveland Clinic cohort, and substantial heterogeneity in RNA-sequencing processing pipeline of Ivy GAP cohort). While we compared the clinical parameters (age, gender), molecular (MGMT, IDH status) and EOR with RRS to the extent possible, due to the limited training cohort, the development of RRS in this work could not be controlled for clinical parameters, molecular status, and EoR. This will be part of our future study involving large multi-site evaluation of RRS. Further work is also mandated to develop a radiogenomic approach that incorporates transcriptomic data coming from different processing platforms, and thus reducing the influence of batch effects in RNA-sequencing data. In future, our work will also focus on incorporating complementary imaging parameters (PET, perfusion, DWI) which may further improve survival prediction using radiogenomic analysis, while also accounting for follow-up treatment. Additionally, we also plan to expand our analysis with corresponding gene expression data available across all patients, and eventually using prospectively collected scans.
In summary, we developed and independently evaluated a prognostic RRS using features obtained from the tumor habitat on Gd-T1w MRI, which could potentially be used as a non-invasive imaging based surrogate marker of PFS in primary GBM. We further demonstrated that the Gd-T1w RRS, with other clinical and molecular features can improve patient stratification when compared to using Gd-T1w RRS alone. Further, we identified the biological processes of cell differentiation, cell adhesion, and angiogenesis to be associated with Laws energy features from the enhancing tumor and shape features of infiltrative peri-tumoral edema respectively, thus providing a biological basis for the radiomic features driving the RRS with possible underlying processes that dictate tumor behavior in GBM patients. Following additional large-scale prospective validation, our Gd-T1w RRS could potentially be used as a surrogate endpoint in clinical trials,56 predict onset of tumor recurrence,40,41 and for evaluating the efficacy of chemo-radiation therapy in primary GBM patients.57,58
Supplementary Material
Acknowledgments
Financial Support
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers
1U24CA199374-01,
R01CA202752-01A1
R01CA208236-01A1
R01 CA216579-01A1
R01 CA220581-01A1
1U01 CA239055-01
1P20 CA233216-01
National Center for Research Resources under award number 1 C06 RR12463-01
VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service
the DOD Prostate Cancer Idea Development Award (W81XWH-15-1-0558)
the DOD Lung Cancer Investigator-Initiated Translational Research Award (W81XWH-18-1-0440)
the DOD Peer Reviewed Cancer Research Program (W81XWH-16-1-0329)
National Institute of Diabetes and Digestive and Kidney Diseases (1K25 DK115904-01A1)
the Ohio Third Frontier Technology Validation Fund
the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering and the Clinical and Translational Science Award Program (CTSA) at Case Western Reserve University
Department of Defense Peer Reviewed Cancer Research Program (PRCRP) Career Development Award
Dana Foundation David Mahoney Neuroimaging Program
The V Foundation Translational Grant
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, or the United States Government.
Conflicts of interest
Dr. Madabhushi is an equity holder in Elucid Bioimaging and in Inspirata Inc. He is also a scientific advisory consultant for Inspirata Inc. In addition, he currently serves or has served as a scientific advisory board member for Inspirata Inc, Astrazeneca, Bristol-Meyers Squibb, Boehringer-Ingelheim and Merck. He also has sponsored research agreements with Philips and Inspirata Inc. His technology has been licensed to Elucid Bioimaging and Inspirata Inc. He is also involved in a NIH U24 grant with PathCore Inc, and 3 different R01 grants with Inspirata Inc.
Dr Vinay Varadan is involved in sponsored research with Curis Inc. and Philips Healthcare. In addition, he currently serves or has served as a scientific advisory board member for Curis Inc.
REFERENCES
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N Engl J Med. 2005. March 10;352(10):987–996. doi: 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: A Cancer Journal for Clinicians. 2019. January;69(1):7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 3.Hu LS, Ning S, Eschbacher JM, Baxter LC, Gaw N, Ranjbar S, Plasencia J, Dueck AC, Peng S, Smith KA, Nakaji P, Karis JP, Quarles CC, Wu T, Loftus JC, Jenkins RB, Sicotte H, Kollmeyer TM, O’Neill BP, Elmquist W, Hoxworth JM, Frakes D, Sarkaria J, Swanson KR, Tran NL, Li J, Mitchell JR. Radiogenomics to characterize regional genetic heterogeneity in glioblastoma. Neuro Oncol. 2017. January;19(1):128–137. doi: 10.1093/neuonc/now135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qazi MA, Vora P, Venugopal C, Sidhu SS, Moffat J, Swanton C, Singh SK. Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann Oncol. 2017. July 1;28(7):1448–1456. doi: 10.1093/annonc/mdx169 [DOI] [PubMed] [Google Scholar]
- 5.Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N Engl J Med. 2015. June 10;372(26):2481–2498. doi: 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivera AL, Pelloski CE, Gilbert MR, Colman H, De La Cruz C, Sulman EP, Bekele BN, Aldape KD. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010. February;12(2):116–121. doi: 10.1093/neuonc/nop020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skardelly M, Dangel E, Gohde J, Noell S, Behling F, Lepski G, Borchers C, Koch M, Schittenhelm J, Bisdas S, Naumann A, Paulsen F, Zips D, von Hehn U, Ritz R, Tatagiba MS, Tabatabai G. Prolonged Temozolomide Maintenance Therapy in Newly Diagnosed Glioblastoma. Oncologist. 2017. May;22(5):570–575. doi: 10.1634/theoncologist.2016-0347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Huang Y, Hong S, Yang Y, Yu G, Jia J, Peng P, Wu X, Lin Q, Xi X, Peng J, Xu M, Chen D, Lu X, Wang R, Cao X, Chen X, Lin Z, Xiong J, Lin Q, Xie C, Li Z, Pan J, Li J, Wu S, Lian Y, Yang Q, Zhao C. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: a multicentre, randomised, open-label, phase 3 trial. Lancet. 2016. October 15;388(10054):1883–1892. doi: 10.1016/S0140-6736(16)31388-5 [DOI] [PubMed] [Google Scholar]
- 9.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Diéras V, Hegg R, Im S-A, Shaw Wright G, Henschel V, Molinero L, Chui SY, Funke R, Husain A, Winer EP, Loi S, Emens LA, IMpassion130 Trial Investigators. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med. 2018. 29;379(22):2108–2121. doi: 10.1056/NEJMoa1809615 [DOI] [PubMed] [Google Scholar]
- 10.Lemée J-M, Clavreul A, Menei P. Intratumoral heterogeneity in glioblastoma: don’t forget the peritumoral brain zone. Neuro-oncology. 2015. October;17(10):1322–1332. doi: 10.1093/neuonc/nov119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakas S, Akbari H, Pisapia J, Martinez-Lage M, Rozycki M, Rathore S, Dahmane N, O’Rourke DM, Davatzikos C. In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the φ index. Clin Cancer Res. 2017. August 15;23(16):4724–4734. doi: 10.1158/1078-0432.CCR-16-1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thawani R, McLane M, Beig N, Ghose S, Prasanna P, Velcheti V, Madabhushi A. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer. 2018. January 1;115(Supplement C):34–41. doi: 10.1016/j.lungcan.2017.10.015 [DOI] [PubMed] [Google Scholar]
- 13.Prasanna P, Patel J, Partovi S, Madabhushi A, Tiwari P. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: Preliminary findings. Eur Radiol. 2016. October 24; doi: 10.1007/s00330-016-4637-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zinn PO, Singh SK, Kotrotsou A, Hassan I, Thomas G, Luedi MM, Elakkad A, Elshafeey N, Idris T, Mosley J, Gumin J, Fuller GN, Groot JF de, Baladandayuthapani V, Sulman EP, Kumar AJ, Sawaya R, Lang FF, Piwnica-Worms D, Colen RR. A Coclinical Radiogenomic Validation Study: Conserved Magnetic Resonance Radiomic Appearance of Periostin-Expressing Glioblastoma in Patients and Xenograft Models. Clin Cancer Res. 2018. December 15;24(24):6288–6299. doi: 10.1158/1078-0432.CCR-17-3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kickingereder P, Neuberger U, Bonekamp D, Piechotta PL, Götz M, Wick A, Sill M, Kratz A, Shinohara RT, Jones DTW, Radbruch A, Muschelli J, Unterberg A, Debus J, Schlemmer H-P, Herold-Mende C, Pfister S, von Deimling A, Wick W, Capper D, Maier-Hein KH, Bendszus M. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro-oncology. 2018. 18;20(6):848–857. doi: 10.1093/neuonc/nox188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000. May;25(1):25–29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017. 04;45(D1):D331–D338. doi: 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Fröhling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, Hahn WC. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009. November 5;462(7269):108–112. doi: 10.1038/nature08460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. Journal of Digital Imaging. 2013. December;26(6):1045–1057. doi: 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scarpace L, Mikkelsen T, Cha S, Rao S, Tekchandani S, Gutman D, Saltz JH, Erickson BJ, Pedano N, Flanders AE, Barnholtz-Sloan J, Ostrom Q, Barboriak D, Pierce LJ. Radiology Data from The Cancer Genome Atlas Glioblastoma Multiforme [TCGA-GBM] collection [Internet]. The Cancer Imaging Archive; 2016. [cited 2018 Oct 17]. Available from: https://wiki.cancerimagingarchive.net/x/sgAe doi: 10.7937/K9/TCIA.2016.RNYFUYE9 [DOI] [Google Scholar]
- 21.Shah N, Feng X, Lankerovich M, Puchalski R, Keogh B. Ivy GAP - The Cancer Imaging Archive (TCIA) Public Access - Cancer Imaging Archive Wiki. The Cancer Imaging Archive. 2016; doi: 10.7937/K9/TCIA.2016.XLwaN6nL [DOI] [Google Scholar]
- 22.Puchalski RB, Shah N, Miller J, Dalley R, Nomura SR, Yoon J-G, Smith KA, Lankerovich M, Bertagnolli D, Bickley K, Boe AF, Brouner K, Butler S, Caldejon S, Chapin M, Datta S, Dee N, Desta T, Dolbeare T, Dotson N, Ebbert A, Feng D, Feng X, Fisher M, Gee G, Goldy J, Gourley L, Gregor BW, Gu G, Hejazinia N, Hohmann J, Hothi P, Howard R, Joines K, Kriedberg A, Kuan L, Lau C, Lee F, Lee H, Lemon T, Long F, Mastan N, Mott E, Murthy C, Ngo K, Olson E, Reding M, Riley Z, Rosen D, Sandman D, Shapovalova N, Slaughterbeck CR, Sodt A, Stockdale G, Szafer A, Wakeman W, Wohnoutka PE, White SJ, Marsh D, Rostomily RC, Ng L, Dang C, Jones A, Keogh B, Gittleman HR, Barnholtz-Sloan JS, Cimino PJ, Uppin MS, Keene CD, Farrokhi FR, Lathia JD, Berens ME, Iavarone A, Bernard A, Lein E, Phillips JW, Rostad SW, Cobbs C, Hawrylycz MJ, Foltz GD. An anatomic transcriptional atlas of human glioblastoma. Science. 2018. May 11;360(6389):660–663. doi: 10.1126/science.aaf2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, Davatzikos C. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific Data. 2017. September 5;4:170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bakas S, Zeng K, Sotiras A, Rathore S, Akbari H, Gaonkar B, Rozycki M, Pati S, Davatzikos C. GLISTRboost: Combining Multimodal MRI Segmentation, Registration, and Biophysical Tumor Growth Modeling with Gradient Boosting Machines for Glioma Segmentation. Brainlesion (2015). 2016;9556:144–155. doi: 10.1007/978-3-319-30858-6_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davatzikos C, Rathore S, Bakas S, Pati S, Bergman M, Kalarot R, Sridharan P, Gastounioti A, Jahani N, Cohen E, Akbari H, Tunc B, Doshi J, Parker D, Hsieh M, Sotiras A, Li H, Ou Y, Doot RK, Bilello M, Fan Y, Shinohara RT, Yushkevich P, Verma R, Kontos D. Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J Med Imaging (Bellingham). 2018. January;5(1):011018. doi: 10.1117/1.JMI.5.1.011018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohlfing T, Zahr NM, Sullivan EV, Pfefferbaum A. The SRI24 multichannel atlas of normal adult human brain structure. Hum Brain Mapp. 2010. May;31(5):798–819. doi: 10.1002/hbm.20906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010. June;29(6):1310–1320. doi: 10.1109/TMI.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith SM, Brady JM. SUSAN—A New Approach to Low Level Image Processing. International Journal of Computer Vision. 1997. May 1;23(1):45–78. doi: 10.1023/A:1007963824710 [DOI] [Google Scholar]
- 29.Madabhushi A, Udupa JK. New methods of MR image intensity standardization via generalized scale. In: Fitzpatrick JM, Reinhardt JM, editors. San Diego, CA; 2005. [cited 2018 Dec 25]. p. 1143 Available from: http://proceedings.spiedigitallibrary.org/proceeding.aspx?doi=10.1117/12.595925 doi: 10.1117/12.595925 [DOI] [PubMed] [Google Scholar]
- 30.Bradley WG. MR appearance of hemorrhage in the brain. Radiology. 1993. October;189(1):15–26. doi: 10.1148/radiology.189.1.8372185 [DOI] [PubMed] [Google Scholar]
- 31.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012. November;30(9):1323–1341. doi: 10.1016/j.mri.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010. October 1;26(19):2363–2367. doi: 10.1093/bioinformatics/btq431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Analysis-ready standardized TCGA data from Broad GDAC Firehose 2016_01_28 run. Broad Institute of MIT and Harvard. 2016;(Broad Institute TCGA Genome Data Analysis Center; ). doi: 10.7908/C11G0KM9 [DOI] [Google Scholar]
- 34.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008. October;455(7216):1061–1068. doi: 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005. October 25;102(43):15545. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011. June 15;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017. July 3;45(W1):W130–W137. doi: 10.1093/nar/gkx356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han K, Ren M, Wick W, Abrey L, Das A, Jin J, Reardon DA. Progression-free survival as a surrogate endpoint for overall survival in glioblastoma: a literature-based meta-analysis from 91 trials. Neuro-oncology. 2014. May;16(5):696–706. doi: 10.1093/neuonc/not236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heiland DH, Masalha W, Franco P, Machein MR, Weyerbrock A. Progression-free and overall survival in patients with recurrent Glioblastoma multiforme treated with last-line bevacizumab versus bevacizumab/lomustine. J Neurooncol. 2016. February;126(3):567–575. doi: 10.1007/s11060-015-2002-z [DOI] [PubMed] [Google Scholar]
- 41.Kelly C, Majewska P, Ioannidis S, Raza MH, Williams M. Estimating progression-free survival in patients with glioblastoma using routinely collected data. J Neurooncol. 2017;135(3):621–627. doi: 10.1007/s11060-017-2619-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Li Y, Qian Z, Sun Z, Xu K, Wang K, Liu S, Fan X, Li S, Zhang Z, Jiang T, Wang Y. A radiomic signature as a non-invasive predictor of progression-free survival in patients with lower-grade gliomas. NeuroImage: Clinical. 2018. January 1;20:1070–1077. doi: 10.1016/j.nicl.2018.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beig N, Patel J, Prasanna P, Hill V, Gupta A, Correa R, Bera K, Singh S, Partovi S, Varadan V, Ahluwalia M, Madabhushi A, Tiwari P. Radiogenomic analysis of hypoxia pathway is predictive of overall survival in Glioblastoma. Scientific Reports. 2018. January 8;8(1):7. doi: 10.1038/s41598-017-18310-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zinn PO, Majadan B, Sathyan P, Singh SK, Majumder S, Jolesz FA, Colen RR. Radiogenomic Mapping of Edema/Cellular Invasion MRI-Phenotypes in Glioblastoma Multiforme. PLOS ONE. 2011. October 5;6(10):e25451. doi: 10.1371/journal.pone.0025451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jain AK, Farrokhnia F. Unsupervised texture segmentation using Gabor filters. 1990 IEEE International Conference on Systems, Man, and Cybernetics Conference Proceedings 1990. p. 14–19. doi: 10.1109/ICSMC.1990.142050 [DOI] [Google Scholar]
- 46.Beig N, Khorrami M, Alilou M, Prasanna P, Braman N, Orooji M, Rakshit S, Bera K, Rajiah P, Ginsberg J, Donatelli C, Thawani R, Yang M, Jacono F, Tiwari P, Velcheti V, Gilkeson R, Linden P, Madabhushi A. Perinodular and Intranodular Radiomic Features on Lung CT Images Distinguish Adenocarcinomas from Granulomas. Radiology. 2019. March;290(3):783–792. doi: 10.1148/radiol.2018180910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Penzias G, Singanamalli A, Elliott R, Gollamudi J, Shih N, Feldman M, Stricker PD, Delprado W, Tiwari S, Böhm M, Haynes A-M, Ponsky L, Fu P, Tiwari P, Viswanath S, Madabhushi A. Identifying the morphologic basis for radiomic features in distinguishing different Gleason grades of prostate cancer on MRI: Preliminary findings. Ahmad A, editor. PLOS ONE. 2018. August 31;13(8):e0200730. doi: 10.1371/journal.pone.0200730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braman N, Prasanna P, Whitney J, Singh S, Beig N, Etesami M, Bates DDB, Gallagher K, Bloch BN, Vulchi M, Turk P, Bera K, Abraham J, Sikov WM, Somlo G, Harris LN, Gilmore H, Plecha D, Varadan V, Madabhushi A. Association of Peritumoral Radiomics With Tumor Biology and Pathologic Response to Preoperative Targeted Therapy for HER2 (ERBB2)–Positive Breast Cancer. JAMA Netw Open. 2019. April 5;2(4):e192561–e192561. doi: 10.1001/jamanetworkopen.2019.2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ling M-T, Wang X, Zhang X, Wong Y-C. The multiple roles of Id-1 in cancer progression. Differentiation. 2006. December;74(9-10):481–487. doi: 10.1111/j.1432-0436.2006.00083.x [DOI] [PubMed] [Google Scholar]
- 50.Soroceanu L, Murase R, Limbad C, Singer E, Allison J, Adrados I, Kawamura R, Pakdel A, Fukuyo Y, Nguyen D, Khan S, Arauz R, Yount GL, Moore DH, Desprez P-Y, McAllister SD. Id-1 is a key transcriptional regulator of glioblastoma aggressiveness and a novel therapeutic target. Cancer Res. 2013. March 1;73(5):1559–1569. doi: 10.1158/0008-5472.CAN-12-1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu Y-Y, Gao P, Sun Y, Duan Y-R. Development of targeted therapies in treatment of glioblastoma. Cancer Biol Med. 2015. September;12(3):223–237. doi: 10.7497/j.issn.2095-3941.2015.0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong X, Guan J, Ma W, Li Y, Xing B, Yang Y, Wang Y, Gao J, Wei J, Yao Y, Xu Z, Dou W, Lian W, Su C, Ren Z, Wang R. CD34 Over-Expression is Associated With Gliomas’ Higher WHO Grade. Medicine (Baltimore). 2016. February 18;95(7). doi: 10.1097/MD.0000000000002830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonavia R, Inda M-M, Cavenee W, Furnari F. Heterogeneity Maintenance in Glioblastoma: a social network. Cancer Res. 2011. June 15;71(12):4055–4060. doi: 10.1158/0008-5472.CAN-11-0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fatai AA, Gamieldien J. A 35-gene signature discriminates between rapidly- and slowly-progressing glioblastoma multiforme and predicts survival in known subtypes of the cancer. BMC Cancer. 2018. April 3;18. doi: 10.1186/s12885-018-4103-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ismail M, Hill V, Statsevych V, Huang R, Prasanna P, Correa R, Singh G, Bera K, Beig N, Thawani R, Madabhushi A, Aahluwalia M, Tiwari P. Shape Features of the Lesion Habitat to Differentiate Brain Tumor Progression from Pseudoprogression on Routine Multiparametric MRI: A Multisite Study. Am J Neuroradiol. 2018. November 1; doi: 10.3174/ajnr.A5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han K, Ren M, Wick W, Abrey L, Das A, Jin J, Reardon DA. Progression-free survival as a surrogate endpoint for overall survival in glioblastoma: a literature-based meta-analysis from 91 trials. Neuro Oncol. 2014. May;16(5):696–706. doi: 10.1093/neuonc/not236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mallick S, Kunhiparambath H, Gupta S, Benson R, Sharma S, Laviraj MA, Upadhyay AD, Julka PK, Sharma D, Rath GK. Hypofractionated accelerated radiotherapy (HART) with concurrent and adjuvant temozolomide in newly diagnosed glioblastoma: a phase II randomized trial (HART-GBM trial). J Neurooncol. 2018. October;140(1):75–82. doi: 10.1007/s11060-018-2932-3 [DOI] [PubMed] [Google Scholar]
- 58.Syed M, Liermann J, Verma V, Bernhardt D, Bougatf N, Paul A, Rieken S, Debus J, Adeberg S. Survival and recurrence patterns of multifocal glioblastoma after radiation therapy. Cancer Manag Res. 2018;10:4229–4235. doi: 10.2147/CMAR.S165956 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.