Abstract
The Mucin 1 (MUC1) protein is overexpressed in various cancers and mediates chemotherapy resistance. However, the mechanism is not fully understood. Given that most chemotherapeutic drugs disrupt ER homeostasis as part of their toxicity, and MUC1 expression is regulated by proteins involved in ER homeostasis, we investigated the link between MUC1 and ER homeostasis. MUC1 knockdown in pancreatic cancer cells enhanced unfolded protein response (UPR) signaling and cell death upon ER stress induction. Transcriptomic analysis revealed alterations in the pyrimidine metabolic pathway and cytidine deaminase (CDA). ChIP and CDA activity assays showed that MUC1 occupied CDA gene promoter upon ER stress induction correlating with increased CDA expression and activity in MUC1-expressing cells as compared to MUC1 knockdown cells. Inhibition of either the CDA or pyrimidine metabolic pathway diminished survival in MUC1-expressing cancer cells upon ER stress induction. Metabolomic analysis demonstrated that MUC1-mediated CDA activity corresponded to deoxycytidine to deoxyuridine metabolic reprogramming upon ER stress induction. The resulting increase in deoxyuridine mitigated ER stress-induced cytotoxicity. Additionally, given 1) the established roles of MUC1 in protecting cells against reactive oxygen species (ROS) insults, 2) ER stress-generated ROS further promote ER stress and 3) the emerging anti-oxidant property of deoxyuridine, we further investigated if MUC1 regulated ER stress by a deoxyuridine-mediated modulation of ROS levels. We observed that deoxyuridine could abrogate ROS-induced ER stress to promote cancer cell survival. Taken together, our findings demonstrate a novel MUC1-CDA axis of the adaptive UPR that provides survival advantage upon ER stress induction.
Keywords: MUC1, ER Stress, Pyrimidine Metabolism, CDA
Introduction
The Endoplasmic Reticulum (ER) is a vital organelle in cells involved in protein synthesis, synthesis of cell membrane lipids such as cholesterol, calcium storage and glucose metabolism (1–6). As such a multi-functional organelle, ER homeostasis is critical for the survival of cells particularly tumor cells that exhibit ER stress due to the hostile tumor microenvironment. Stress in the ER activates the unfolded protein response (UPR) of which the glucose-regulated protein 78 (GRP78) is the major regulator (7). In the absence of stress, binding of GRP78 to the UPR upstream signaling transducers PERK, IRE1α and ATF6, sequesters them. However, dissociation of GRP78 during stress signaling activates the UPR.
Upon activation by dimerization and auto-phosphorylation, PERK phosphorylates the eukaryotic initiation factor 2 alpha (eIF2α) leading to inhibition of cap-dependent mRNA translation and a decrease in the load of proteins entering the ER. However, a cap-independent translation of specific ER stress response genes, such as ATF4, ensues. ATF4 is a transcription factor that activates CCAAT-enhancer-binding protein homologous protein (CHOP) which sensitizes cells to ER stress-induced cell death by multiple mechanisms including a down-regulation of pro-survival B-cell lymphoma 2 (Bcl-2) protein and a surge in the levels of reactive oxygen species (ROS), mediated by the ER oxidoreductase-1-beta (ERO1B) protein, followed by a release of cytochrome c into the cytoplasm and activation of the caspase cascade (8–13).
Like PERK, IRE1α is activated by dimerization and auto-phosphorylation. With its RNAse activity, it splices the X-box binding protein 1 (XBP-1) to generate an active transcription factor which translocates to the nucleus to activate ER stress response genes including ERDJ4 (14, 15). With the kinase function, IRE1α recruits TNF-receptor-associated factor 2 (TRAF2) to activate the apoptosis signal-regulating kinase 1 (ASK1) and the c-Jun N-terminal kinase (JNK), leading to caspase cascade and cell death (16–18). Conversely, ATF6α translocates to the Golgi apparatus where it is proteolytically cleaved by the Golgi-resident site-1 and −2 proteases (S1P and S2P) to generate an active transcription factor which is then transported to the nucleus to activate ER stress-induced expression of chaperones, including GRP78 (19).
Tumor microenvironment induces ER stress and activates UPR (20). Accordingly, UPR-related proteins are up-regulated in various cancers (21).The outcome of the UPR varies from adaption to apoptosis. Although pro-apoptotic ER stress signaling proteins such as CHOP and ASK1 are up-regulated in cancer (21, 22) and stress-activated apoptosis-regulated pathways such as JNK are constitutively active (23, 24), the mechanisms by which cancer cells mitigate ER stress are not well understood and therefore are the subject of studies in cancer etiology research. In that context, we investigated the link between the Mucin1 (MUC1) oncoprotein and the survival UPR signaling.
MUC1 oncoprotein is overexpressed in various cancers (25) and is associated with poor chemotherapy response and poor patient survival (26, 27). Given that chemotherapeutic drugs disrupt the ER homeostasis as part of their cyto-toxicity (28), we surmised that a resistance to these drugs may involve adaptive signaling pathways to protect against their stress-related toxicity. Significantly, studies have implicated MUC1 in tolerance against stress such as oxidative stress (29, 30). Oxidative stress and ER stress are intertwined, as ROS-induced oxidative stress perturbs the redox balance of the ER environment and induces ER stress while, reciprocally, ERO1B-mediated generation of ROS during ER stress signaling promotes oxidative stress (31, 32). Of note, cellular metabolism such as glucose metabolism modulates ROS levels and affects the outcome of chemotherapeutic drugs (33–35). Germane to that are studies implicating MUC1 in glucose metabolism with the latest research demonstrating that MUC1-dependent reprogramming of glucose metabolism altered pyrimidine nucleosides levels to confer resistance to pyrimidine nucleoside analogue drugs such as gemcitabine (36, 37). Together, these studies strongly suggest potential implication of MUC1 in stress regulation via metabolic reprogramming. Additional studies demonstrated that inhibition of the pyrimidine metabolic pathway with leflunomide (38) induces ER stress (39) while ER stress induction up-regulated a key pyrimidine salvage pathway protein, activation-induced cytidine deaminase (AID) (40), involved in the conversion of deoxycytidine/cytidine to deoxyuridine/uridine (41, 42). These reports indicate that the pyrimidine metabolic pathway is critical for ER homeostasis and hint to the interconnection between the UPR signaling and the pyrimidine metabolic pathway. Lastly, MUC1 expression is up-regulated by the ER stress signaling protein, the anterior gradient-2 (AGR2), an ER-resident protein and a member of the protein disulfide isomerase family protein involved in ER homeostasis (43–46), indicating that MUC1 may be involved in ER homeostasis. In the present study, we demonstrate that MUC1 modulates the outcome of ER stress signaling. MUC1 increased the activity of the pyrimidine salvage pathway enzyme cytidine deaminase, CDA, to reprogram the pool of the pyrimidine nucleosides deoxycytidine and deoxyuridine upon ER stress. These effects correlated with a decrease in ROS levels, a reduced thapsigargin-induced ER stress-related toxicity and an increased adaptive capacity of cells indicated by increased cell survival.
Results
MUC1 Deficiency Exacerbates ER Stress upon Induction
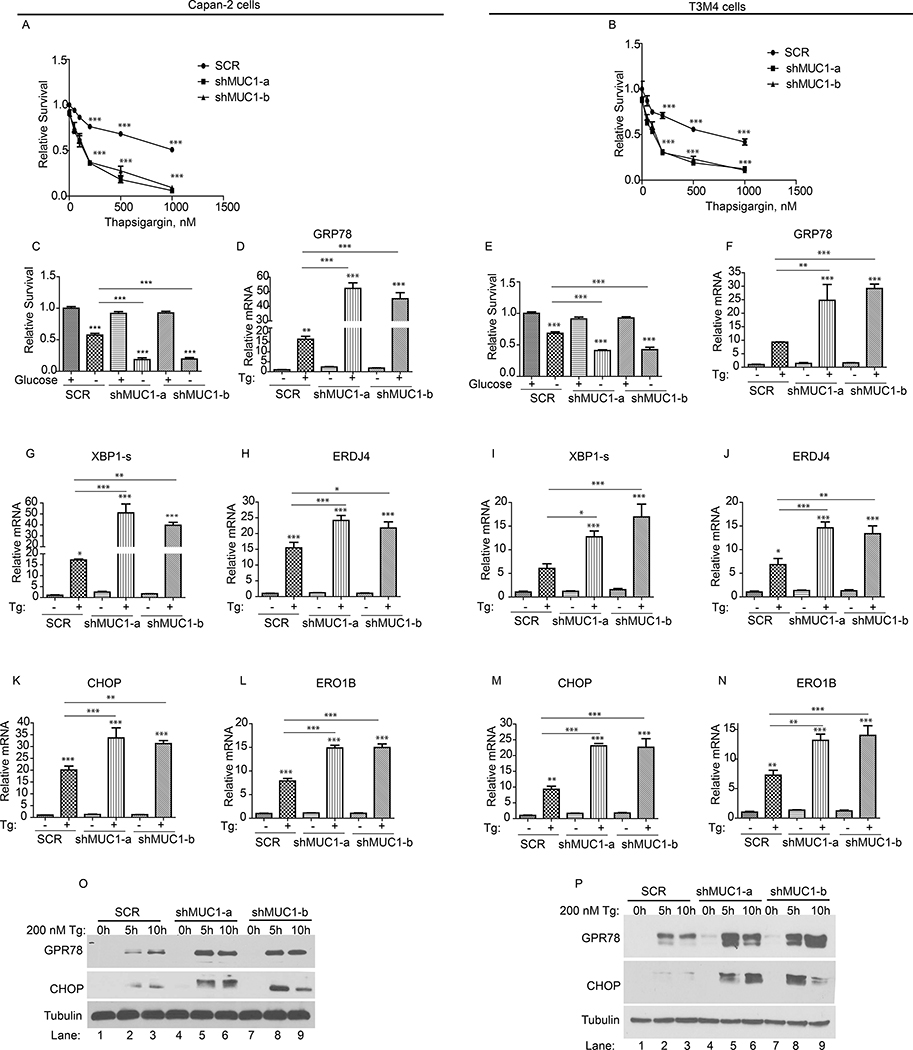
To assess the involvement of MUC1 in ER stress response, we generated stable MUC1 knockdown in a panel of four pancreatic cancer cell lines (Capan-2, PATU8902, CFPAC, and T3M4), by utilizing a scrambled hairpin (SCR; as a control) or two short hairpin RNA (shRNA), herein designated as shMUC1-a and shMUC1-b, targeting different regions of MUC1 mRNA. The SCR and MUC1 knockdown cells were then exposed to the UPR-inducing pharmacological agent thapsigargin, that inhibits ER calcium pump (47), or glucose starvation, a physiological UPR-inducer (47). MUC1 knockdown was confirmed by western blotting with antibody against the cytoplasmic tail of MUC1 protein (Fig. S1A). Assessment of the cell survival showed a thapsigargin-dependent decrease in survival of SCR cells (Fig. 1A–B and S1 B–C). Similarly, a decrease in survival upon glucose starvation was also observed in SCR cells (Fig. 1C and 1E; S1 D–E). Importantly, MUC1 knockdown cells exhibited a more robust and significant decrease in survival upon thapsigargin treatment or glucose starvation as compared to SCR cells (Fig. 1A–C, E and S1B–E). Because thapsigargin treatment and glucose deprivation, two ER stress-inducing conditions, produced similar effects on cell survival in four cell lines, downstream experiments were carried out using thapsigargin and two cell lines (Capan-2 and T3M4). To determine whether the decrease in cell survival was due to increased apoptosis, we performed caspase 3/7 activity assays using the SCR control and MUC1 knockdown cells, cultured with or without thapsigargin treatment, and noted that MUC1 knockdown cells showed increased caspase 3/7 activity relative to SCR cells (S1 F–G). Next, we evaluated the thapsigargin-induced expression of the UPR-related genes in SCR and MUC1 knockdown cells. We noted induction of GRP78, the major regulator of the UPR (Fig. 1D and F), and spliced XBP-1 (Fig. 1G and I) along with its downstream target ERDJ4 (Fig. 1H and J) in SCR cells (14). Likewise, CHOP, the transcription factor that activates the expression of ER stress-induced apoptosis-related pathway genes (8–13), was also induced in SCR cells (Fig. 1K and M) along with its downstream target ERO1B (8) (Fig. 1L and N). Significantly, the expression of all of these genes was higher in MUC1 knockdown cells (Fig. 1D and F–N), indicating more ER stress. Finally, we validated GRP78 and CHOP proteins expression in all four cell lines (Capan-2, T3M4, CFPAC and PATU8902) by western blotting that showed greater GRP78 and CHOP expression in MUC1 knockdown cells (Fig. 1O–P and S1 H–I). Altogether, knockdown of MUC1 enhances UPR signaling and cell death upon ER stress induction.
Figure 1: MUC1 deficiency exacerbates ER stress upon induction.
(A-C; E): Cell survival in SCR and MUC1 knockdown cells, in response to the indicated doses of thapsigargin (Tg, A-B) or glucose-starvation (C and E) for 48 hours, by MTT assays. Values were normalized to SCR. (D, F-N): The mRNA levels relative to SCR. Indicated cells were treated with thapsigargin (Tg, 200 nM) for 6 hours followed by total RNA isolation and qPCR with primers for indicated genes. (O-P): Expression levels of UPR marker proteins in cells treated with thapsigargin, by western blotting. Immunoblotting with anti-Tubulin antibody was used as a loading control. P-values are indicated as: p<0.05, *; p<0.01, **; p<0.001, ***.
Transcriptomic Analysis Reveals Alterations in the Pyrimidine Salvage Pathway and Cytidine Deaminase (CDA) Gene Expression upon ER Stress Induction
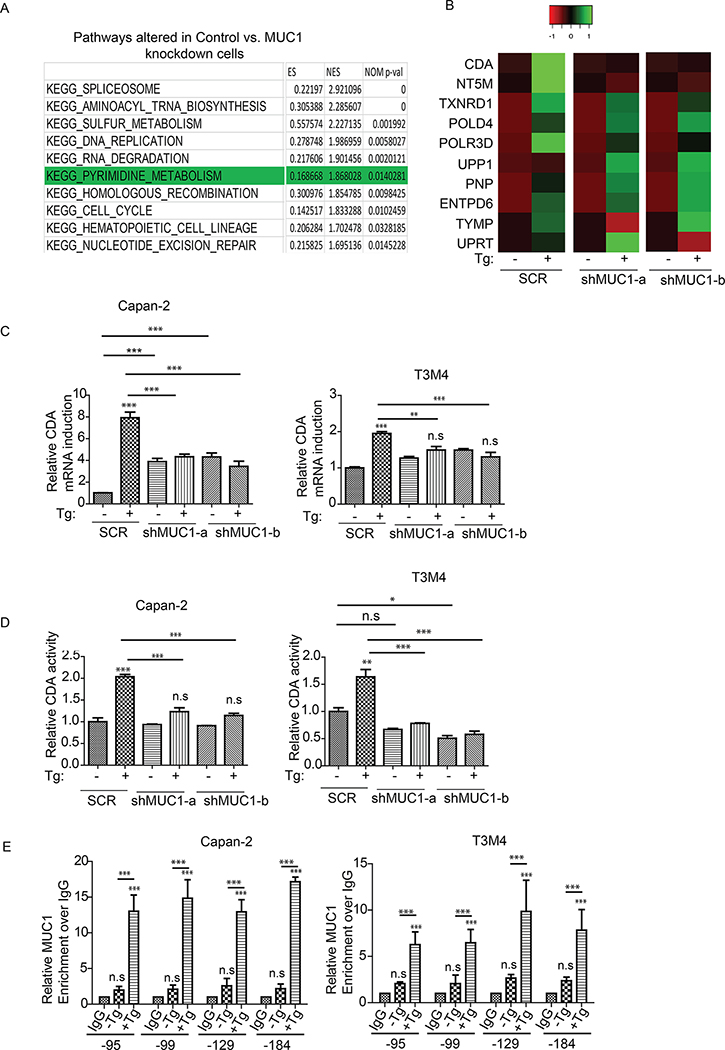
The results above showed that knockdown of MUC1 exacerbated UPR upon ER stress induction. Thus, we next investigated the mechanistic basis of MUC1-mediated suppression of UPR. To achieve that, we performed RNA-seq-based comparative transcriptomic studies of SCR and MUC1 knockdown cells with or without thapsigargin treatment. Pathway analysis by gene set enrichment analysis (GSEA) algorithm revealed that spliceosome, aminoacyl tRNA biosynthesis, sulfur metabolism, DNA replication, RNA degradation and the pyrimidine salvage were among the top altered pathways upon ER stress induction (Fig. 2A). Previous studies indicated that the pyrimidine metabolic pathway is essential for ER homeostasis as its inhibition induced ER stress (39). Given the recent roles of MUC1 in promoting chemotherapy resistance in pancreatic cancer cells by up-regulating the pyrimidine metabolic pathway (36, 48), we explored a potential link between the UPR and the pyrimidine pathway. Cytidine deaminase (CDA) was among the top genes altered in the pyrimidine metabolic pathway (Fig. 2B). A previous study reported that ER stress upregulated a CDA family protein, activation-induced CDA (40), an enzyme that catalyzes the irreversible deamination of deoxycytidine/cytidine to deoxyuridine/uridine to balance the pool of deoxyuridine/uridine (41, 42). This suggests not only the involvement of CDA in ER homeostasis but also that CDA may be at the interface of ER stress response and the pyrimidine metabolic pathway (39). Additional studies indicated that MUC1 is a potential regulator of CDA family proteins in response to other form of stresses such as genotoxic stress (49, 50). Accordingly, we further pursued CDA. To assess the role of CDA in the UPR, we first determined the relative CDA gene expression upon Thapsigargin treatment in SCR and MUC1 knockdown cells by qPCR analysis. Upon Thapsigargin-induced ER stress, CDA gene transcript levels increased in SCR cells as compared to MUC1 knockdown cells (Fig. 2C), corresponding to higher CDA protein levels in SCR cells (S2 A). We next measured the activity of the CDA enzyme in SCR and MUC1 knockdown cells and observed that MUC1 knockdown significantly abolished ER stress-induced CDA activity (Fig. 2D). MUC1 cytoplasmic tail is well established as a transcriptional co-activator that regulates genes involved in metabolism, apoptosis, and drug resistance (26, 37, 51, 52). We next asked whether MUC1 transcriptionally regulates CDA upon Thapsigargin treatment to affect the outcome of UPR signaling. To address that question, we performed ChIP assays with anti-MUC1 cytoplasmic tail antibody in cells treated with Thapsigargin, followed by qPCR with primers against various regions of the CDA promoter. For the positive control, we investigated MUC1 cytoplasmic tail occupancy on the CTGF promoter (51) (Fig. S2 B). We noted significant enrichment of MUC1 occupancy at various regions of the CDA gene promoter upon Thapsigargin treatment (Fig. 2E).
Figure 2: Transcriptomic analysis reveals alterations in the pyrimidine salvage pathway and Cytidine Deaminase (CDA) upon UPR induction.
(A): Top altered pathways in Capan-2 SCR vs. MUC1 knockdown (KD) cells upon ER stress induction with thapsigargin (Tg, 200 nM) for 6 hours. (B): Heatmap of genes altered in the pyrimidine pathway upon Tg treatment. The absolute values in Tg-treated samples were normalized over their base line levels in vehicle-treated cells. (C): CDA mRNA fold change, relative to SCR, upon treatment with thapsigargin (Tg, 200 nM) for 6 hours in indicated cells (SCR and MUC1 knockdown) by qPCR analysis. (D): CDA activity in SCR and MUC1 knockdown cells relative to SCR. Indicated cells were treated with thapsigargin (Tg, 200 nM) for 5 hours and the lysates were then utilized for CDA assay, using equal amounts of protein for each sample. (E): Bar charts representing the occupancy of MUC1 on the indicated regions of CDA promoter by ChIP assays upon Thapsigargin treatment. Cells were treated with thapsigargin (Tg, 200 nM) for 5 hours followed by cross-linking and preparation of protein and chromatin complex. Associated chromatin fragments were then eluted and amplified by qPCR using primers for various CDA promoter regions. IgG antibody pulldowns were used as negative controls. Enrichment is over IgG. P-values are indicated as: p<0.05, *; p<0.01, **; p<0.001, ***.
Inhibition of Either the CDA Enzyme Activity or the Pyrimidine Metabolic Pathway Sensitized to ER stress
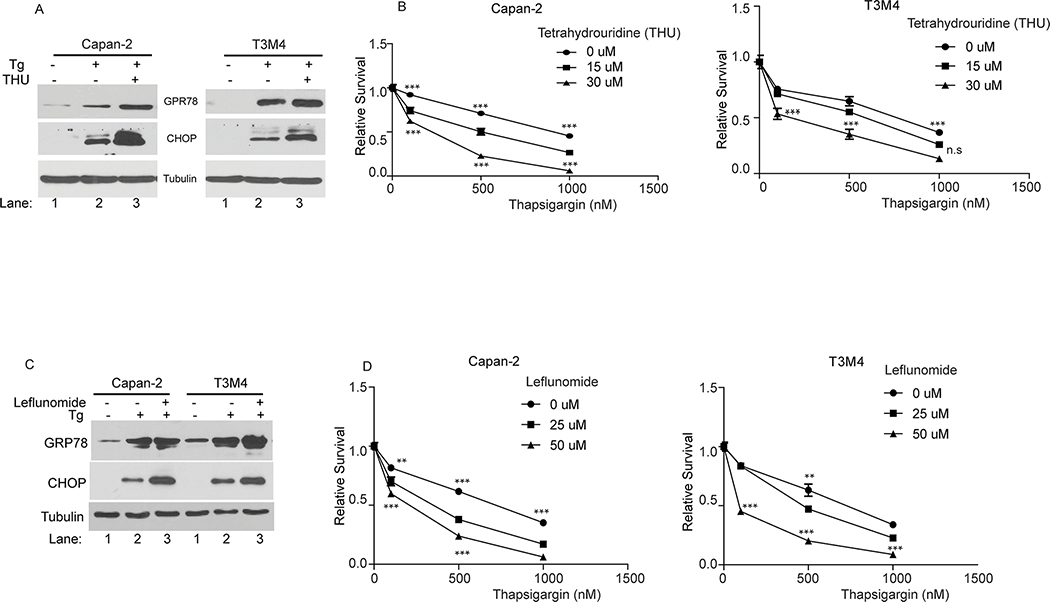
To explore the potential link between the CDA enzyme in the pyrimidine salvage pathway, and UPR signaling, we used Tetrahydrouridine (THU), a specific CDA inhibitor (53, 54). Chemical inhibition of the CDA protein with THU enhanced thapsigargin-induced ER stress as indicated by increased expression of UPR markers GRP78 and CHOP with THU and thapsigargin co-treatment (Fig. 3A). Moreover, the increased UPR signaling correlated with decreased cell survival in CDA-inhibited cells (Fig. 3B). Likewise, co-treatment of thapsigargin with the pyrimidine metabolic pathway inhibitor leflunomide (38) enhanced UPR signaling upon ER stress induction as shown by increased GRP78 and CHOP expression (Fig. 3C), consistent with the role of the pyrimidine metabolic pathway in ER homeostasis (39). Like CDA inhibition, pyrimidine metabolic pathway inhibition also greatly decreased cell survival upon ER stress induction (Fig. 3D). Taken together, these results indicate that the CDA-mediated pyrimidine salvage pathway, a previously uncharacterized axis of the UPR, is critical for adaptive ER stress response.
Figure 3: Inhibition of either the CDA enzyme activity or the pyrimidine pathway sensitizes cancer cells to ER stress.
(A and C): UPR marker protein levels in the indicated cells treated with thapsigargin (Tg; 200 nM) alone or in combination with either tetrahydrouridine (THU; 50 μM) or Leflunomide (50 μM) for 10 hours, by western blotting. (B and D): Cell survival in the indicated cells treated with thapsigargin alone or in combination with THU or Leflunomide for 48 hours, by MTT assays relative to untreated cells. P-values are indicated as: p<0.05, *; p<0.01, **; p<0.001, ***.
MUC1-mediated CDA Activity Correlates with Deoxycytidine to Deoxyuridine Reprogramming upon ER Stress Induction
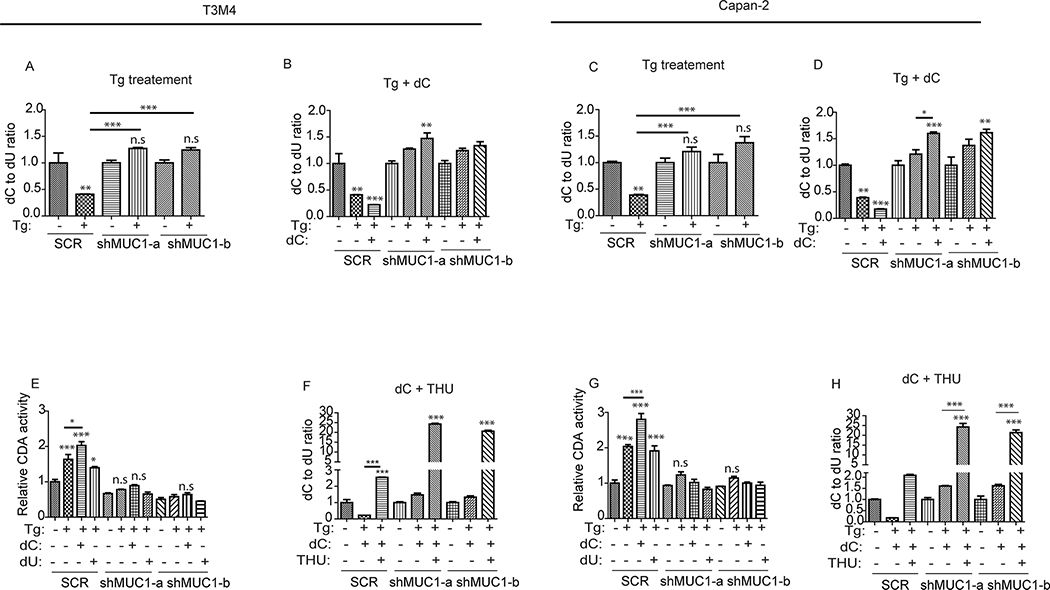
The CDA’s main function in cells is to maintain cellular deoxyuridine/uridine pool by deaminating deoxycytidine/cytidine (41, 42). As such, CDA has been known for its association with chemo-therapy resistance against nucleotide analogue-based therapies (55). The increased CDA activity in the SCR MUC1-expressing cells upon ER stress induction (Fig. 2D) associated with their resistance to ER stress (Fig. 1) on one hand, and their sensitivity to ER stress induction upon inhibition of CDA enzyme or the pyrimidine metabolic pathway (Fig. 3) on the other hand, prompted the question as to whether the pools of deoxycytidine and deoxyuridine are reprogrammed upon ER stress induction. To address this question, we measured deoxycytidine (dC) and deoxyuridine (dU) levels in SCR and MUC1 knockdown cells, treated or not with thapsigargin, by quantitative mass spectrometry. Polar metabolites extracted from the respective cells were subjected to LC-MS/MS analysis and quantitation analysis. Upon thapsigargin treatment, the ratio of dC to dU decreased in SCR cells (Fig. 4A and 4C), indicating an increase in dU levels. Notably, the dC to dU ratio not only did not decrease but in fact increased in MUC1 knockdown cells (Fig. 4A and 4C), indicating an accumulation of dC in MUC1 knockdown cells. Addition of exogenous dC to the cell culture media further decreased dC to dU ratio in SCR cells upon thapsigargin treatment when compared to thapsigargin alone (Fig. 4B and D), suggesting a further increase in dU levels in SCR cells when supplemented with dC, the substrate of CDA. On the other hand, dC supplementation to MUC1 knockdown cells caused dC to further accumulate in those cells indicated by a further increase in dC to dU ratio (Fig. 4B and 4D), suggesting that MUC1 knockdown cells are impaired in their ability to convert dC to dU even in the presence of abundant dC. However, supplementation with dU increased dU levels in MUC1 knockdown cells, indicated by a decreased dC to dU ratio, but not so much so in SCR cells (Fig. S3. A–B). Assessment of CDA activity showed that the enzyme activity further increased in the presence of its substrate dC, but not in the presence of dU (Fig 4E and G). Finally, we inhibited CDA activity using its specific inhibitor THU (53, 54), to determine whether CDA mediated the dC to dU alteration. Remarkably, inhibition of CDA abrogated the increase in dU levels and caused an accumulation of dC as indicated by an increased dC to dU ratio in SCR cells (Fig. 4F and H). This demonstrates that active CDA enzyme is required for dC to dU conversion in MUC1-expressing cells. In MUC1 knockdown cells where the CDA enzyme is less active (Fig. 2D), dC supplementation upon CDA inhibition further accumulated dC in the cells indicated by a further increased dC to dU ratio (Fig. 4F and H). Taken together, these results support the conclusion that MUC1 regulates CDA activity to promote dC to dU reprogramming upon ER stress induction.
Figure 4: MUC1-mediated CDA activity correlates with deoxycytidine to deoxyuridine reprogramming upon UPR induction.
(A-D, F and H): LC-MS/MS showing change in the levels of intracellular deoxycytidine (dC) and deoxyuridine (dU) assessed by dC to dU ratio relative to respective untreated cells. Indicated cells were treated with thapsigargin, (Tg; 200 nM) alone or in combination with dC (100 μM) or tetrahydrouridine (THU; 10 μM) for 16 hours. Experiments were performed in triplicates. All experiments were run in parallel with same control. (E and G): CDA activity, relative to SCR, is represented by the bar charts. Indicated cells were treated with thapsigargin (Tg, 200 nM) alone for 5 hours or in combination with dC (100 μM) or dU (100 μM). Experiments were done in parallel and used same control as in Fig. 2D. P-values are indicated as: p<0.05, *; p<0.01, **; p<0.001, ***.
Deoxyuridine Mitigated ER Stress and Provided Survival Advantage
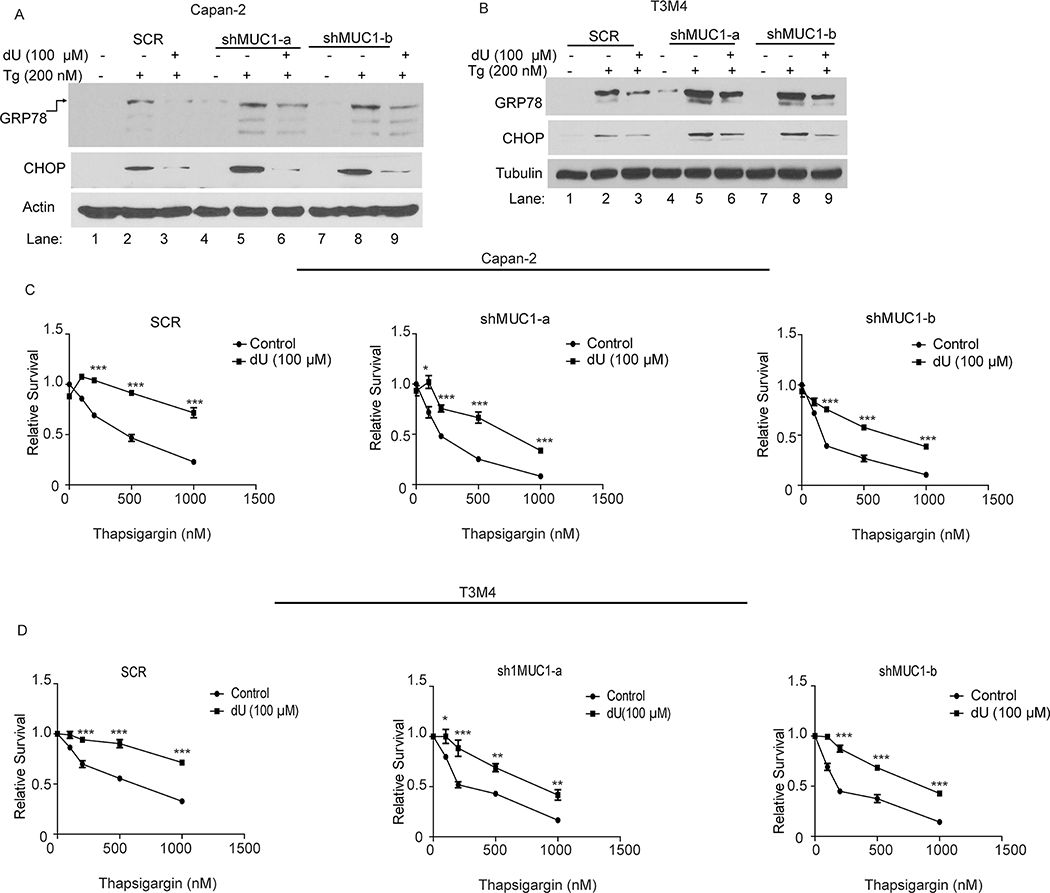
The dynamic deoxycytidine (dC) and deoxyuridine (dU) pools led us to assess the functional role of the CDA activity during ER stress induction. Hence, we investigated if dU, the product of the CDA enzymatic activity, can rescue cells from ER stress. To achieve this, we treated SCR and MUC1 knockdown cells with thapsigargin alone or in combination with dU and determined the expression levels of the UPR signaling markers GRP78 and CHOP by western blotting. Thapsigargin alone induced UPR signaling, indicated by increased expression of GRP78 and CHOP (Fig. 5A–B). However, addition of dU suppressed thapsigargin-induced expression of GRP78 and CHOP in both SCR and MUC1 knockdown cells (Fig. 5A–B). Similarly, the decreased cell survival due to thapsigargin treatment was reversed by the addition of dU in both SCR and MUC1 knockdown cells (Fig. 5C–D), indicating that dU mitigated ER stress. Furthermore, a significant rescue from ER stress-induced UPR response and cell survival was observed in MUC1-expressing cells supplemented with dC (Fig. S4A–D). However, dC-mediated rescue from ER stress was minimal in MUC1 knockdown cells (Fig. S4A–D). Taken together, supplementation with dU, the product of the CDA enzyme activity, rescued cells from ER stress.
Figure 5: Deoxyuridine rescues cancer cells from ER stress and provides survival advantage.
(A-B): UPR marker protein levels in the indicated cells treated with deoxyuridine (dU, 100 μM) for 5 hours and subsequently with thapsigargin (Tg; 200 nM) for additional 5 hours, by western blotting. Immunoblotting for Tubulin was used as a loading control. (C-D): Cell survival in the indicated cells treated with thapsigargin alone or in combination with dU for 48 hours, by MTT assays. The values are relative to their respective untreated cells. P-values are indicated as: p<0.05, *; p<0.01, **; p<0.001, ***.
Deoxyuridine Modulates ER Stress-induced Reactive Oxygen Species (ROS) Generation and Oxidative Stress
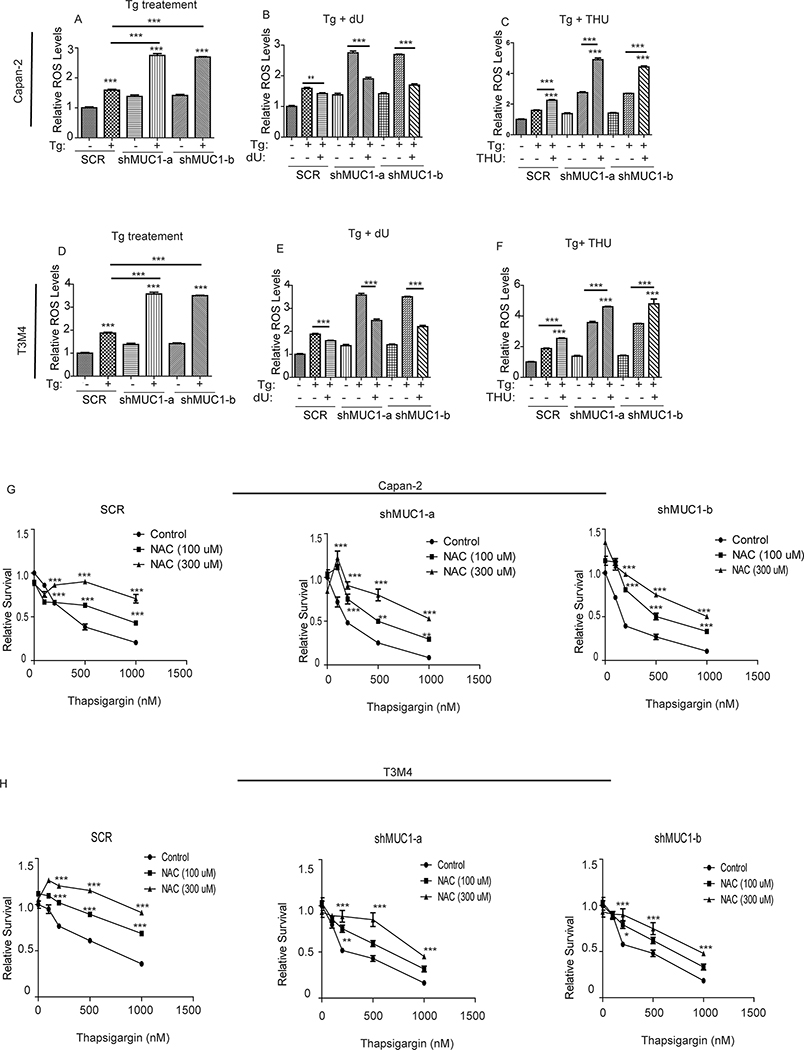
To gain further mechanistic insights into the regulatory role of the MUC1-CDA axis of the UPR signaling, we first evaluated if MUC1 could mediate induction of O-GlcNAcylation, a protein post-translational modification, to contribute to protection from ER stress. Protein O-GlcNacylation is known to confer tolerance to some forms of stresses (56–58) and is generated by covalent addition of N-Acetyl-Glucosamine (N-GlcNAc), derived from Uridine Diphosphate (UDP)-N-Acetyl-Glucosamine, to serine/threonine residues of some proteins. Previous studies demonstrated that MUC1 over-expression increased the levels of the UDP-conjugated sugar moieties such as UDP-N-Acetyl-Glucosamine, UDP-glucose and UDP-galactose (37). High levels of UDP-conjugated sugars not only imply high levels of uridine, but also suggest a potentially elevated O-GlcNacylation in MUC1-expressing cells upon ER stress induction. However, contrastingly, SCR cells exhibited lower O-GlcNAcylation levels upon Thapsigargin treatment compared to MUC1 knockdown cells (Fig. S5 A–B). Therefore, we pursued other mechanisms. Beside the pyrimidine salvage pathway, the sulfur metabolic pathway was another one of the top significantly altered pathways in the cells upon thapsigargin (Fig. 2A), which piqued our curiosity in assessing how this might affect ROS levels during ER stress induction given that 1) sulfur metabolism is critical for ROS regulation and oxidative stress response (59, 60), 2) ROS induces ER stress (31, 32, 61) and 3) MUC1 is a known regulator of ROS and oxidative stress (29, 30). Notably, uridine also has emerging anti-oxidants properties (62–64). Initial evidence for the involvement of ROS is provided by the increased expression of the ROS-regulated protein, the nuclear factor erythroid 2-related factor 2 (NRF2) (65), in MUC1 knockdown cells compared to SCR cells upon thapsigargin treatment (Fig. S5 C), which suggested an increased ROS levels in MUC1 knockdown cells. To assess ROS levels, we used the ROS sensitive 2′,7′–dichlorofluorescin diacetate (DCFDA) dye that fluoresces in the presence of ROS. Treatment with H2O2, used as a positive control, increased ROS levels as expected (Fig. S5 D–E). Likewise, thapsigargin treatment increased ROS in SCR cells (Fig. 6A and D). However, the increase in ROS levels was more pronounced in MUC1 knockdown cells (Fig. 6A and D). Supplementation with dU reversed the thapsigargin-induced increase in ROS production in both SCR and MUC1 knockdown cells (Fig. 6B and E), indicating a rescue from ROS-mediated ER stress. CDA inhibition with THU treatment further increased ROS levels in both SCR and MUC1 knockdown cells (Fig. 6C and F). Finally, we assessed whether the anti-oxidant N-Acetyl-Cysteine (NAC) could rescue survival in cells treated with thapsigargin. We noted that NAC significantly abolished the thapsigargin-induced decrease in survival in both SCR and MUC1 knockdown cells (Fig. 6G and H), mimicking the effect of deoxyuridine (Fig. 5C and D). Taken together, these results demonstrate that CDA-mediated deoxyuridine salvage may function as anti-oxidant, in a MUC1-dependent manner, to protect pancreatic cancer cells against ROS generation during ER stress.
Figure 6: Deoxyuridine modulates ER stress-induced Reactive Oxygen Species (ROS) generation and Oxidative stress.
(A-F): ROS levels in the indicated cells treated with thapsigargin (Tg, 200nM) alone (A and D) or in combination with deoxyuridine (dU; B and E) or tetrahydrouridine (THU; C and F) for 12 hours. ROS levels were assessed by utilizing the fluorescent dye 2′,7′–dichlorofluorescin diacetate (DCFDA). All experiments were run in parallel and used same controls. The values were relative to SCR. (G and H): Relative cell survival in cells treated with thapsigargin alone or in combination with N-Acetyl Cysteine (NAC) for 48 hours, as measured by MTT assays. P-values are indicated as: p<0.05, *; p<0.01, **; p<0.001, ***.
Discussion
Tumor cells prevail in a very hostile microenvironment featuring various forms of stresses including stress in the endoplasmic reticulum (20, 21). Accordingly, proteins related to the ER stress response, UPR, are up-regulated in various cancers (20, 21). The UPR signaling in cancer is a paradox as both pro-survival and pro-apoptotic pathways are concomitantly up-regulated and yet the balance is tilted toward an aggressive growth and survival of cancer cells. This inconsistency argues that the tumor microenvironment harbors pro-adaptive signaling pathways that protect tumor cells against the deleterious effects of the various forms of stresses encountered in tumor, including chemotherapeutic drugs that disrupt ER homeostasis. In the present study, we demonstrated a novel MUC1-CDA pro-adaptive axis of the UPR.
Knockdown of MUC1 sensitized cancer cells to ER stress-inducing conditions, thapsigargin treatment or glucose starvation, as indicated by the increased UPR signaling and cell death in MUC1 knockdown cells upon ER stress induction (Fig. 1 and S1). Those results validate the pro-survival role of MUC1 in cancer cells (66). Transcriptomic analysis revealed that the pyrimidine salvage pathway was among the top altered pathways (Fig. 2A) upon thapsigargin treatment along with the CDA gene (Fig. 2B–C). This is consistent with studies indicating that 1) the pyrimidine metabolic pathway is critical for ER homeostasis (39), and that 2) CDA may be at the interface of ER stress response and the pyrimidine metabolic pathway (40). ChIP and CDA activity assays showed that, upon ER stress induction, MUC1, a known transcriptional co-activator (37, 51) and a potential regulator of CDA family proteins (49, 50), occupied CDA gene promoter (Fig. 2E) and transcriptionally increased the expression and activity of CDA (Fig 2C–D and S2 A). Metabolomic analysis demonstrated that these effects correlated with a reprogramming of deoxycytidine to deoxyuridine (Fig 4) upon ER stress induction. Abundant deoxyuridine, a metabolite with emerging anti-oxidative properties (62–64), mitigated ER stress by reducing ROS levels as evidenced by the rescue from ER stress (Fig. 5 and 6). Similarly, deoxycytidine, the substrate of CDA enzymatic activity, alleviated ER stress in MUC1-expressing cells when co-treated with thapsigargin, but negligible rescuing effect was observed in MUC1 knockdown cells (Fig. S4). This can be explained by the CDA enzyme being active in MUC1-expressing cells to convert deoxycytidine to deoxyuridine upon Thapsigargin treatment. However, in MUC1 knockdown cells, CDA enzyme activity is impaired upon ER stress induction (Fig. 2D). The increase in the basal level of CDA gene transcript seen in untreated Capan-2 MUC1 knockdown cells (Fig. 2C), though not associated with an increased CDA basal activity (Fig. 2D), may be due to some forms of compensatory responses to MUC1 knock down. However, such compensatory mechanism is not sustained in stress condition as SCR cells expressed more CDA than MUC1 knock down cells upon thapsigargin treatment (Fig. 2C). It may also reflect a cell type difference when compared to the response in T3M4 MUC1 knock down cells where CDA basal levels did not change.
Our findings agree with previous studies demonstrating that MUC1 is critical for cancer cell survival in oxidative stress (29, 30). However, our findings present a novel distinct metabolic mechanism by which MUC1 regulates cancer cell response to stress via the CDA protein. Consistently, past studies linked MUC1 to chemotherapy and radiotherapy resistance via upregulation of pyrimidine nucleotide biosynthesis (36, 67). MUC1-mediated resistance to pyrimidine analogue-based chemotherapies is, at least in part, due to increased pyrimidine biosynthesis (36). CDA is also known to deaminate pyrimidine analogue drugs and deactivate them (68). In line with that, other labs have shown that MUC1 knockdown, which as per our data impairs CDA activity (Fig. 2D), sensitizes cancer cells to pyrimidine analogue drugs such as gemcitabine (26). Like MUC1, CDA is also linked to the development of chemotherapy resistance in cancer (69).
The alterations in the pyrimidine metabolic pathway, DNA replication, spliceosome, aminoacyl tRNA biosynthesis, nucleotide excision repair, and homologous recombination pathways (Fig. 2A) further argue that MUC1 and CDA may integrate and coordinate stress responses that implicate a network of pathways to affect the outcome of stress response. That idea is supported by studies demonstrating that CDA family proteins not only promote DNA replication by balancing cellular pyrimidine nucleotides levels (70, 71), but also are involved in mRNA editing and translation in hypoxic stress adaptation and survival (72). Our data demonstrate that inhibition of either the CDA or pyrimidine metabolic pathway sensitized MUC1-expressing cells to ER stress induction and cell death (Fig. 3), further linking CDA to stress adaptation and survival via pyrimidine metabolic alterations. Additionally, nucleotide pools can also regulate DNA damage response, a function that directly affects cells survival in response to genotoxic stress. ROS, ER stress and chemotherapeutic drugs induce genotoxic stress (73–75). Interestingly, MUC1 is a known player in genotoxic stress response (49) and a potential regulator of CDA family proteins upon genotoxic stress induction (49, 50). Like CDA family proteins, MUC1 is also a known epigenetic regulator in cancer (72, 76–79).
Finally, MUC1-mediated regulation of ER stress response may, in part, be mediated by calcium signaling. One of the primary roles of ER is maintaining the intracellular calcium storage and signaling (1). Along the same lines, our current findings showing the role of MUC1 in ER homeostasis and ER stress signaling are supported by previous publications establishing a role of MUC1 in calcium signaling (80, 81). It has been shown that thapsigargin induces ER stress-related cytotoxicity by inhibiting calcium flow to the ER (47). Therefore, the increased sensitivity of MUC1 knock down cells to ER stress/thapsigargin (Fig. 1 and S1) suggests that the flow of calcium from the cytosol to the ER may be impaired in MUC1 knock down cells. Our studies also demonstrate increased apoptosis in MUC1 knock down cells in response to ER stress, as assessed by caspase 3/7 activity assay (S1 F–G). These results are further supported by previous studies showing that cytosolic calcium contributes to ER stress-related cell death by activating the caspase cascade (82, 83). Future studies will substantiate calcium-related mechanisms for MUC1-mediated mitigation of ER stress. However, remarkably, not only does calcium cross-talk with ROS (84) but it also regulates the expression of CDA family protein members (85). Notably, our data also demonstrates that MUC1, which controls the expression of CDA family proteins (49, 50), indeed regulates CDA gene expression and activity upon ER stress signaling (Fig. 2C–E and S2 A). Taken together, MUC1 facilitates a novel pro-adaptive axis of stress response through a combination of converging mechanisms, including ROS alterations, CDA-mediated deoxycytidine-to-deoxyuridine metabolic reprogramming, and potentially a calcium-related mechanism. Considering the role of ER stress in tumor cell survival and chemotherapy response (86) and the role of MUC1 in intracellular signaling and metabolic reprogramming (67, 87–92), these studies provide novel avenues for targeting aggressiveness in cancer.
Materials and Methods
Cell Culture and Reagents
Pancreatic cancer cell lines Capan-2, T3M4, PATU8902 and CFPAC have been already described (36). Cell lines were validated by STR profiling by the Genetics Core at the University of Arizona. All cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM) (Sigma-Aldrich, St Louis, MO USA) supplemented with 10% fetal bovine serum (FBS), 100 I.U/ml penicillin, 100 μg/ml streptomycin, and incubated at 37°C in a humidified incubator with 5% CO2. Thapsigargin was purchased from Cell Signaling Technology (Danvers, MA USA) and stocks were diluted in DMSO. Tetrahydrouridine (THU) was purchased from Cayman Chemical (Ann Arbor, MI USA), Leflunomide was from Enzo Life Sciences (Farmingdale, NY USA). Deoxycytidine and deoxyuridine were obtained from MP Biomedical (Irvine, CA USA) and Sigma-Aldrich, respectively.
Lentivirus Transfection
For the generation of MUC1 knockdown cells, short hairpin RNA (shRNA) constructs, with scrambled SCR or targeting two independent regions of MUC1 mRNA, were obtained from Millipore Sigma (Burlington, MA USA). These lentiviral constructs were used to generate packaged lentiviruses by transfecting the constructs with packaging constructs into HEK293T cell to produce supernatants which were used to transduce indicated cells. Cells were selected at 48 hours post infection using puromycin for 72 hours or until complete death of un-infected cells.
MTT Cytotoxicity Assays
Cell viability was determined by MTT assays as described previously (36). Experiments were repeated at least three times in triplicates.
RNA Isolation and qRT-PCR
Total RNA isolation and qPCR were carried out as previously described (36). Gene-specific primers are included in the Primers Table in supplementary materials. Experiments were repeated at least three times in triplicates.
Chromatin immunoprecipitation assay (ChIP)
Chromatin immunoprecipitation (ChIP) assay was carried out two times using the Thermo Fisher Scientific kit. In brief, cells were treated or left un-treated with thapsigargin for 5 hours. Fragmented chromatin lysate was subjected to immunoprecipitation with anti-MUC1 antibodies followed by quantitative PCR with primer sets specific to various regions of the CDA promoter. The qPCR data were analyzed using the percent input method and normalized against the SCR IgG. The percent input was calculated as 100 × 2 to the power of (adjusted input − CT of IP), where the adjusted input is the CT (threshold cycle) of the input minus log2(dilution factor). The results are represented as a fold increase over enrichment detected using IgG.
Immunoblotting
For protein isolation and western blotting, cells were washed with 1X PBS and lysed in radio-immunoprecipitation assay (RIPA) lysis buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium Deoxycholate, 0.1% SDS supplemented with protease inhibitor tablets). Protein quantification and western blotting were carried out as previously described (36). Antibody against MUC1 has been previously described (51). Antibodies against GRP78, CHOP, NRF2 and O-GlcNAc were from Cell Signaling Technology. CDA antibody was from Santa Cruz Technology (Dallas, TX USA).
CDA and Caspase 3/7 Activity Assays
For CDA activity assay, 1 million cells were treated as indicated and processed using the CDA activity assay kit as per the manufacturer’s protocol (Biovision, Milpitas, CA USA). Caspase 3/7 activity assay was carried as per the manufacturer’s protocol (Promega, Madison, WI USA). Experiments were repeated two times.
Reactive Oxygen Species (ROS) Assay
For measuring ROS, 15000 cells were plated in a clear flat bottom black 96-well plate in triplicate. 12 hours later, the cells were treated with the indicated chemicals. Then, the media was replaced with fresh media containing 20 μM of the fluorescent dye carboxy-H2DCFDA) or carboxy-DCFDA (negative SCR), for 30 min after which the cells were first washed with PBS before additional 100 μl of PBS was added to each well. DCFDA fluorescence was measured at 485 nm excitation wavelength and 529 nm emission wavelength using the Biotek Cytation 3 plate reader. H2O2 treatment was used as a positive control for ROS generation. Experiments were repeated three times.
RNA-seq Analysis
SCR and MUC1 knockdown (shMUC1-a and shMUC1-b) cells were left un-treated or treated with Thapsigargin followed by total RNA isolation using Qiagen RNAeasy mini kit (Germantown, MD USA), and library was preparation for sequencing. The library quality was evaluated using an Agilent 2100 Bioanalyzer before sequencing through an Illumina system. TopHat2 was used for alignment and differential expression was done through DESeq2 R package. Gene set enrichment analysis (GSEA) was performed using the Broad Institute algorithm (93). Heatmaps were generated through ggplot2 R package.
Quantitative Mass Spectrometry
Indicated cells were left un-treated or treated as indicated in three replicates. Then, the media was removed, and the cells were quickly washed with Milli-Q water to remove residual media. Polar metabolites were extracted by lysing the cells in pre-chilled 80% mass spectrometry-grade methanol on dry ice. After pipetting up and down, the extracts were transferred into 1.5 ml Eppendorf tubes and incubated on dry ice for 1 hour before being vortexed to allow for complete lysis. Following centrifugation at 15000 RPM for 15 minutes, the supernatant was collected into fresh Eppendorf tubes and dried in speed vacuum. The pellet was then re-suspended in 50% mass spectrometry-grade methanol. The resulting cell extract was then analyzed with LC-MS/MS. Peak areas were integrated using MassLynx (Water Inc.) and normalized to the respective protein concentrations. The resulting peak areas were subjected to relative quantification analyses.
Statistical Analysis
For the assessment of statistical significance, one-way or two-way ANOVA was performed to compare differences in and between treatment groups. Statistical tests were performed using GraphPad Prism5 software. P < 0.05 was considered significant.
Supplementary Material
Acknowledgment
This work was supported in part by funding from the National Institutes of Health (R01 CA216853, R01 CA163649, R01 CA210439, NCI) to PKS, the Specialized Programs of Research Excellence (SPORE, 2P50 CA127297, NCI) to PKS, P01 CA2117798 (NCI) to PKS, and a supplement to NIH grant (R01CA216853-01) to OAA. We would also like to acknowledge the Fred & Pamela Buffett Cancer Center Support Grant (P30CA036727, NCI) for supporting shared resources.
Footnotes
Conflicts of Interest: The authors declare that they have no conflict of interest.
References
- 1.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nature reviews Molecular cell biology. 2003;4(7):517–29. [DOI] [PubMed] [Google Scholar]
- 2.Fagone P, Jackowski S. Membrane phospholipid synthesis and endoplasmic reticulum function. Journal of lipid research. 2009;50 Suppl:S311–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikonen E Cellular cholesterol trafficking and compartmentalization. Nature reviews Molecular cell biology. 2008;9(2):125–38. [DOI] [PubMed] [Google Scholar]
- 4.Ron D, Hampton RY. Membrane biogenesis and the unfolded protein response. The Journal of cell biology. 2004;167(1):23–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jun HS, Lee YM, Cheung YY, McDermott DH, Murphy PM, De Ravin SS, et al. Lack of glucose recycling between endoplasmic reticulum and cytoplasm underlies cellular dysfunction in glucose-6-phosphatase-beta-deficient neutrophils in a congenital neutropenia syndrome. Blood. 2010;116(15):2783–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marini C, Ravera S, Buschiazzo A, Bianchi G, Orengo AM, Bruno S, et al. Discovery of a novel glucose metabolism in cancer: The role of endoplasmic reticulum beyond glycolysis and pentose phosphate shunt. Sci Rep. 2016;6:25092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2(6):326–32. [DOI] [PubMed] [Google Scholar]
- 8.Li G, Mongillo M, Chin KT, Harding H, Ron D, Marks AR, et al. Role of ERO1-alpha-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J Cell Biol. 2009;186(6):783–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y, Brewer JW, Diehl JA, Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol. 2002;318(5):1351–65. [DOI] [PubMed] [Google Scholar]
- 10.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21(4):1249–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song B, Scheuner D, Ron D, Pennathur S, Kaufman RJ. Chop deletion reduces oxidative stress, improves beta cell function, and promotes cell survival in multiple mouse models of diabetes. J Clin Invest. 2008;118(10):3378–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorp E, Li G, Seimon TA, Kuriakose G, Ron D, Tabas I. Reduced apoptosis and plaque necrosis in advanced atherosclerotic lesions of Apoe−/− and Ldlr−/− mice lacking CHOP. Cell Metab. 2009;9(5):474–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12(7):982–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23(21):7448–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–6. [DOI] [PubMed] [Google Scholar]
- 16.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287(5453):664–6. [DOI] [PubMed] [Google Scholar]
- 17.Zeng T, Peng L, Chao H, Xi H, Fu B, Wang Y, et al. IRE1alpha-TRAF2-ASK1 complex-mediated endoplasmic reticulum stress and mitochondrial dysfunction contribute to CXC195-induced apoptosis in human bladder carcinoma T24 cells. Biochem Biophys Res Commun. 2015;460(3):530–6. [DOI] [PubMed] [Google Scholar]
- 18.Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16(11):1345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–64. [DOI] [PubMed] [Google Scholar]
- 20.Song M, Sandoval TA, Chae CS, Chopra S, Tan C, Rutkowski MR, et al. IRE1alpha-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature. 2018;562(7727):423–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014;14(9):581–97. [DOI] [PubMed] [Google Scholar]
- 22.Hayakawa Y, Hirata Y, Nakagawa H, Sakamoto K, Hikiba Y, Kinoshita H, et al. Apoptosis signal-regulating kinase 1 and cyclin D1 compose a positive feedback loop contributing to tumor growth in gastric cancer. Proc Natl Acad Sci U S A. 2011;108(2):780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hui L, Zatloukal K, Scheuch H, Stepniak E, Wagner EF. Proliferation of human HCC cells and chemically induced mouse liver cancers requires JNK1-dependent p21 downregulation. J Clin Invest. 2008;118(12):3943–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang Q, Chen J, Beezhold KJ, Castranova V, Shi X, Chen F. JNK1 activation predicts the prognostic outcome of the human hepatocellular carcinoma. Mol Cancer. 2009;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau SK, Weiss LM, Chu PG. Differential expression of MUC1, MUC2, and MUC5AC in carcinomas of various sites: an immunohistochemical study. Am J Clin Pathol. 2004;122(1):61–9. [DOI] [PubMed] [Google Scholar]
- 26.Nath S, Daneshvar K, Roy LD, Grover P, Kidiyoor A, Mosley L, et al. MUC1 induces drug resistance in pancreatic cancer cells via upregulation of multidrug resistance genes. Oncogenesis. 2013;2:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jing X, Liang H, Hao C, Yang X, Cui X. Overexpression of MUC1 predicts poor prognosis in patients with breast cancer. Oncol Rep. 2019;41(2):801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M, Law ME, Castellano RK, Law BK. The unfolded protein response as a target for anticancer therapeutics. Crit Rev Oncol Hematol. 2018;127:66–79. [DOI] [PubMed] [Google Scholar]
- 29.Hiraki M, Suzuki Y, Alam M, Hinohara K, Hasegawa M, Jin C, et al. MUC1-C Stabilizes MCL-1 in the Oxidative Stress Response of Triple-Negative Breast Cancer Cells to BCL-2 Inhibitors. Sci Rep. 2016;6:26643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin L, Li Y, Ren J, Kuwahara H, Kufe D. Human MUC1 carcinoma antigen regulates intracellular oxidant levels and the apoptotic response to oxidative stress. J Biol Chem. 2003;278(37):35458–64. [DOI] [PubMed] [Google Scholar]
- 31.Farooqi AA, Li KT, Fayyaz S, Chang YT, Ismail M, Liaw CC, et al. Anticancer drugs for the modulation of endoplasmic reticulum stress and oxidative stress. Tumour Biol. 2015;36(8):5743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eletto D, Chevet E, Argon Y, Appenzeller-Herzog C. Redox controls UPR to control redox. J Cell Sci. 2014;127(Pt 17):3649–58. [DOI] [PubMed] [Google Scholar]
- 33.Blackburn RV, Spitz DR, Liu X, Galoforo SS, Sim JE, Ridnour LA, et al. Metabolic oxidative stress activates signal transduction and gene expression during glucose deprivation in human tumor cells. Free Radic Biol Med. 1999;26(3–4):419–30. [DOI] [PubMed] [Google Scholar]
- 34.Lee YJ, Galoforo SS, Berns CM, Chen JC, Davis BH, Sim JE, et al. Glucose deprivation-induced cytotoxicity and alterations in mitogen-activated protein kinase activation are mediated by oxidative stress in multidrug-resistant human breast carcinoma cells. J Biol Chem. 1998;273(9):5294–9. [DOI] [PubMed] [Google Scholar]
- 35.Spitz DR, Sim JE, Ridnour LA, Galoforo SS, Lee YJ. Glucose deprivation-induced oxidative stress in human tumor cells. A fundamental defect in metabolism? Ann N Y Acad Sci. 2000;899:349–62. [DOI] [PubMed] [Google Scholar]
- 36.Shukla SK, Purohit V, Mehla K, Gunda V, Chaika NV, Vernucci E, et al. MUC1 and HIF-1alpha Signaling Crosstalk Induces Anabolic Glucose Metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer Cell. 2017;32(1):71–87 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaika NV, Gebregiworgis T, Lewallen ME, Purohit V, Radhakrishnan P, Liu X, et al. MUC1 mucin stabilizes and activates hypoxia-inducible factor 1 alpha to regulate metabolism in pancreatic cancer. Proc Natl Acad Sci U S A. 2012;109(34):13787–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruckemann K, Fairbanks LD, Carrey EA, Hawrylowicz CM, Richards DF, Kirschbaum B, et al. Leflunomide inhibits pyrimidine de novo synthesis in mitogen-stimulated T-lymphocytes from healthy humans. J Biol Chem. 1998;273(34):21682–91. [DOI] [PubMed] [Google Scholar]
- 39.Ren Z, Chen S, Qing T, Xuan J, Couch L, Yu D, et al. Endoplasmic reticulum stress and MAPK signaling pathway activation underlie leflunomide-induced toxicity in HepG2 Cells. Toxicology. 2017;392:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kriss CL, Pinilla-Ibarz JA, Mailloux AW, Powers JJ, Tang CH, Kang CW, et al. Overexpression of TCL1 activates the endoplasmic reticulum stress response: a novel mechanism of leukemic progression in mice. Blood. 2012;120(5):1027–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung SJ, Fromme JC, Verdine GL. Structure of human cytidine deaminase bound to a potent inhibitor. J Med Chem. 2005;48(3):658–60. [DOI] [PubMed] [Google Scholar]
- 42.Vu LT, Tsukahara T. C-to-U editing and site-directed RNA editing for the correction of genetic mutations. Biosci Trends. 2017;11(3):243–53. [DOI] [PubMed] [Google Scholar]
- 43.Norris AM, Gore A, Balboni A, Young A, Longnecker DS, Korc M. AGR2 is a SMAD4-suppressible gene that modulates MUC1 levels and promotes the initiation and progression of pancreatic intraepithelial neoplasia. Oncogene. 2013;32(33):3867–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dumartin L, Alrawashdeh W, Trabulo SM, Radon TP, Steiger K, Feakins RM, et al. ER stress protein AGR2 precedes and is involved in the regulation of pancreatic cancer initiation. Oncogene. 2017;36(22):3094–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chevet E, Fessart D, Delom F, Mulot A, Vojtesek B, Hrstka R, et al. Emerging roles for the pro-oncogenic anterior gradient-2 in cancer development. Oncogene. 2013;32(20):2499–509. [DOI] [PubMed] [Google Scholar]
- 46.Zweitzig DR, Smirnov DA, Connelly MC, Terstappen LW, O’Hara SM, Moran E. Physiological stress induces the metastasis marker AGR2 in breast cancer cells. Mol Cell Biochem. 2007;306(1–2):255–60. [DOI] [PubMed] [Google Scholar]
- 47.Oslowski CM, Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods in enzymology. 2011;490:71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Zhang Y, Wang W, Zhu Y, Chen Y, Tian B. Gemcitabine treatment induces endoplasmic reticular (ER) stress and subsequently upregulates urokinase plasminogen activator (uPA) to block mitochondrial-dependent apoptosis in Panc-1 cancer stem-like cells (CSCs). PLoS One. 2017;12(8):e0184110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei X, Xu H, Kufe D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell. 2005;7(2):167–78. [DOI] [PubMed] [Google Scholar]
- 50.Menendez D, Nguyen TA, Snipe J, Resnick MA. The Cytidine Deaminase APOBEC3 Family Is Subject to Transcriptional Regulation by p53. Molecular cancer research : MCR. 2017;15(6):735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Behrens ME, Grandgenett PM, Bailey JM, Singh PK, Yi CH, Yu F, et al. The reactive tumor microenvironment: MUC1 signaling directly reprograms transcription of CTGF. Oncogene. 2010;29(42):5667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agata N, Ahmad R, Kawano T, Raina D, Kharbanda S, Kufe D. MUC1 oncoprotein blocks death receptor-mediated apoptosis by inhibiting recruitment of caspase-8. Cancer Res. 2008;68(15):6136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen RM, Wolfenden R. Cytidine deaminase from Escherichia coli. Purification, properties and inhibition by the potential transition state analog 3,4,5,6-tetrahydrouridine. J Biol Chem. 1971;246(24):7561–5. [PubMed] [Google Scholar]
- 54.Stoller RG, Myers CE, Chabner BA. Analysis of cytidine deaminase and tetrahydrouridine interaction by use of ligand techniques. Biochem Pharmacol. 1978;27(1):53–9. [DOI] [PubMed] [Google Scholar]
- 55.Weizman N, Krelin Y, Shabtay-Orbach A, Amit M, Binenbaum Y, Wong RJ, et al. Macrophages mediate gemcitabine resistance of pancreatic adenocarcinoma by upregulating cytidine deaminase. Oncogene. 2014;33(29):3812–9. [DOI] [PubMed] [Google Scholar]
- 56.Alejandro EU, Bozadjieva N, Kumusoglu D, Abdulhamid S, Levine H, Haataja L, et al. Disruption of O-linked N-Acetylglucosamine Signaling Induces ER Stress and beta Cell Failure. Cell Rep. 2015;13(11):2527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Denzel MS, Storm NJ, Gutschmidt A, Baddi R, Hinze Y, Jarosch E, et al. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell. 2014;156(6):1167–78. [DOI] [PubMed] [Google Scholar]
- 58.Ferrer CM, Reginato MJ. Sweet connections: O-GlcNAcylation links cancer cell metabolism and survival. Mol Cell Oncol. 2015;2(1):e961809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dickinson DA, Forman HJ. Glutathione in defense and signaling: lessons from a small thiol. Ann N Y Acad Sci. 2002;973:488–504. [DOI] [PubMed] [Google Scholar]
- 60.Kannan N, Nguyen LV, Makarem M, Dong Y, Shih K, Eirew P, et al. Glutathione-dependent and -independent oxidative stress-control mechanisms distinguish normal human mammary epithelial cell subsets. Proc Natl Acad Sci U S A. 2014;111(21):7789–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeeshan HM, Lee GH, Kim HR, Chae HJ. Endoplasmic Reticulum Stress and Associated ROS. Int J Mol Sci. 2016;17(3):327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lebrecht D, Vargas-Infante YA, Setzer B, Kirschner J, Walker UA. Uridine supplementation antagonizes zalcitabine-induced microvesicular steatohepatitis in mice. Hepatology. 2007;45(1):72–9. [DOI] [PubMed] [Google Scholar]
- 63.Lebrecht D, Deveaud C, Beauvoit B, Bonnet J, Kirschner J, Walker UA. Uridine supplementation antagonizes zidovudine-induced mitochondrial myopathy and hyperlactatemia in mice. Arthritis Rheum. 2008;58(1):318–26. [DOI] [PubMed] [Google Scholar]
- 64.Castellvi A, Crespo I, Crosas E, Camara-Artigas A, Gavira JA, Aranda MAG, et al. Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation. Sci Rep. 2019;9(1):3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma Q Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raina D, Kharbanda S, Kufe D. The MUC1 oncoprotein activates the anti-apoptotic phosphoinositide 3-kinase/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J Biol Chem. 2004;279(20):20607–12. [DOI] [PubMed] [Google Scholar]
- 67.Gunda V, Souchek J, Abrego J, Shukla SK, Goode GD, Vernucci E, et al. MUC1-Mediated Metabolic Alterations Regulate Response to Radiotherapy in Pancreatic Cancer. Clin Cancer Res. 2017;23(19):5881–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Serdjebi C, Milano G, Ciccolini J. Role of cytidine deaminase in toxicity and efficacy of nucleosidic analogs. Expert Opin Drug Metab Toxicol. 2015;11(5):665–72. [DOI] [PubMed] [Google Scholar]
- 69.Ye FG, Song CG, Cao ZG, Xia C, Chen DN, Chen L, et al. Cytidine Deaminase Axis Modulated by miR-484 Differentially Regulates Cell Proliferation and Chemoresistance in Breast Cancer. Cancer Res. 2015;75(7):1504–15. [DOI] [PubMed] [Google Scholar]
- 70.Chabosseau P, Buhagiar-Labarchede G, Onclercq-Delic R, Lambert S, Debatisse M, Brison O, et al. Pyrimidine pool imbalance induced by BLM helicase deficiency contributes to genetic instability in Bloom syndrome. Nat Commun. 2011;2:368. [DOI] [PubMed] [Google Scholar]
- 71.Gemble S, Buhagiar-Labarchede G, Onclercq-Delic R, Biard D, Lambert S, Amor-Gueret M. A balanced pyrimidine pool is required for optimal Chk1 activation to prevent ultrafine anaphase bridge formation. J Cell Sci. 2016;129(16):3167–77. [DOI] [PubMed] [Google Scholar]
- 72.Sharma S, Wang J, Alqassim E, Portwood S, Cortes Gomez E, Maguire O, et al. Mitochondrial hypoxic stress induces widespread RNA editing by APOBEC3G in natural killer cells. Genome Biol. 2019;20(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD. ROS and the DNA damage response in cancer. Redox Biol. 2018:101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodriguez-Vargas JM, Ruiz-Magana MJ, Ruiz-Ruiz C, Majuelos-Melguizo J, Peralta-Leal A, Rodriguez MI, et al. ROS-induced DNA damage and PARP-1 are required for optimal induction of starvation-induced autophagy. Cell Res. 2012;22(7):1181–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamamori T, Meike S, Nagane M, Yasui H, Inanami O. ER stress suppresses DNA double-strand break repair and sensitizes tumor cells to ionizing radiation by stimulating proteasomal degradation of Rad51. FEBS Lett. 2013;587(20):3348–53. [DOI] [PubMed] [Google Scholar]
- 76.Rajabi H, Hiraki M, Tagde A, Alam M, Bouillez A, Christensen CL, et al. MUC1-C activates EZH2 expression and function in human cancer cells. Sci Rep. 2017;7(1):7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rajabi H, Tagde A, Alam M, Bouillez A, Pitroda S, Suzuki Y, et al. DNA methylation by DNMT1 and DNMT3b methyltransferases is driven by the MUC1-C oncoprotein in human carcinoma cells. Oncogene. 2016;35(50):6439–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tagde A, Rajabi H, Stroopinsky D, Gali R, Alam M, Bouillez A, et al. MUC1-C induces DNA methyltransferase 1 and represses tumor suppressor genes in acute myeloid leukemia. Oncotarget. 2016;7(26):38974–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamamoto M, Jin C, Hata T, Yasumizu Y, Zhang Y, Hong D, et al. MUC1-C Integrates Chromatin Remodeling and PARP1 Activity in the DNA Damage Response of Triple-Negative Breast Cancer Cells. Cancer Res. 2019;79(8):2031–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guang W, Kim KC, Lillehoj EP. MUC1 mucin interacts with calcium-modulating cyclophilin ligand. The international journal of biochemistry & cell biology. 2009;41(6):1354–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rahn JJ, Shen Q, Mah BK, Hugh JC. MUC1 initiates a calcium signal after ligation by intercellular adhesion molecule-1. The Journal of biological chemistry. 2004;279(28):29386–90. [DOI] [PubMed] [Google Scholar]
- 82.Juin P, Pelletier M, Oliver L, Tremblais K, Gregoire M, Meflah K, et al. Induction of a caspase-3-like activity by calcium in normal cytosolic extracts triggers nuclear apoptosis in a cell-free system. The Journal of biological chemistry. 1998;273(28):17559–64. [DOI] [PubMed] [Google Scholar]
- 83.Sharma AK, Rohrer B. Calcium-induced calpain mediates apoptosis via caspase-3 in a mouse photoreceptor cell line. The Journal of biological chemistry. 2004;279(34):35564–72. [DOI] [PubMed] [Google Scholar]
- 84.Feno S, Butera G, Vecellio Reane D, Rizzuto R, Raffaello A. Crosstalk between Calcium and ROS in Pathophysiological Conditions. Oxidative medicine and cellular longevity. 2019;2019:9324018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hauser J, Sveshnikova N, Wallenius A, Baradaran S, Saarikettu J, Grundstrom T. B-cell receptor activation inhibits AID expression through calmodulin inhibition of E-proteins. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(4):1267–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tadros S, Shukla SK, King RJ, Gunda V, Vernucci E, Abrego J, et al. De Novo Lipid Synthesis Facilitates Gemcitabine Resistance through Endoplasmic Reticulum Stress in Pancreatic Cancer. Cancer Res. 2017;77(20):5503–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gebregiworgis T, Purohit V, Shukla SK, Tadros S, Chaika NV, Abrego J, et al. Glucose Limitation Alters Glutamine Metabolism in MUC1-Overexpressing Pancreatic Cancer Cells. J Proteome Res. 2017;16(10):3536–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goode G, Gunda V, Chaika NV, Purohit V, Yu F, Singh PK. MUC1 facilitates metabolomic reprogramming in triple-negative breast cancer. PLoS One. 2017;12(5):e0176820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mehla K, Singh PK. MUC1: a novel metabolic master regulator. Biochim Biophys Acta. 2014;1845(2):126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singh PK, Behrens ME, Eggers JP, Cerny RL, Bailey JM, Shanmugam K, et al. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J Biol Chem. 2008;283(40):26985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh PK, Hollingsworth MA. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16(9):467–76. [DOI] [PubMed] [Google Scholar]
- 92.Singh PK, Wen Y, Swanson BJ, Shanmugam K, Kazlauskas A, Cerny RL, et al. Platelet-derived growth factor receptor beta-mediated phosphorylation of MUC1 enhances invasiveness in pancreatic adenocarcinoma cells. Cancer Res. 2007;67(11):5201–10. [DOI] [PubMed] [Google Scholar]
- 93.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.