Highlights
-
•
ACE2 are highly expressed in human small intestinal enterocytes.
-
•
2019-nCoV may have same tissue tropism such as small intestine with SARS-CoV.
-
•
The gastrointestinal tract may be an alternative route for 2019-nCoV.
-
•
Distribution pattern of ACE2 gene insinuates the possibility of a fecal–oral transmission for COVID-19.
Keywords: 2019-nCoV, COVID-19, ACE2, Small intestine, Diarrhea
Abstract
The coronavirus disease 2019 (COVID-19) was first reported in Wuhan, China and rapidly spread in other countries in December 2019. The infected patients presented with fever, respiratory symptoms, sometimes with digestive and other systemic manifestations, and some progressed with a severe acute respiratory syndrome or even death. Associated digestive symptoms were frequently observed in the patients, with an unknown significance and mechanism. ACE2, as the major known functional receptor of the 2019 novel coronavirus (2019-nCoV) attracted our attention. We collected the clinical data of the 2019-nCoV-infected patients from published studies and extracted the data about the incidence of gastrointestinal symptoms. Furthermore, we used online datasets to analyze ACE2 expression in different human organs, especially in the small intestine, to explore the relationship between ACE2 expression patterns and clinical symptoms. We found that diarrhea accounted for a notable proportion of COVID-19 patients, ranging from 8.0% to 12.9%. The results reveal that ACE2 mRNA and protein are highly expressed in the small intestinal enterocytes but not in the goblet cells or intestinal immune cells. High expression of ACE2 on the surface cells in the digestive tract may lead to gastrointestinal symptoms and inflammation susceptibility. Overall, digestive symptoms were common in the COVID-19 patients. ACE2 expression on surface cells of the small intestine may mediate the invasion and amplification of the virus and activation of gastrointestinal inflammation. It is a possible mechanism of digestive symptoms in the COVID-19 patients and explains the presence of the virus in patients’ stool samples. The study also highlights the necessity of taking stool samples for suspected patients to help in early diagnosis and assessment of disease status.
Introduction
Coronavirus Disease 2019 (COVID-19), which was first reported in Wuhan, China during December 2019, is a newly emerged viral infection caused by the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), also known as the 2019 novel coronavirus (2019-nCoV) (Zhu et al., 2020). This disease is characterized by fever, dry cough, fatigue and lymphopenia, which can also lead to gastrointestinal symptoms, organ dysfunction, rapid progression to a severe acute respiratory syndrome and even death (Huang et al., 2020, Kui et al., 2020). According to a daily report by the World Health Organization (WHO), as of April 4th, 2020, 1,051,635 cases were confirmed globally, involving 207 countries and regions, including 82,875 cases in China, causing 56,985 deaths in total. The coronavirus COVID-19 pandemic is considered a global health crisis of our time and the greatest challenge we have faced since World War Two.
Two other unusual coronavirus outbreaks occurred in the 21st century, including the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV), causing high mortality rates. As the seventh member of the coronavirus family that infects humans, the 2019-nCoV genome sequence is reported to be approximately 82% identical to the human SARS-CoV (Chan et al., 2020). The 2019-nCoV has the same cell entry receptor as the SARS-CoV, the angiotensin converting enzyme 2 (ACE2), which regulates the cross-species and human-to-human transmissions of the SARS-CoV (Xu et al., 2020, Wan et al., 2020, Zhou et al., 2020, Letko and Munster, 2020). ACE2 belongs to the ACE family of dipeptidyl carboxy-dipeptidases and plays a critical role in the RAS systems and in some organs (Vuille-dit-Bille et al., 2015). It is distributed broadly in the arterial and the venous endothelial cells, the arterial smooth muscle cells and the cholangiocytes, and especially highly expressed in the renal, the cardiovascular, and the gastrointestinal tissues. This indicates that COVID-19 may involve multiple organs, which may also explain the various extrapulmonary symptoms (Hamming et al., 2004).
Although COVID-19 is characterized by fever and respiratory tract manifestations, data from recent studies suggest that concurrent gastrointestinal symptoms are not uncommon (Song et al., 2020a). Moreover, it has been reported that the SARS-CoV exhibits an intestinal tropism (Leung et al., 2003). It is possible that the 2019-nCoV had the same tissue tropism as that of the SARS-CoV, as they had the same functional cell receptor (Xu et al., 2020). Several studies have found the surface expression pattern of the ACE2 protein in enterocytes of the small intestine, mainly on cells in contact with the external environment. ACE2 is mainly expressed in the endothelium and the vascular smooth muscle cells in the colon (Letko and Munster, 2020, Wang et al., 2020a, Ren et al., 2006). ACE2 expressed on surface cells of the small intestine is a potential primary entrance for the 2019-nCoV to infect the host and cause gastrointestinal symptoms.
This study aimed to determine the expression pattern of ACE2 gene in the small intestine and attempted to determine the potential mechanism of diarrhea and other gastrointestinal manifestations in patients with COVID-19. Meanwhile, the distribution pattern of ACE2 may insinuate a fecal–oral transmission for the COVID-19. A high virus titer in the digestive tract may indicate a high risk of transmission via feces of the patients, which raises new problems in the disposal of feces.
Materials and methods
Clinical data
We summarized the clinical data of four recent studies involving clinical characteristics of the COVID-19 to extract the incidence of gastrointestinal symptoms in patients infected with the 2019-nCoV (Kui et al., 2020, Wang et al., 2020b, Song et al., 2020b, Zhang et al., 2020). Recorded information included the demographic data and the gastrointestinal symptoms during the whole disease process including anorexia, nausea or vomiting, diarrhea, and abdominal pain. All clinical information was directly obtained from the articles cited above.
Public dataset acquisition and processing
We used RNA and protein expression data of the ACE2 gene in different human tissues through the Human Protein Atlas portal (http://www.proteinatlas.org/) (Uhlen et al., 2015). All data are available online. Gene expression matrix and cell type annotation of single cell RNA-seq (scRNA-seq) data from biopsies of the human terminal ileum across 13 children diagnosed with functional gastrointestinal disease (without inflammation) and ranging in age 6–18 years old. Different cell types were identified by mapping the canonical marker genes in the two-dimensional t-distributed stochastic neighbor embedding (tSNE) map. The TOP principle components were used to project the data using tSNE. Cell labels were used for cell-type annotations. The visualisation results were obtained from the scRNA-seq Single Cell Portal (https://singlecell.broadinstitute.org/single_cell) and the data was uploaded by Ziegler et al. (2020).
Results
We analyzed data from the 4 most recent studies focused on the clinical features of COVID-19 patients (Table 1 ). In these 4 cohort studies, the number of confirmed patients was 138, 137, 51 and 140, respectively. The data from three relatively large sample studies indicated that approximately 8–12.9% of the COVID-19 patients had diarrhea. Cohort1 showed anorexia in 39.9%, and cohort 4 found nausea in 17.3% and vomiting in 5% of the 2019-nCoV-infected patients. Meanwhile, multiple clinical reports confirmed positive presence of the 2019-nCoV in the stool of patients. The clinical outcomes indicated that the gastrointestinal tract may be an alternative route for the 2019-nCoV infection apart from the respiratory tract.
Table 1.
Summary of the renal function characteristics of COVID-19 patients in 4 cohorts.
| Characteristics | Cohort 1 | Cohort 2 | Cohort 3 | Cohort 4 |
|---|---|---|---|---|
| Patient number | 138 | 137 | 51 | 140 |
| Age (years), median (range) | 56 (22–92) | 57 (20–83) | 49*(16–76) | 57 (25–87) |
| Sex | ||||
| Male | 75 (54.3%) | 61 (44.5%) | 25 (49%) | 71 (50.7%) |
| Female | 63 (45.7%) | 76 (55.5%) | 26 (51%) | 69 (49.3%) |
| Digestive symptoms | ||||
| Anorexia | 55 (39.9%) | NA | 9 (18%) | 17/139 (12.2%) |
| Nausea or vomiting | 14 (10.1%)/5 (3.6%) | NA | 3 (6%) | 24/139 (17.3%) & 7/139 (5.0%) |
| Diarrhea | 14 (10.1%) | 11(8.0%) | 5(10%) | 18/139 (12.9%) |
| Abdominal pain | 3 (2.2%) | NA | NA | 8/139 (5.8%) |
NA: not available.
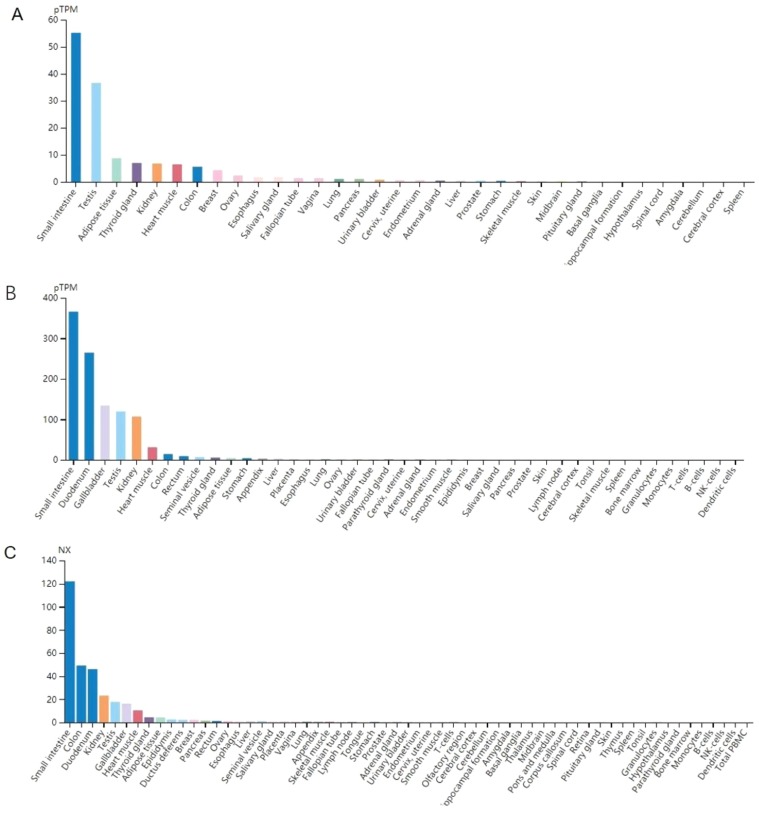
ACE2 is considered a major functional receptor of the 2019-nCoV in humans. We analyzed the published data from the online datasets to explore the expression level of ACE2 gene in human digestive system. The data from the Human Protein Atlas portal indicated that ACE2 mRNA expression level was highest in the small intestine of all tissues (Figure 1 ).
Figure 1.
Data of mRNA expression level of ACE2 in different human tissues from online datasets, data from (A) GTEx portal, (B) The Human Protein Atlas dataset, (C) Consensus dataset. pTPM, transcripts per million.
The RNA-seq details section shows detailed information about the individual samples used for the transcript profiling and results of the RNA-seq analysis. Information about some individual samples has been listed in Table 2 from The Human Protein Atlas, including sex, age and estimated fractions of cell types.
Table 2.
The individual samples used for the transcript profiling of ACE2 in the human small intestinal tissues from the Human Protein Atlas portal.
| Small intestine samples | Case 1 | Case 2 | Case 3 |
|---|---|---|---|
| Age (years) | 65 | 39 | 68 |
| Gender | Male | Male | Male |
| Estimated fractions of cell typespTPM (%) | |||
| Glandular cells | 55 | 40 | 30 |
| Smooth muscle cells | 5 | 5 | 15 |
| Other cell types | 40 | 55 | 55 |
Information about each individual sample has been listed below, including sex, age, tissue section image, and estimated fractions of cell types. pTPM, transcripts per million. pTPM values provide a quantification of the ACE2 gene abundance, which is comparable between different samples.
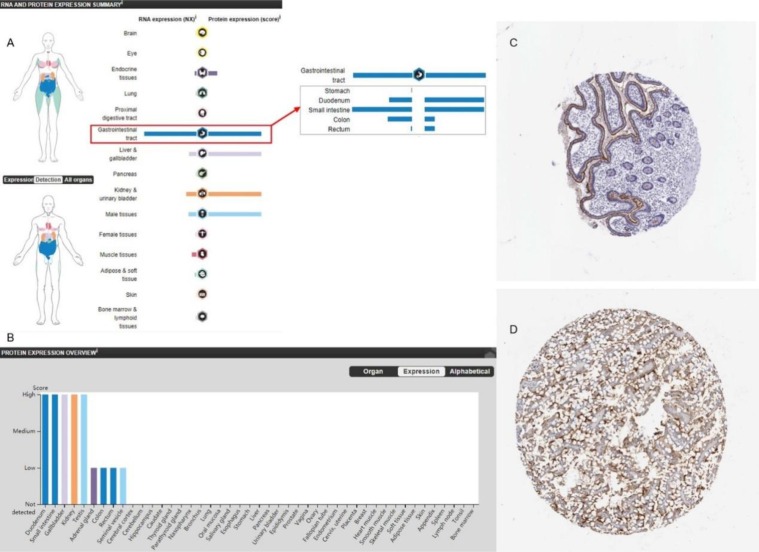
Furthermore, the protein expression level of ACE2 was relatively higher in the small intestine than that in the other tissues (Figure 2A and B), and the results of immunohistochemistry (IHC) also indicated that the protein expression level of ACE2 was significantly higher in the small intestine than in the other tissues (Figure 2C and D). These results suggest that cells of the small intestine may be potential target-organ of the 2019-nCoV.
Figure 2.
ACE2 protein expression levels in different human tissues from the Human Protein Atlas portal. (A, B) ACE2 protein expression levels in different tissues. (C,D) IHC staining of ACE2 in the small intestinal tissues.
To assess the cell type-specific expression of ACE2, we analyzed datasets containing the scRNA-seq data of the non-inflamed terminal ileum tissue from 13 children diagnosed with functional gastrointestinal disease. We divided single cells into sub-clusters based on the canonical markers and cell classification in the original literature (Figure 3A), and the results revealed specific ACE2 expression in the small intestinal epithelium cells (absorptive and crypt enterocytes) (Figure 3B). In contrast, ACE2 expression was not observed in goblet, paneth, tuft or enteroendocrine cells. This observation partially matched with that of the previous data based on ACE2 protein expression (Hamming et al., 2004), where small intestinal enterocytes were ACE2-positive and other cells in the small intestine were ACE2-negative. Therefore, ACE2 expression in the small intestinal enterocytes may be a potential mechanism of infection and direct an intestinal epithelial injury by the 2019-nCoV-binding ACE2 as the host cell receptors.
Figure 3.
Single cell analysis of human small intestinal cells using a single cell portal. (A) Small intestinal cell atlas visualised by tSNE. (B) The violin plot showing ACE2 gene expression of major intestinal epithelium cell types.
Discussion
Since COVID-19 emerging in Asia late last year, the virus has spread to every continent except Antarctica. Cases are rising daily in Africa, the Americas, and Europe. COVID-19 has mainly gained attention for its rapid contagious ability and severe respiratory manifestations. The symptoms of COVID-19 are complex and diverse, ranging from an asymptomatic infection to ARDS with multiple organ dysfunction, making it difficult to recognize at a very early stage. In addition to the respiratory system, COVID-19 showed varying degrees of involvement in the digestive system, evidenced by diarrhea, nausea and/or vomiting, anorexia, and abdominal pain, the mechanisms and implications of which have not been determined. It is reported that diarrhea may be underestimated in COVID-19 patients (Liang et al., 2020). Xiao et al. found that more than half of their hospitalized patients were tested SARS-CoV-2 RNA positive in their stool samples, while no more than 20.0% of confirmed patients were reported with digestive symptoms in other cohorts (Xiao et al., 2020). The research found positive staining of ACE2 and 2019-nCoV in gastrointestinal epithelium from patients with positive 2019-nCoV RNA in their stool, which added convincing evidence that the 2019-nCoV can infect digestive system. 2019-nCoV patient with onset symptom of diarrhea was also reported recently, reminding us of the importance of digestive symptoms in 2019-nCoV infection (Song et al., 2020a). In our study, we focused on the digestive manifestation and possible mechanism, aiming to offer adjuvant therapeutic methods for patients infected with the 2019-nCoV and exhibiting gastrointestinal symptoms.
ACE2 has been suggested to be the functional receptor for the 2019-nCoV, similar to that for the SARS-CoV. Although ACE2 mRNA is known to be present in virtually all organs, its protein expression is largely unknown. Here, we evaluated the expression pattern of ACE2 in the small intestine of healthy human and mice using online datasets and bioinformatics methods. The results revealed that the expressions of mRNA and protein of the ACE2 gene are extremely high in the intestinal cells especially in enterocytes. Xiao et al. had observed plasma cells and lymphocytes infiltration in 2019-nCoV patients’ lamina propria of stomach, duodenum and rectum, accompanied with interstitial edema, instead of direct damage of mucous epithelium (Xiao et al., 2020). Therefore, the 2019-nCoV spike protein may enter the small intestinal enterocytes by binding to ACE2, which induces immune cells infiltration and abnormal intestinal function.
Notably, the results of the study by Hamming et al. indicate that ACE2 is present in all parts of the small intestinal epithelium cells, including the duodenum, the jejunum, and the ileum, but not in enterocytes of the colon (Hamming et al., 2004). Furthermore, the 2019-nCoV may replicate in the enterocytes of the small intestine because the viral RNA can be detected in stool samples of the patients. The high expression of ACE2 protein as a functional receptor for the 2019-nCoV in enterocytes of the small intestine, combined with the presence of the virus in stool samples of the patients, suggests that a fecal–oral transmission may exist. It may also necessitate the collection of stool samples of the suspected patients to help in an early diagnosis and assessment of the disease status. A cluster of cases infected with SARS reported in late March, 2003 in Amoy Garden, a populated residential area in Hong Kong, China should also be noted that the patients were believed to get infected as a result of the unscientific drainage system, which indicated that there were possibilities that virus transmit by a fecal–oral way. We should be wary of the coming warmer seasons, which is also the season when digestive infectious diseases happened with higher incidence, the 2019-nCoV could spread again.
ACE2 is identified as an enzyme that negatively regulates the RAS by converting Ang II and the main bioactive molecule of the RAS. In addition, ACE2 is essential for neutral amino acid transporters in the gut (Vuille-dit-Bille et al., 2015, Jando et al., 2017). A study showed that amino acid malnutrition is related to an intestinal inflammation via ACE2, which plays a significant role in amino acid homeostasis, innate immunity, and maintaining intestinal microbiota (Hashimoto et al., 2012). The ACE2 knockout mice model showed a decline in the uptake of tryptophan, resulting in a decrease in the expression of antimicrobial peptides from small intestinal paneth cells and the change of the intestinal microbiota, leading to a high sensitivity to colitis, which was restored by tryptophan supplementation (He et al., 2018). It explains how the aberrant absorption of tryptophan in the small intestine leads to manifestations of colitis, such as diarrhea (Hashimoto et al., 2012, Perlot and Penninger, 2013). Through this way, ACE2 modulates innate immune responses and influences the composition of the gut microbiota, which may further explain diarrhea observed in the 2019-nCoV patients. As ACE2 being known to be the receptor of the 2019-nCoV, there may be competitive combination of the 2019-nCoV and ACE2, which may inhibit the absorption of dietary tryptophan in the small intestine and end up in the change of the intestinal microbiota and susceptibility to colitis. The absorption function of small intestinal epithelial cells may be interrupted by immune reactions induced by viral infection at the same time. Therefore, intake of foods rich in amino acids, especially tryptophan, may be beneficial for the 2019-nCoV patients. Our study also provides a theoretical basis for the treatment of COVID-19 with microecological preparations.
Conclusion
Digestive symptoms were common in the 2019-nCoV-infected patients. ACE2 expression on the surface cells of the small intestine may mediate the invasion and amplification of the virus and activation of a gastrointestinal inflammation. It is a possible mechanism of digestive symptoms in the 2019-nCoV infected patients and explains the presence of the virus in stool samples of the patients. The study also necessitates the collection of stool samples of suspected patients to help in an early diagnosis and assessment of the disease status.
Funding
This research received no external funding.
Ethical approval
No ethical approval was obtained because this study did not involve a clinical evaluation, did not involve laboratory animals and invasive procedures.
Authors’ contribution
H.-B.L. and H.Z. contribute equally to the work. Z.-M.D. designed the study. Z.-M.D., J.L. and H.Z. wrote the main manuscript text, H.Z., H.-B.L., X.-M.L. and W.L. prepared figures and tables, Z.-M.D., J.-R.L., G.W., and J.L. reviewed the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgements
The authors acknowledge The Human Protein Atlas portal (http://www.proteinatlas.org/), Single Cell Portal (https://singlecell.broadinstitute.org/single_cell) and Ziegler et al. for facilitating open data sharing.
References
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T., Perlot T., Rehman A., Trichereau J., Ishiguro H., Paolino M. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487(7408):477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Wu C., Li P., Li N., Zhang D., Zhu Q. Functions and signaling pathways of amino acids in intestinal inflammation. Biomed Res Int. 2018;2018:9171905. doi: 10.1155/2018/9171905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jando J., Camargo S.M.R., Herzog B., Verrey F. Expression and regulation of the neutral amino acid transporter B0AT1 in rat small intestine. PLOS ONE. 2017;12(9):e0184845. doi: 10.1371/journal.pone.0184845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kui L., Fang Y.Y., Deng Y., Liu W., Wang M.F., Ma J.P. Clinical characteristics of novel coronavirus cases in tertiary hospitals in Hubei Province. Chin Med J (Engl) 2020 doi: 10.1097/CM9.0000000000000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Munster V. Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV. bioRxiv. 2020 doi: 10.1101/2020.01.22.915660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W.K., To K.F., Chan P.K., Chan H.L., Wu A.K., Lee N. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology. 2003;125(4):1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Feng Z., Rao S., Xiao C., Xue X., Lin Z. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020 doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- Perlot T., Penninger J.M. ACE2 - from the renin-angiotensin system to gut microbiota and malnutrition. Microbes Infect. 2013;15(13):866–873. doi: 10.1016/j.micinf.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Glende J., Al-Falah M., de Vries V., Schwegmann-Wessels C., Qu X. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J Gen Virol. 2006;87(Pt 6):1691–1695. doi: 10.1099/vir.0.81749-0. [DOI] [PubMed] [Google Scholar]
- Song Y., Liu P., Shi X.L., Chu Y.L., Zhang J., Xia J. SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut. 2020 doi: 10.1136/gutjnl-2020-320891. [DOI] [PubMed] [Google Scholar]
- Song F., Shi N., Shan F., Zhang Z., Shen J., Lu H. Emerging coronavirus 2019-nCoV pneumonia. Radiology. 2020;6:200274. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Vuille-dit-Bille R.N., Camargo S.M., Emmenegger L., Sasse T., Kummer E., Jando J. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids. 2015;47(4):693–705. doi: 10.1007/s00726-014-1889-6. [DOI] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol. 2020 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhao S., Liu M., Zhao Z., Xu Y., Wang P. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. medRxiv. 2020 doi: 10.1101/2020.02.05.20020545. [DOI] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen P., Wang J., Feng J., Zhou H., Li X. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020 doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.J., Dong X., Cao Y.Y., Yuan Y.D., Yang Y.B., Yan Y.Q. Clinical characteristics of 140 patients infected by SARS-CoV-2 in Wuhan, China. Allergy. 2020 doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 2020 doi: 10.1101/2020.01.22.914952. [DOI] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C., Allon S.J., Nyquist S.K., Mbano I., Miao V.N., Cao Y. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is enriched in specific cell subsets across tissues. Cell. 2020 doi: 10.2139/ssrn.3555145. Available at SSRN: https://ssrn.com/abstract=3555145. [DOI] [PMC free article] [PubMed] [Google Scholar]