Abstract
Endoplasmic Reticulum (ER) stress underlies the pathogenesis of numerous kidney diseases. A better care of patients with kidney disease involves the identification and validation of ER stress biomarkers in the early stages of kidney disease. For the first time to our knowledge, we demonstrate that the prion protein PrPC is secreted in a conventional manner by ER-stressed renal epithelial cell under the control of the transcription factor x-box binding protein 1 (XBP1) and can serve as a sensitive urinary biomarker for detecting tubular ER stress. Urinary PrPC elevation occurs in patients with chronic kidney disease. In addition, in patients undergoing cardiac surgery, detectable urine levels of PrPC significantly increase after cardiopulmonary bypass, a condition associated with activation of the IRE1-XBP1 pathway in the kidney. In conclusion, our study has identified PrPC as a novel urinary ER stress biomarker with potential utility in early diagnosis of ongoing acute or chronic kidney injury.
Subject terms: Prions, Diagnostic markers
Introduction
Kidneys have to cope with a wide array of injuries that translate into elementary stressors at the cellular level. Adaptive responses to these stresses are molecular systems that primarily aim to eradicate or reduce stress intensity and promote metabolic reprograming to maintain cellular homeostasis and other vital functions1. Adaptive stress responses shape the endogenous repair and scarring equilibrium in tissues. As such, they are critical for tissue remodeling, and can impact the functional outcomes of the injured kidney, ultimately leading to chronic kidney disease (CKD)2,3. The molecular reprogramming circuitries and cellular adaptive responses that occur in renal epithelial cells early after the initiation of injury, before cell death and the engagement of the maladaptive repair process, offer the opportunity for a non-invasive monitoring and the implementation of potential nephroprotective strategies. For this reason, the characterization of the “secretome” produced by tubular cells is critical for the design of urine markers of kidney injury.
Endoplasmic reticulum (ER) stress and its adaptive response, the unfolded protein response (UPR), are archetypal examples of these adaptive stress responses, and the detection of its activation constitutes an opportunity for an early diagnosis of ongoing tissue injury. Of particular interest in the UPR is the IRE1α-XBP1 axis. IRE1α catalyzes the unconventional processing of the mRNA encoding the transcriptional factor X-Box binding protein-1 (XBP1) and creates a transcriptionally active XBP1 (sXBP1) that contains a potent transactivation domain and enters the nucleus. Genes that are regulated by sXBP1 enhance protein folding, transport, and degradation, as well as the expansion of the protein secretory pathways. XBP1 is critical for cellular adaptation in stressful conditions. Whether sXBP1 activity is specifically implicated in the pathophysiology of kidney disease is mostly unknown, but recent studies have indicated that sXBP1 is required for podocytes, mesangial and tubular cells maintenance under stressful conditions4–7, or, on the contrary, contributes to renal inflammation and injury8. The clinical consequence of the activation of this pathway would be that the detection of sXBP1 or its target genes may have prognostic value. Since XBP1 splicing depends exclusively on IRE1α activity, sXBP1 is only produced if and when ER stress occurs (with a rare exception), therefore reflecting the presence of ER stress with high sensitivity.
Although multiple lines of evidence from clinical and experimental studies have demonstrated that a maladaptive ER response is mechanistically linked to the pathogenesis of kidney diseases, a lack of robust biomarkers for monitoring renal ER stress has hampered early detection and effective therapeutic intervention to restore ER homeostasis. Here, we investigated if the soluble form of the ER stress-inducible protein, prion protein (PrPC) is also a candidate urinary ER stress biomarker. The cellular prion protein PrPC, which is involved in the pathophysiology of some neurodegenerative diseases under its pathogenic scrapie isoform, is a ubiquitous protein mainly found as a GPI-anchored cell surface molecule and present in the extracellular space as a soluble isoform9. The aim of this study was to determine whether PrPC could serve as a urinary ER stress biomarker for ER stress-mediated kidney diseases by studying a combination of cellular models and human patient samples.
Material and methods
Reagents
Tunicamycin (Tun), Thapsigargin (Tg), Brefeldin A (BFA), Etoposide (Eto), DL-Dithiothreitol (DTT) and mouse monoclonal antibody against α-tubulin were from Sigma Aldrich (St. Louis, MO, USA). Mouse monoclonal antibodies against PrPC (Sha31) were from SPI-Bio (Montigny Le Bretonneux, France). Goat polyclonal antibodies against BIP (GRP78) were from Santa Cruz (SC-1050 Santa Cruz, CA, USA). Polyclonal rabbit antibodies against PARP (#9542) and PERK (#3192) were from Cell Signalling Technology (Danvers, MA, USA).
Cell culture and treatments
Human Renal Epithelial Cells (HREC) of proximal origin immortalized with the HPF 16 E6/E7 genes (HK-2) were purchased from American Type Culture Collection (lot #710257641). Primary HREC were harvested from a human nephrectomy specimen. This cellular model has been previously molecularly characterized confirming the proximal descent of the vast majority of the cultured tubular epithelial cells10. HREC were cultured at 37 °C in an atmosphere containing 5% CO2 in Dulbecco’s Modified Eagle’s Medium (41965–039 Gibco, MD, USA) supplemented with 1% fetal bovine serum (HyClone, SV30160.03 GE lifesciences), 0.5 µg/ml hydrocortisone (Sigma-Aldrich), 1X Insulin Transferrin Selenium (Sigma-Aldrich), 10 ng/mL Epithelial Growth Factor (Sigma-Aldrich), 6.5 ng/mL Triiodothyronine (Sigma-Aldrich), and 1X of Penicillin-Streptomycin mix (Gibco). Mycoplasma-free cells (Mycoalert Mycoplasma Detection Kit, Lonza) were subcultured before complete confluence using 0.05% Trypsine-EDTA (Gibco) and experiments were not performed with cells beyond the third passage since it has been shown that no phenotypic changes occur up to this passage number.
Sub-confluent cells were grown in six-wells plates for the indicated times under the indicated conditions. Unless otherwise stated, Tun was used at 250 ng/ml, Tg at 250 nM, DTT at 1 µM, BFA at 5 µg/ml and Etoposide at 100 µM. For glucose deprivation, cells were grown during 48 hours in glucose free Dulbecco’s Modified Eagle’s Medium (11966–025 Gibco) with control cells supplemented with 4.5 g/L glucose (Gibco). Cell culture media were collected and cleared through centrifugation at 5000 g during 5 minutes then culture supernatants were immediately stored at −80 °C prior analysis. For transient siRNA-mediated silencing, cells were transfected with siRNA sequences (50 nM) using the Lipofectamine 2000 reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Specific siRNA sequences used were: 5′-CAGUACAGCAACCAGAACATT-3′ (sense si-PRNP). siRNA against XBP1 (Hs_XBP1_7 and Hs_XBP1_7) was from Qiagen. AllStars Negative Control siRNA (5′-AACGAUGACACGAACACACTT-3′) has no homology to any known mammalian gene, and validation has been performed using Affymetrix GeneChip arrays and a variety of cell-based assays to ensure minimal nonspecific effects on gene expression and phenotype.
Cell proliferation and viability assays
For cell counting experiments, 2.5 × 105 cells were seeded in six-well plates and transfected with siRNA against PRNP 24 h post seeding. After another 24 h, culture medium was replaced by fresh medium containing siRNA and Tun or Tg. After 24 h of incubation, cells from three wells per group were harvested and total cell numbers (cells/well) were determined using a CASY.TT cell counter (Schärfe System GmbH, Reutlingen, Germany). Cells transfected with control siRNA and treated with vehicle (DMSO) were used as reference for normalization.
Immunoprecipitation
Immunoprecipitation of PrPC was performed using Dynabeads Protein G magnetic beads (Life technology), according to manufacturer’s protocol. Briefly, 50 µL of washed beads were incubated with 1 µg of anti-PrPC antibody under rotation for 1 h at 4 °C. Crosslinking of antibody to the beads was performed with the BS3 (bis(sulfosuccinimidyl)suberate) linker (Thermofischer). Immunoprecipitation was performed with 250 µL of culture supernatant of HK-2 cells overnight under rotation at 4 °C. The immuno-precipitated material was eluted in elution buffer and analyzed by western blot.
Protein extraction and western blot analysis
Cells were washed in PBS and incubated for 30 min at 4 °C in NaDOC lysis buffer [50 mM Tris·HCl (pH 7.4)/150 mM NaCl/5 mM EDTA/0.5% Triton X-100/0.5% sodium deoxycholate] and a mixture of phosphatase (Thermo-Scientific, Waltham, MA, USA) and protease (Roche, Mannheim, Germany) inhibitors. Extracts were centrifuged at 14,000 g for 15 min and protein concentration in supernatants were measured using the bicinchoninic acid method (Pierce, Rockford, IL, USA). Deglycosylation was performed on 15 µg of proteins with 500U PNGaseF (New England Biolabs, Ipswich, MA, USA) for 1 hour at 37 °C. In all, 25 µg of protein extracts were resolved by 4–12% SDS-PAGE (Invitrogen) and transferred to nitrocellulose membranes (iBlot, Invitrogen). Membranes were blocked with SEABLOCK blocking buffer (Thermo-Scientific) for 1 h at room temperature and then incubated overnight at 4 °C with primary antibody diluted in blocking buffer. After washings in PBS-Tween buffer, membranes were incubated with secondary antibody coupled to IRDye fluorophores. Infrared signal of membranes was revealed using an Odyssey detection system (Li-Cor biosciences, Lincoln, NE, USA).
RNA extraction and real-time qPCR analysis
Total RNA was extracted using the RNeasy Mini Kit® (Qiagen) following the manufacturer’s protocol. The yield and purity of RNA were measured using a NanoDrop ND-1000® spectrophotometer (Nanodrop Technologies). For reverse transcriptase-polymerase chain reaction (RT-PCR) analysis, first-strand cDNA was synthesized on 1 µg of RNA with oligo(dT) primer and random 6mers, using the High-capacity cDNA Reverse Transcription (Applied Biosystems) according to the manufacturer’s protocol. Real-time PCR was performed using Absolute QPCR SYBR Green ROX Mix (Thermo-Scientific, Waltham, MA, USA) on a ABI PRISM 7900HT (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA). Real-time PCR analyzes were performed with the SDS software 2.3 (Applied biosystems). Primers used for the PCR reactions are shown on Supplementary Table 1. Results are expressed as a relative quantification of a target gene transcript normalized to the RPL13A (human samples) or Tbp (mouse samples) housekeeping gene using the ΔΔCt method.
Enzyme-linked immunosorbent assays
Soluble PrPC was quantified in cell culture supernatant or urinary samples using the BetaPrion Human ELISA immunoassay (Analytikjena Leipzig, Germany), according to the manufacturer’s protocol. Soluble Angiogenin and neutrophil gelatinase-associated lipocalin (NGAL) were quantified in urinary samples using the Quantikine® human ANG and NGAL immunoassays (RD Systems), according to the manufacturer’s protocol.
Animal studies
Analyses were carried out on kidneys from 4 month-old-male C57BL/6 mice exposed to one injection of Tunicamycin (intraperitoneal injection, 1 mg/kg) or Vehicle (DMSO) and collected 48 h after injection11. Samples were analysed under blinded conditions.
Human studies
Urinary PrPC concentration measurements in individuals with Chronic Kidney Disease
In all, 55 consecutive patients who were referred to the Nephrology Department at the Georges Pompidou European Hospital (Paris, France) for kidney biopsy from 7 December 2017 to 22 February 2018 were included. Indications for biopsy were estimated GFR < 60 ml/min and/or proteinuria >0.5 g/L. Kidney biopsies were not performed for the purpose of this non-interventional study, but only for patient care. At the time of biopsy, urine samples were collected for routine clinical chemistry analyses and stored at −80 °C. Samples were analyzed under blinded conditions. Patients provided informed consent that their urine specimen could be used for research purpose.
Urinary PrPC concentration measurements in individuals with cardiopulmonary bypass
To monitor PrPC in the setting of ischemia-reperfusion injury (IRI), we took advantage of the biocollection of a previous study12. From 17 February 2017 to 26 April 2017, 42 patients undergoing scheduled cardiac surgery with CPB were enrolled. The exclusion criteria were: an eGFR <30 ml/min/1.73 m2, infusion of a radio contrast agent within the 24 h before surgery, a preoperative left ventricular ejection fraction <40%, age <18 years, pregnancy, and the inability to provide consent. AKI was diagnosed according to the KDIGO Clinical Practice Guideline for AKI criteria (http://kdigo.org/) using serum creatinine levels and urine output after the surgery. This single-center, prospective, pilot study was approved by the French ethical committee on 7 February 2017 (CPP Sud Est III n° 2016–072 B) and registered under the EudraCT n° 2016-A01871–50. All patients provided written consent for study participation and for the biological analysis before inclusion. Urine samples were collected from a urinary catheter at three different times: (1) after the induction of anesthesia and before the start of cardiopulmonary bypass (CBP), (2) at the end of the CPB procedure, and (3) on the day after surgery in the ICU. Urine samples were collected in Corning 50-ml conical tubes and centrifuged at 2000 g for 20 min within 4 hours of collection. Cell pellets were conserved in 350 μL of RLT® buffer (Qiagen™, France) and stored until mRNA extraction. Supernatants and cell pellets were stored at −80 °C until analysis. Clinical data were prospectively extracted from the hospital’s electronic medical records. All clinical data and samples were de-identified. Urine PrPC monitoring has been performed in a subgroup of 19 patients in whom urine samples were available at the three time-points for PCR and ELISA analyses. Experiments were all carried out under blinded conditions.
Immunohistochemistry of human kidney biopsies with chronic kidney disease
Three kidney biopsies from patients explored for chronic kidney disease were retrospectively analyzed for PrPC immunohistochemistry studies. Kidney biopsies were not performed for the purpose of this non-interventional study, but only for patient care. Patients provided informed consent that these kidney biopsy specimen could be used for research purpose.
Fragment analysis
RNA was extracted from the pellets of urinary cells using a RNeasy Mini Kit® (Qiagen) and reverse-transcribed into cDNAs using TaqMan® Reverse Transcription Reagents (Applied Biosystems). Basically, the fragment analysis involved the following 3 steps: (1) Amplification of the sXBP1 and XBP1 cDNAs by PCR using fluorogenic oligonucleotide primers. Several differently colored fluorescent dyes are detectable in one sample. We designed the following fluorogenic oligonucleotide primers (tagged with hexachloro-fluorescein, HEX, green) for fluorometric detection of the sXBP1 and XBP1 mRNA levels: forward primer: (5′-HEX)GGAGTTAAGACAGCGCTTGG-3′ and reverse primer: 5′-GAGATGTTCTGGAGGGGTGA-3′. We performed PCR using HotStart Taq® DNA polymerase on a thermal cycler with the following program: 95 °C for 10 min; 40 cycles of 94 °C for 30 s, 59 °C for 30 s, and 72 °C for 30 s; and a final step of 72 °C for 10 min. (2) Labeled fragments (amplicons) were separated by size using capillary electrophoresis, and the fluorescence intensity was measured using the Applied Biosystems™ 3730xl DNA Analyzer. One of the dye colors (GENESCAN® ROX 400 HD size standard, Applied Biosystems™, red) was used to detect a labeled size standard in each sample. Fragments and ROX 400 HD were mixed with HiDi™ Formamide (Applied Biosystems) prior to capillary electrophoresis. (3) The data were analyzed using GeneMapper® Software to determine the relative size of each dye‐labeled fragment in the sample by comparing fragments with the standard curve for that specific sample.
Immunohistochemistry
Kidney biopsies were fixed in alcohol-formalin-acetic acid, dehydrated with ethanol and xylene, embedded in paraffin, and cut into 3 μm sections. Samples were then deparaffinized, rehydrated and heated for 20 min at 97 °C in citrate buffer. Endogenous peroxidase was inactivated by incubation for 10 min at room temperature in 0.3% H2O2. Sections were incubated with PBS containing 1:1000 anti-PrPC antibody (Sha31, SPI-Bio) or control isotype (mouse IgG1κ, Biolegend, San Diego, CA, USA). Next, sections were incubated with anti-rabbit conjugated with peroxydase labeled polymer (Dako), visualized with a peroxydase kit (Dako). Finally, the tissue sections were counterstained with hematoxylin.
Statistical analysis
All data are represented as individual values and means ± SEM of at least two independent experiments, unless otherwise specified. Graphs were generated using GraphPad Prism 7 Software (GraphPad Software, Inc.). According to current discussion and criticisms on the performance and interpretation of statistical tests in experimental settings, and in particular the misuse and misinterpretation of p values13–15, we choose not to systematically perform comparison of biological data using statistic tests to compute p values for significance. When deemed necessary, ie when the size effect appeared small, we provided mean and 95% confidence intervals of the two groups of values for interpretation.
Results
ER stressors promote PRNP mRNA and PrPC protein expression in HREC
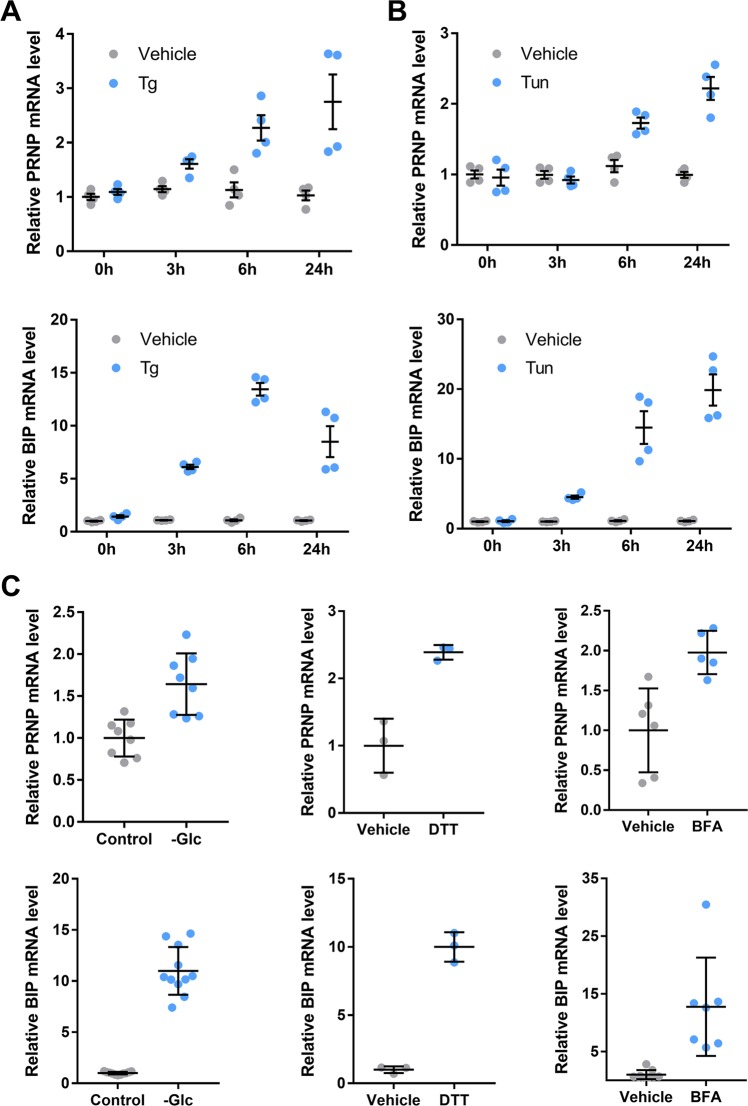
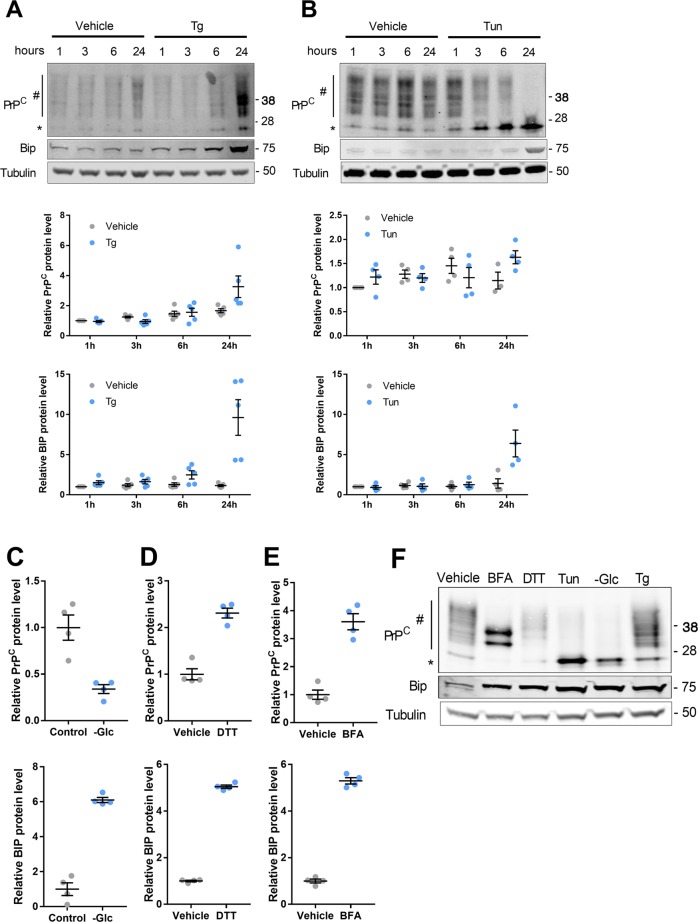
To assess whether the PrPC-encoding PRNP transcripts are sensitive to ER stressors in human renal epithelium, we monitored PRNP mRNA expression during a 24-h kinetics of exposure of Human Renal Epithelial Cells (HREC) to 250 nM Thapsigargin (Tg) or 250 ng/ml Tunicamycin (Tun). As shown in Fig. 1a, b, both treatments induced PRNP (top panels) and BIP (bottom panels) transcripts expression as measured through RT-qPCR. PRNP induction was mild after 3 h for Tg and induction started after 6 h for Tun. Twenty-four hours after ER Stress induction, PRNP expression reached over 200% of its basal levels. As depicted in Fig. 1c, such increase in PRNP (top panels) and BIP (bottom panels) transcription extended to ER stress resulting from a 48-hour glucose deprivation (-Glc), as well as a 24-h treatment with 1 mM of Dithiothreitol (DTT) or 5 µg/ml of Brefeldin-A (BFA) (Fig. 1c, bottom panels). Likewise, robust inductions of UPR transcripts and PRNP mRNA were found in primary HREC exposed for 24 h to Tg, Tun or BFA (Supplementary Fig. 1A). Corroborating these data, we observed an upregulation of PRNP mRNA together with UPR transcripts in the kidneys of mice collected 48 h after an intraperitoneal injection of Tunicamycin (1 mg/kg) (Supplementary Fig. 2), an established experimental animal model of ER stress11. At the protein level, a 24-h Tg treatment induced a robust increase in PrPC expression in HREC (Fig. 2a), in line with the kinetics of PRNP mRNA induction previously described. With Tun, which inhibits the N-linked glycosylation step of protein maturation16, PrPC progressively resolved into a single immunoreactive band of low molecular weight (Fig. 2b), corresponding to the non-glycosylated form (Supplementary Fig. 3A). Densitometry analysis revealed a slight but significant increase in PrPC content in cells incubated with Tun for 24 h compared with vehicle-treated cells. As for Tun, the non-glycosylated form of PrPC was overexpressed in cells undergoing glucose deprivation. However, total (Glycosylated plus non-glycosylated) PrPC levels were reduced in glucose-deprived cells (Fig. 2c and Supplementary Fig. 3B). As expected, DTT, which affects disulfide bonds formation, did not affect PrPC glycosylation profile (Fig. 2d and Supplementary Fig. 3C), whereas BFA produced a partially glycosylated protein, consistent with a blockade of transit through the Golgi network (Fig. 2e and Supplementary Fig. 3D). Thus, the pattern of PrPC protein expression and maturation depends on whether the ER stress inducer affects protein maturation or ER function (Fig. 2f). Of note, western blot analysis of PrPC in extracts from primary HREC exposed to Tg, Tun or BFA (Supplementary Fig. 1B) confirmed the quantitative and qualitative changes monitored with immortalized HREC.
Fig. 1. PRNP expression is induced by ER stressors.
a, b Kinetics analysis of PRNP (top panels) or BIP (bottom panels) mRNA expression in HREC exposed to Tg (a) or Tun (b) compared to vehicle-treated cells. c qRT-PCR analysis of PRNP (top panels) or BIP (bottom panels) mRNA expression in HREC submitted to glucose deprivation for 48 h (left panel) incubated with DTT (middle panel) or BFA (right panel) for 24 h. Results are represented as individual values with means of n = 2–3 independent duplicates or triplicates of cell preparations ± SEM.
Fig. 2. Impact of ER stressors on PrPC protein expression.
a, b Representative images and kinetics analysis of PrPC (top panels) or BIP (bottom panels) protein expression in HREC exposed to Tg (a) or Tun (b) compared to vehicle-treated cells. c–e Protein quantification of PrPC (top panels) or BIP (bottom panels) protein expression in HREC submitted to glucose deprivation for 48 h (c), incubated with DTT (d) or BFA (e) for 24 h or (e). Protein levels were normalized to Tubulin. Corresponding Western blot are displayed in Supplementary Fig. 2. f Comparative migration profile of PrPC isoforms in HREC according to ER stressors. Hash indicates mono- or bi-glycosylated PrPC isoforms. Asterisk indicates unglycosylated PrPC. Results are represented as individual values with means of n = 2–3 independent duplicates or triplicates of cell preparations ± SEM.
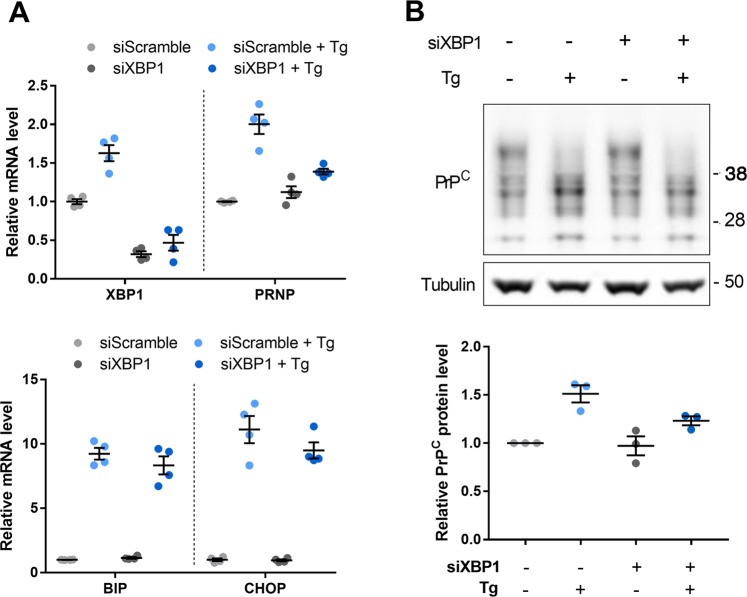
Since the PRNP promoter contains ER stress response elements17,18, we next assessed a potential contribution of XBP1 to the upregulation of PRNP mRNA in response to Tg. XBP1 silencing in HREC did not induce any change in the basal levels of PRNP mRNA and PrPC protein, in the absence of ER stressor. In contrast, the silencing of XBP1 drastically reduced the upregulation of PRNP mRNA and PrPC protein triggered by Tg (Fig. 3a top panel and B), while it did not affect the induction of BIP and CHOP expression, which depend on the activity of the ATF6 and PERK pathways of the UPR respectively (Fig. 3a bottom panel). Together, these results indicate that PRNP mRNA is induced in response to a variety of ER stressors in HREC and that this induction is XBP1-dependent at least in the case of Tg.
Fig. 3. ER stress-induced PRNP expression is dependent on XBP1.
a qRT-PCR analysis of the expression of XBP1 and PRNP (top panel) as well as BIP and CHOP (bottom panel) in XBP1-silenced vs. control HREC exposed or not to Tg for 24 h. b Western blot analysis of PrPC protein expression in XBP1-silenced vs. control HREC exposed or not to Tg for 24 h. Protein levels were normalized to Tubulin. Results are represented as individual values with means of n = 2 independent duplicates of cell preparations or n = 3 independent experiments ± SEM. Mean and 95% IC relative PrPC expression levels: 200% [160–240] in the Tg+siXBP1− condition, and 139% [127–150] in the Tg+siXBP1+ condition.
ER stress promotes PrPC secretion by HREC
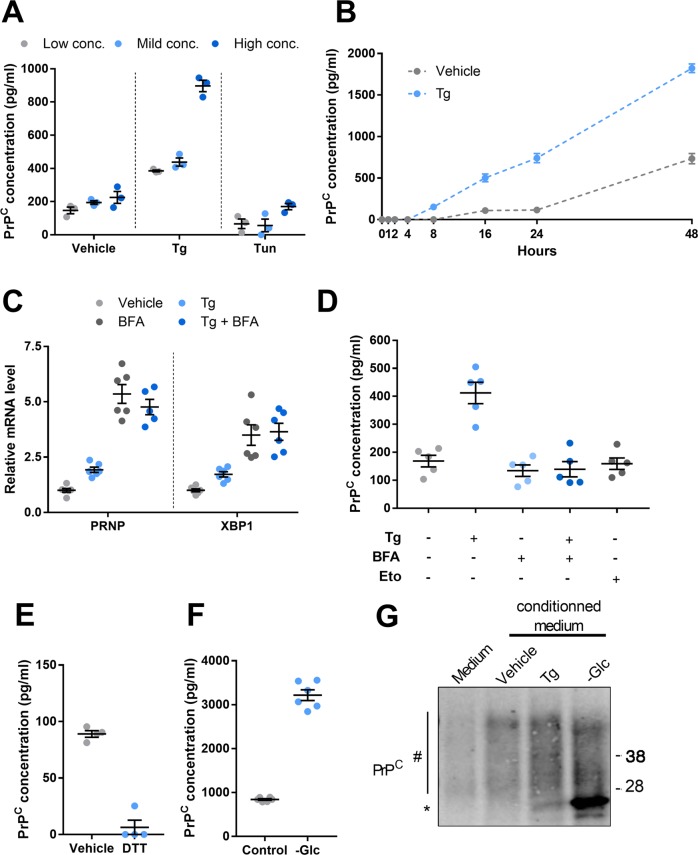
Since PrPC can be found as a soluble form in addition to its major location at the cell surface9, we examined whether it could be secreted by renal epithelial cells in response to ER stress. To this aim, HREC were exposed for 24 h to various concentrations of Tg, which induce dose-dependent increases in UPR-associated transcripts (Supplementary Fig. 4A) and soluble PrPC was quantified by ELISA in the extracellular medium. We observed a dose-dependent increase in soluble PrPC in the supernatant of cells exposed to Tg, reaching almost 400% of control for the maximal dose (Fig. 4a). While the epitope recognized by the ELISA PrPC quantification test is not disclosed, our preliminary data (not shown) suggest that soluble PrPC species in the supernatant of HREC mainly correspond to N-terminally truncated isoforms19. To shed some light on the kinetics of soluble PrPC accumulation, we collected supernatants from Tg- or vehicle-incubated HREC over a 48 h-time-frame. Soluble PrPC concentration increased between 24 hours and 48 h in vehicle-exposed cells, supporting the occurrence of a basal shedding of PrPC in the absence of stress (Fig. 4b). Under exposure to Tg at 250 nM, the levels of soluble PrPC steadily accumulated from 8 h of drug addition onwards, reaching about 1800 pg/ml corresponding to 250% of control at 48 h (Fig. 4b).
Fig. 4. Impact of ER stressors on soluble PrPC.
a, b Soluble PrPC was measured in the conditioned medium of HREC exposed to increasing doses of Tg (Low:100 nM, Mild: 250 nM and High: 2.5 µM) or Tun (Low:1.25 µg/ml, Mild: 2.5 µg/ml and High: 5 µg/ml) for 24 h (a) or treated with 250 nM Tg during a 48-hour kinetics (b). c qRT-PCR analysis of PRNP mRNA expression in HREC exposed to 250 nM Tg in combination with 5 µg/ml BFA for 24 h. d, e Measurement of soluble PrPC in the conditioned medium of HREC exposed to 250 nM Tg in combination with 5 µg/ml BFA or to 100 µM Etoposide for 24 h (d), in the conditioned medium of HREC exposed 1 µM DTT for 24 h (e) or submitted to glucose deprivation for 48 h (f). g Comparative migration profile of PrPC protein immunoprecipitated from 200 µl of fresh culture medium or conditioned media from HREC treated with Tg for 24 h or vehicle or submitted to glucose deprivation for 48 h. Hash indicates mono- or bi-glycosylated PrPC isoforms. Asterisk indicates unglycosylated PrPC. Results are represented as individual values with means of n = 2 independent duplicates or triplicates of cell preparations or n = 3 independent experiments ± SEM.
As a GPI-anchored molecule, PrPC carries a signal sequence for translocation in the ER lumen and follows the conventional secretory pathway20. The hallmark of conventional protein secretion is the trafficking through the ER-Golgi network, a process inhibited by Brefeldin A (BFA)21. Because of the disruption of the secretory pathway, BFA is also an ER stress inducer. In agreement, as observed with Tg and other ER stressors, BFA induced the expression of PRNP mRNA (Figs. 1c and 4c) and UPR-associated transcripts among which XBP1 (Fig. 4c and Supplementary Fig. 4B). However, when we monitored PrPC secretion by HREC incubated with BFA, PrPC could not be detected in the extracellular medium, suggesting that its secretion is inhibited when ER stress is generated by BFA (Fig. 4d). Importantly, Tg-induced PrPC secretion was inhibited by BFA despite the fact that PrPC transcripts were still expressed under Tg exposure (Fig. 4c, d). These results indicate that the intracellular trafficking of PrPC during ER stress is altered by BFA, and that PrPC is likely secreted through the canonical path under ER stress. In line with this, Etoposide, which promotes DNA damage and cell death without ER stress in our experimental system22, did not induce PrPC secretion, indicating that PrPC release is not a non-specific consequence of cell death and membrane permeabilization, and corroborates our finding supporting that PrPC secretion is selectively controlled by ER stress (Fig. 4d).
When cells were exposed to Tun (Fig. 4a) or DTT (Fig. 4e) instead of Tg, there was no increase in soluble PrPC whatever the dose, despite the induction of PRNP and UPR-associated transcripts in ER-stressed cells (Supplementary Fig. 4C, D). This is in line with the observed impact of Tun (Fig. 2b) on PrPC maturation and may be accounted for by the necessity of PrPC to undergo proper glycosylation to reach the cell surface. In addition, the reported effect of DTT on PrPC maturation23 and the lack of impact of DTT on PrPC glycosylation (Fig. 2f) indicate that PrPC must undergo disulfide bond formation to exit the ER toward the Golgi apparatus, and that PrPC lacking disulfide bond is likely directed towards the proteasome through the ER associated degradation pathway24. However, 48 hours of glucose deprivation, which induces ER stress and prevents PrPC glycosylation in HREC (Supplementary Fig. 3B, F) was associated with a prominent increase (3.8-fold, 3200 pg/ml) in soluble PrPC level compared to the control condition (Fig. 4f). The apparent discrepancy between the effects of Tun and glucose deprivation on PrPc secretion may be partly due to quantitatively different effects on PrPc glycosylation. Tun blocks N-linked glycosylation by inhibiting N-acetylglucosamine phosphotransferase, which catalyzes the first step of the glycosylation process25. On the other hand, glucose starvation also affects glycosylation by reducing the pool of substrates available for N-acetylglucosamine-1-phosphate production26, but gluconeogenesis (from glucogenic aminoacids for example) might mitigate the global effects of glucose deprivation, thus enabling residual N-linked glycosylation activity. However, immunoblotting of culture-medium immunoprecipitated PrPC suggested that a proportion of secreted PrPC upon glucose deprivation was non/underglycosylated, suggesting that under this condition, non/underglycosylated proteins can escape ER quality controls and still be secreted (Fig. 4g).
PrPC silencing does not affect cell viability in ER-stressed HREC
We next sought to determine whether the induction of PrPC upon ER stress affects cell viability. To this purpose, PrPC expression was silenced in HREC by siRNA-mediated RNA interference for 24 h and cells were further exposed to Tg or Tun in the presence of siPRNP for another 24 h. PrPC-silenced HREC, which expressed only 20% PRNP mRNA and <35 % PrPC protein (Supplementary Fig. 5A, B), retained the ability to mount a robust ER-stress response (Supplementary Fig. 5C). We nevertheless found that PrPC silencing mitigated the Tun- or Tg-induced increase in CHOP mRNA levels in HREC (Fig. 5a). However, PERK phosphorylation, reflecting activation of the Integrated Stress Response that mediate CHOP expression was not affected by PrPC silencing, suggesting that PrPC could affect CHOP expression downstream PERK (Fig. 5b). Since CHOP is associated with the pro-apoptotic branch of the UPR27, this would suggest that PrPC favors a pro-apoptotic fate in ER-stressed cells. This prompted us to compare the impact of ER stress on cell viability in PrPC-silenced HREC versus control HREC using a CASY.TT cell counter. In basal conditions, i.e. in the absence of Tun or Tg, we observed a significant reduction in cell viability upon PrPC depletion (Fig. 5c), in line with the physiological pro-survival role of PrPC9. As expected, ER stressors reduced cell viability, but the extent of cell viability reduction in PrPC-silenced HREC incubated with Tun or Tg was not different from PrPC-expressing HREC (Fig. 5c). These results were corroborated by the cleavage of PARP, a proxy of apoptotic cell death28. Indeed, in non-treated cells, PrPC silencing promoted an increase in cleaved PARP to total PARP, and ER stressors induced PARP cleavage that was comparable whether cells expressed PrPC or not (Fig. 5d). Altogether, these results lend support to a pro-survival effect of PrPC under basal conditions that would not necessarily be maintained upon cell stress, which is reminiscent of the ambivalent role described for this protein29.
Fig. 5. Impact of PRNP silencing on the response of HREC to ER stress.
a qRT-PCR analysis of the expression of CHOP in PRNP-silenced vs. control HREC exposed or not to Tg or Tun for 24 h. Mean and 95% IC of relative CHOP mRNA levels: Tg+ siScramble: 4500% [2241–6760]; and Tg+ siPRNP: 3064% [1946–4183]. Tn+ siScramble: 1463% [849–2075] and Tn+ siPRNP: 851% [482–1219]. b Western blot analysis of the expression of PERK in PRNP-silenced vs. control HREC exposed or not to Tg or Tun for 24 h. c Relative cell viability of PRNP-silenced vs. control HREC exposed or not to Tg or Tun for 24 h. Mean and 95% IC of % cell viability: vehicle siScramble: 100% [96–103] and vehicle siPRNP: 89% [93–97]: Tg+ siScramble: 88%[93–94], and Tg+ siPRNP: 84 [78–89]; Tn+ siScramble: 91% [84–97] and Tn+ siPRNP: 87% [81–93]. d Western blot analysis of the expression of PARP (left) and quantification of the ratio cleaved PARP to total PARP (right) in PRNP-silenced vs. control HREC exposed or not to Tg or Tun for 24 h. Protein levels were normalized to Tubulin. Results are expressed as individual values with means of n = 5 (a), n = 6 (B,C) or n = 10 (d) independent experiments ± SEM.
Urinary PrPC as a potent non-invasive biomarker of AKI
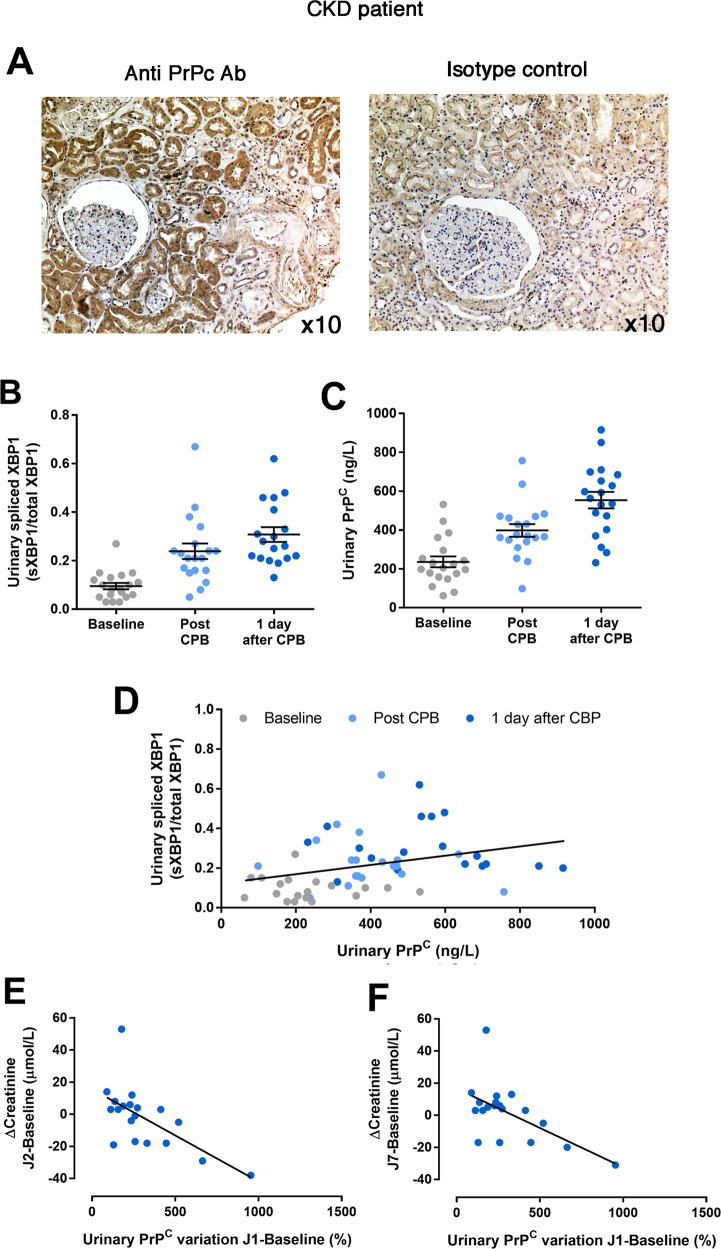
Since PrPC is secreted by the renal epithelium under stressful conditions in vitro, we wondered if it could be detected and quantified in urines of individuals with a renal tubular epithelial injury. Supporting this possibility, we found that PrPC expression was expressed only in epithelial cells, and accumulated beneath the apical membrane of the cells, a feature suggestive of secretion process (Fig. 6a). We then measured the concentrations of PrPC in urines of individuals with a kidney disease. To demonstrate the biological and clinical relevance of the detection of PrPC in urine, we monitored ER stress markers in urine of 19 patients undergoing scheduled cardiac surgery with cardiopulmonary bypass, which promote IRI in the kidney, a condition associated with ER stress (Supplementary Table 2)12. We repeatedly measured the urine angiogenin concentrations (an indirect marker of the activity of the IRE1α/sXBP1 axis22), and NGAL, which is regulated by the PERK/ATF4 pathway30, to obtain information about the activation of the UPR during the CPB procedure. In addition, we measured CHOP and sXBP1 mRNA in urine cell pellets. Urine angiogenin concentrations and the ratio of sXBP1/(sXBP1 + XBP1) increased significantly over time (before, immediately after and one day after CPB), confirming that hemodynamic impairment initiates ER stress and sXBP1 expression (Supplementary Fig. 6A and Fig. 5b). NGAL concentrations, and CHOP or BIP transcripts (Supplementary Fig. 6B, C and D) did not vary, indicating that the IRE1α/sXBP1 axis, rather than the PERK/ATF4 axis of the UPR, appears to be activated in the kidneys under these specific hemodynamic conditions. Finally, we observed an incremental increase in the concentrations of PrPC after CPB, and a positive correlation between urinary sXBP1 transcripts levels and PrPc concentrations in urines (Fig. 6b, c), in line with the activation of the IRE1-sXBP1 pathway, which regulate PrPC expression upon ER stress. Moreover, patients with the highest amplitude of PrPC urinary concentrations between baseline and the day after CBP were those with the greatest recovery in serum creatinine, suggesting that the sXBP1-PrPC pathway is likely cyto and nephroprotective (Fig. 6d). Together, these results indicate that PrPC is a potent non-invasive marker of kidney injury reflecting ongoing ER stress in the kidney.
Fig. 6. PrPC is expressed in kidney and is released in urines of patients with hemodynamic impairment.
a Representative photomicrograph of PrPC expression evaluated by immunohistochemistry in a kidney from individual with chronic kidney disease (left panel). Staining with the isotype control antibody is shown in the right panel. Original magnification, ×10. b–f Measurement of relative XBP1 splicing (b) and soluble PrPC (c) in the urines of n = 19 patients undergoing CBP before, at the end and the day after surgery. c Linear regression curve between urinary PrPC concentrations and sXBP1 mRNA levels in urines before, after and the day after CBP. d, e Correlation between PrPC increase rate between the day 1 after CBP and baseline, and the variation of serum creatinine at day 2 (d) and day 7 (e) compared to baseline (before CBP).
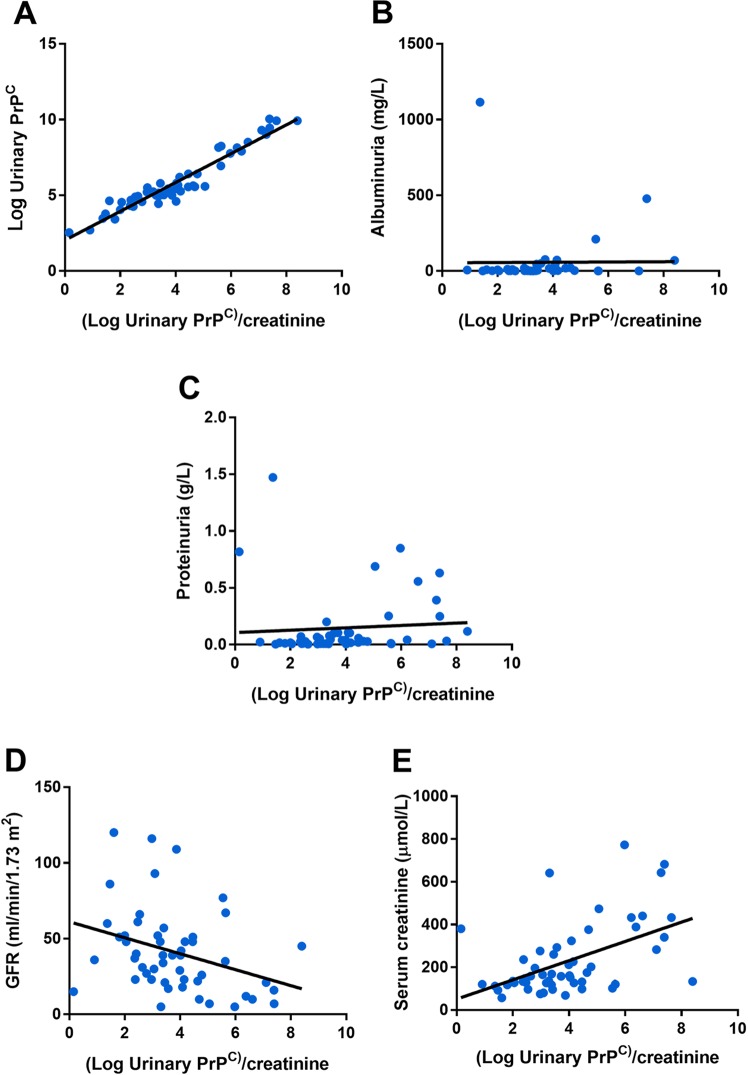
Urinary PrPC as a potent non-invasive biomarker of CKD
We next aimed at providing insights into the specificity of urinary PrPC in regards with proteinuria and renal function. To this end, we measured the concentrations of PrPC in the urines of 55 consecutive patients referred to a nephrology department for a CKD (defined by presence of significant proteinuria and/or an estimated Glomerular filtration rate < 60 mL/min/1.73 m2) (Supplementary Table 3). PrPC concentration was reported to the urinary concentrations of creatinine to avoid bias related to variations in urine concentration (Fig. 7a). Because the distribution of PrPC was skewed, we log-transformed the values to obtain a Gaussian distribution and compared PrPC concentration with other parametric variables. We tested the correlation between the levels of urinary PrPC, the high molecular weight protein albumin (a marker of glomerular injury), and total proteinuria. Indeed, the presence of PrPC in urines could be a non-specific consequence of increased glomerular permeability, and therefore a component of total proteinuria. We found that the urinary concentrations of PrPC did not correlate with total urinary proteins and albuminuria (Fig. 7b, c). However, urinary PrPC concentrations were positively correlated with serum creatinine (R2, 0.25, p < 0.0001) and inversely correlated with estimated glomerular filtration rates (R2, 0.11, p < 0.01), two markers of renal function (Fig. 7d, e), suggesting that in patients with CKD, chronically injured tubules secrete PrPC and that PrPC concentrations could reflect the severity or the extent of the injury. These results indicate that the detection and quantification of PrPC in urine could provide information on renal function.
Fig. 7. Urinary PrPC is elevated in patients with CKD.
a–f Log-transformed values of urinary PrPC reported to urinary creatinine concentrations and plotted as a function of urinary PrPC (R2 = 0.92) (a) albuminuria (R2 = 0) (b), proteinuria (R2 = 0) (c), glomerular filtration rate (R2 = 0.11) (d) or serum creatinine (R2 = 0.24) (e) in a cohort of n = 55 patients with CKD.
Discussion
This proof-of-concept study is the first to demonstrate that PrPC is a potent non-invasive, and mechanistic biomarker for ER stress-mediated kidney diseases. Aberrant ER protein proteostasis and ER dysfunction underpin the initiation and development of a variety of kidney diseases, and it is of outmost importance to identify and validate ER stress biomarkers that can be applied in human kidney disease patients. Here, we demonstrate that PrPC is rapidly upregulated and secreted when renal cells were subjected to ER stress. Moreover, PrPC is detectable and easily quantifiable in urine of patients with CKD, and correlates with renal function. Finally, we validated the potent clinical utility of PrPC as a marker of ER stress in patients with kidney IRI.
ER-stressed cells produce signals that alert neighboring cells of the presence of a stressor, and an emerging concept is that the secretome produced by stressed cells, including the mediators secreted in urines by the renal epithelial cells upon kidney injury, can constitute the fingerprint ER stress. Hence, a deep molecular characterization of this secretome appears as a valuable approach for the identification of soluble markers of an ongoing tissue injury associated with ER stress. The use of biomarkers of ER stress in the kidney has promising applications. Since ER stress is associated with the underlying pathogenesis of kidney diseases, mechanistic ER stress markers could allow the initiation of treatments early on in the disease, which will reduce the follow-up time to identify kidney disease with serum creatinine alone. Indeed, serum creatinine elevations occur when irreversible changes such as scarring and fibrosis of the kidney are often present, and treatment is unlikely to restore full kidney function.
Critically, monitoring urinary PrPC allows an early detection of tubular ER stress, an underlying causative mechanism for ischemic AKI. Approximately 45% of critically ill patients and 20% of hospitalized patients develop AKI, which leads to increased hospital stays, and increased morbidity and mortality31. Our translational studies in patients undergoing CPB surgery suggest that PrPC could be utilized to stratify the risk of developing AKI after IRI, which remains to be demonstrated further investigations. The ultimate utility of PrPC may be to identify an early therapeutic window during which tubular ER stress modulators can be applied before a detectable change in renal function occurs. We took advantage of the CBP cohort to assess potential correlations between changes in urinary PrPC levels and clinical correlates in patients. PrPC levels correlate with those of spliced XBP1, in line with our in vitro results demonstrating that the increase in PRNP gene expression in response to ER stress depends on XBP1. Furthermore, we found that the variation of PrPC levels within the first 24 h of CPB is inversely correlated to the variation of creatinine levels within the first 48 h of CPB. This also holds true when considering the variation levels of creatinine within the first week of CPB. This hence suggests that patients who mount a robust PrPC response (high fold change in PrPC levels versus baseline) are those whose kidney function recovers better/faster (highest rate of creatinine reduction). This would fit in with the protective role ascribed to PrPC in the context of various types of stresses (see ref. 9 for review). Conversely, in the context of CKD, our results suggest that high PrPC levels are associated with worse renal function. The reasons for the differences between acute and chronic injuries are probably multiple and remain to be established. However, one can speculate that the production of PrPC during CKD reflects chronic ER stress, which is known to be deleterious since it is associated with the engagement of proapoptotic pathways. In addition, deleterious stress pathways that are unrelated to ER stress could be engaged upon CKD, leading to PrPC production. Finally, PrPC by itself could promote tubular cell death and activate fibrogenesis, that would ultimately drive chronic structural deterioration and CKD progression.
In addition to AKI, autosomal dominant tubular kidney disease (ADTKD) are monogenic conditions in which ER stress is prominent and leads to tubulointerstitial fibrosis and progression of CKD. ADTKD are phenotypically heterogeneous with variable age of disease onset, disease severity, and rate of disease progression among affected individuals within and between families. Remarkably, mutations in the UMOD gene encoding Uromodulin, which is the most abundant protein secreted in normal urine and has multiple roles in kidney physiology, as well as mutations in the MUC1 gene which encodes the transmembrane epithelial mucin 1 protein supporting roles in epithelial barrier protection, lead to mutant proteins retention and aggregation in the ER, likely due to protein misfolding. The activation of the UPR in those inherited diseases leads to progressive tubular damage, triggering inflammation and interstitial fibrosis32,33. The identification and validation of ER stress markers is extremely important in this area, as it will provide a useful tool for diagnostic purpose and monitoring disease activity.
A set of molecules released or secreted by renal cells under ER stress in urine has been recently described, and the proof-of-concept of a non-invasive monitoring of renal ER stress in human has been done for some of them. For example, ERp57 (also known as PDIA3), is secreted by ER stressed cells involved in TGF-β mediated fibrogenesis, and can be detected in urines of patients with diabetic nephropathy with ongoing fibrosis34. Another example is the ER chaperone mesencephalic astrocyte-derived neurotrophic factor (MANF), which is secreted by ER stressed tubular cells and can be quantified in the urine of tunicamycin-treated mice35. The secreted ribonuclease angiogenin is expressed under the control of IRE1α, thereby reflecting the activation of the UPR in the kidney. Urinary angiogenin levels reflect the severity of tissue damage upon acute kidney allograft injury and are predictive of graft failure, independent of histological lesions36. Supporting a more generalized role of the UPR transducers in renal disease, the PERK pathway, through the expression of lipocalin 2, regulates tubular cell viability upon proteinuric stress, and lipocalin 2 when secreted in urines can provide prognostic information30,37. Reflecting the activity of the ATF6 arm of the UPR, cysteine-rich with EGF-like domains 2 (CRELD2) is upregulated and secreted when renal cells are exposed to ER stressors and appears to be a potential urine ER stress biomarker in patients with kidney diseases associated with ER stress including ADTKD-UMOD and ischemic AKI38.
While considerable attention has been paid to PrPC for its involvement in neurodegeneration, very little is known on its contribution to kidney pathophysiology. PrPC is expressed at the apical cell surface of polarized proximal tubule cells and promotes kidney iron uptake39. Interestingly, PrPC is upregulated in the kidney following IRI40, in line with our own data in CBP patients, and the results indicate that PrPC-knockout animals have an increase sensibility to IRI. However, the authors did not assess the link with the UPR nor the levels of urinary PrPC following IRI. In contrast, tissue damage was increased in PrPC-knockout animals under IRI, which could be accounted for by a reduction in Xbp1 transcripts in PrPC-knockout vs. WT animals (our unpublished data). While the induction of PrPC in ER stressed cells has been described18, the impact of ER stress on soluble PrPC had not been investigated. Besides, studies were mainly carried out on neurons or cancer cells and have highlighted a protective role for PrPC against ER stress-induced cell death. Thus, the functional outcome of ER stress-induced PrPC upregulation may depend on the cell type, tissue considered and duration (acute versus chronic). We believe the connection between PrPC and ER stress may be of particular relevance in the context of kidney in view of our data obtained in patients with acute or chronic kidney injury. Furthermore, our unpublished data indicate a strong correlation between PRNP gene expression and the UPR as well as XBP1-dependent transcription in kidney renal clear cell carcinoma. Incidentally, PrPC overexpression has been reported in several types of cancer41 and a consensus view is that increased levels of PrPC endow cells with proliferative42,43 migratory and invasive capacities44–46, which are also features supporting tissue remodeling in CKD3,47. ER stressed cells shape tissue remodeling since cells upon ER stress secrete numerous specific mediators that promote inflammation, such as Interleukins 6 and 8 and Chemokine (C-X-C) Ligand 3 (CXCL3); angiogenesis through Vascular Endothelial Growth Factor (VEGF) and basic Fibroblast Growth Factor (bFGF); tissue remodeling through the production of collagen 1A2 and the metalloprotease ADAMTS3; apoptosis through the production of Prostate Apoptosis Response 4 (Par-4); and proteostasis with the chaperon ERdj3, which is a critical regulator of tissue homeostasis1. Whether PrPC participate in tissue remodeling upon acute and chronic kidney diseases is unknown and is an important research avenue.
In summary, our study has identified PrPC as a mechanistic urine biomarker in ER stress-mediated kidney diseases. The clinical applications of PrPC in ER stress-mediated kidney diseases may include early diagnosis, stratification of patients at risk and monitoring treatment response. Further studies in larger cohorts are required to evaluate the diagnostic accuracy and predictive value of PrPC.
Supplementary information
Acknowledgements
Y.B. was supported by a fellowship from Agence Nationale de la Recherche. Grant support was provided by the following: Agence Nationale de la Recherche, and INSERM.
Author contributions
Y.B. and V.P.: collection and assembly of data, data analysis and interpretation, and final approval of the paper; H.L.: human samples collection; B.P. and J.L.V.: material support; F.D.: collection and assembly of data and final approval of the manuscript. S.M-R. and N.P.: conception and design, financial support, collection and assembly of data, data analysis and interpretation, paper writing.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Stephanou
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yohan Bignon, Virginie Poindessous
These authors jointly supervised this work: Sophie Mouillet-Richard, Nicolas Pallet
Contributor Information
Sophie Mouillet-Richard, Email: sophie.mouillet-richard@parisdescartes.fr.
Nicolas Pallet, Email: nicolas.pallet@aphp.fr.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-2430-3).
References
- 1.Chovatiya R, Medzhitov R. Stress, inflammation, and defense of homeostasis. Mol. Cell. 2014;54:281–288. doi: 10.1016/j.molcel.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duffield JS. Cellular and molecular mechanisms in kidney fibrosis. J. Clin. Invest. 2014;124:2299–2306. doi: 10.1172/JCI72267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferenbach DA, Bonventre JV. Mechanisms of maladaptive repair after AKI leading to accelerated kidney ageing and CKD. Nat. Rev. Nephrol. 2015;11:264–276. doi: 10.1038/nrneph.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fedeles SV, et al. Sec63 and Xbp1 regulate IRE1alpha activity and polycystic disease severity. J. Clin. Invest. 2015;125:1955–1967. doi: 10.1172/JCI78863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao D, et al. Suppression of XBP1S mediates high glucose-induced oxidative stress and extracellular matrix synthesis in renal mesangial cell and kidney of diabetic rats. PLoS ONE. 2013;8:e56124. doi: 10.1371/journal.pone.0056124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madhusudhan T, et al. Defective podocyte insulin signalling through p85-XBP1 promotes ATF6-dependent maladaptive ER-stress response in diabetic nephropathy. Nat. Commun. 2015;6:6496. doi: 10.1038/ncomms7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaufman DR, Papillon J, Larose L, Iwawaki T, Cybulsky AV. Deletion of inositol-requiring enzyme-1alpha in podocytes disrupts glomerular capillary integrity and autophagy. Mol. Biol. Cell. 2017;28:1636–1651. doi: 10.1091/mbc.e16-12-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferre S, et al. Renal tubular cell spliced X-box binding protein 1 (Xbp1s) has a unique role in sepsis-induced acute kidney injury and inflammation. Kidney Int. 2019;96:1359–1373. doi: 10.1016/j.kint.2019.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch TZ, Martin-Lanneree S, Mouillet-Richard S. Functions of the prion protein. Prog. Mol. Biol. Transl. Sci. 2017;150:1–34. doi: 10.1016/bs.pmbts.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Pallet N, et al. Rapamycin inhibits human renal epithelial cell proliferation: effect on cyclin D3 mRNA expression and stability. Kidney Int. 2005;67:2422–2433. doi: 10.1111/j.1523-1755.2005.00350.x. [DOI] [PubMed] [Google Scholar]
- 11.Mami I, et al. Angiogenin mediates cell-autonomous translational control under endoplasmic reticulum stress and attenuates kidney injury. J. Am. Soc. Nephrol. 2016;27:863–876. doi: 10.1681/ASN.2015020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fohlen B, et al. Real-time and non-invasive monitoring of the activation of the ire1alpha-xbp1 pathway in individuals with hemodynamic impairment. EBioMedicine. 2018;27:284–292. doi: 10.1016/j.ebiom.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amrhein V, Greenland S, McShane B. Scientists rise up against statistical significance. Nature. 2019;567:305–307. doi: 10.1038/d41586-019-00857-9. [DOI] [PubMed] [Google Scholar]
- 14.Leek J, et al. Five ways to fix statistics. Nature. 2017;551:557–559. doi: 10.1038/d41586-017-07522-z. [DOI] [PubMed] [Google Scholar]
- 15.Nuzzo R. Scientific method: statistical errors. Nature. 2014;506:150–152. doi: 10.1038/506150a. [DOI] [PubMed] [Google Scholar]
- 16.Tkacz JS, Lampen O. Tunicamycin inhibition of polyisoprenyl N-acetylglucosaminyl pyrophosphate formation in calf-liver microsomes. Biochem. Biophys. Res. Commun. 1975;65:248–257. doi: 10.1016/S0006-291X(75)80086-6. [DOI] [PubMed] [Google Scholar]
- 17.Misiewicz M, et al. Identification of a novel endoplasmic reticulum stress response element regulated by XBP1. J. Biol. Chem. 2013;288:20378–20391. doi: 10.1074/jbc.M113.457242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dery MA, et al. Endoplasmic reticulum stress induces PRNP prion protein gene expression in breast cancer. Breast Cancer Res. 2013;15:R22. doi: 10.1186/bcr3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linsenmeier L, et al. Diverse functions of the prion protein—does proteolytic processing hold the key? Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:2128–2137. doi: 10.1016/j.bbamcr.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 20.Sarnataro D, Pepe A, Zurzolo C. Cell biology of prion protein. Prog. Mol. Biol. Transl. Sci. 2017;150:57–82. doi: 10.1016/bs.pmbts.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 21.Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J. Biol. Chem. 1988;263:18545–18552. [PubMed] [Google Scholar]
- 22.Mami I, et al. A novel extrinsic pathway for the unfolded protein response in the kidney. J. Am. Soc. Nephrol. 2016;27:2670–2683. doi: 10.1681/ASN.2015060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiti NR, Surewicz WK. The role of disulfide bridge in the folding and stability of the recombinant human prion protein. J. Biol. Chem. 2001;276:2427–2431. doi: 10.1074/jbc.M007862200. [DOI] [PubMed] [Google Scholar]
- 24.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 25.Yoo J, et al. GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation. Nat. Struct. Mol. Biol. 2018;25:217–224. doi: 10.1038/s41594-018-0031-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akella NM, Ciraku L, Reginato MJ. Fueling the fire: emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. 2019;17:52. doi: 10.1186/s12915-019-0671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013;15:481–490. doi: 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pallet N, et al. A comprehensive characterization of membrane vesicles released by autophagic human endothelial cells. Proteomics. 2013;13:1108–1120. doi: 10.1002/pmic.201200531. [DOI] [PubMed] [Google Scholar]
- 29.Guillot-Sestier MV, Checler F. alpha-Secretase-derived cleavage of cellular prion yields biologically active catabolites with distinct functions. Neurodegener. Dis. 2012;10:294–297. doi: 10.1159/000333804. [DOI] [PubMed] [Google Scholar]
- 30.El Karoui K, et al. Endoplasmic reticulum stress drives proteinuria-induced kidney lesions via Lipocalin 2. Nat. Commun. 2016;7:10330. doi: 10.1038/ncomms10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chawla LS, Eggers PW, Star RA, Kimmel PL. Acute kidney injury and chronic kidney disease as interconnected syndromes. N. Engl. J. Med. 2014;371:58–66. doi: 10.1056/NEJMra1214243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devuyst O, et al. Autosomal dominant tubulointerstitial kidney disease. Nat. Rev. Dis. Prim. 2019;5:60. doi: 10.1038/s41572-019-0109-9. [DOI] [PubMed] [Google Scholar]
- 33.Dvela-Levitt M, et al. Small molecule targets TMED9 and promotes lysosomal degradation to reverse proteinopathy. Cell. 2019;178:521–535. e523. doi: 10.1016/j.cell.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Dihazi H, et al. Secretion of ERP57 is important for extracellular matrix accumulation and progression of renal fibrosis, and is an early sign of disease onset. J. Cell Sci. 2013;126:3649–3663. doi: 10.1242/jcs.125088. [DOI] [PubMed] [Google Scholar]
- 35.Kim Y, et al. Mesencephalic astrocyte-derived neurotrophic factor as a urine biomarker for endoplasmic reticulum stress-related kidney diseases. J. Am. Soc. Nephrol. 2016;27:2974–2982. doi: 10.1681/ASN.2014100986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavernier Q, et al. Urinary angiogenin reflects the magnitude of kidney injury at the infrahistologic level. J. Am. Soc. Nephrol. 2017;28:678–690. doi: 10.1681/ASN.2016020218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonventre JV. Diagnosis of acute kidney injury: from classic parameters to new biomarkers. Contrib. Nephrol. 2007;156:213–219. doi: 10.1159/000102086. [DOI] [PubMed] [Google Scholar]
- 38.Kim, Y. et al. Elevated urinary CRELD2 is associated with endoplasmic reticulum stress-mediated kidney disease. JCI Insight.10.1172/jci.insight.92896 (2017). [DOI] [PMC free article] [PubMed]
- 39.Haldar S, et al. Prion protein promotes kidney iron uptake via its ferrireductase activity. J. Biol. Chem. 2015;290:5512–5522. doi: 10.1074/jbc.M114.607507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B, et al. Prion protein protects against renal ischemia/reperfusion injury. PLoS ONE. 2015;10:e0136923. doi: 10.1371/journal.pone.0136923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehrpour M, Codogno P. Prion protein: from physiology to cancer biology. Cancer Lett. 2010;290:1–23. doi: 10.1016/j.canlet.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Lopes MH, et al. Disruption of prion protein-HOP engagement impairs glioblastoma growth and cognitive decline and improves overall survival. Oncogene. 2015;34:3305–3314. doi: 10.1038/onc.2014.261. [DOI] [PubMed] [Google Scholar]
- 43.Provenzano L, et al. Cellular prion protein (PrP(C)) in the development of Merlin-deficient tumours. Oncogene. 2017;36:6132–6142. doi: 10.1038/onc.2017.200. [DOI] [PubMed] [Google Scholar]
- 44.Du L, et al. CD44-positive cancer stem cells expressing cellular prion protein contribute to metastatic capacity in colorectal cancer. Cancer Res. 2013;73:2682–2694. doi: 10.1158/0008-5472.CAN-12-3759. [DOI] [PubMed] [Google Scholar]
- 45.de Lacerda TC, et al. Prion protein binding to HOP modulates the migration and invasion of colorectal cancer cells. Clin. Exp. Metastasis. 2016;33:441–451. doi: 10.1007/s10585-016-9788-8. [DOI] [PubMed] [Google Scholar]
- 46.Iglesia RP, et al. Engagement of cellular prion protein with the co-chaperone Hsp70/90 organizing protein regulates the proliferation of glioblastoma stem-like cells. Stem Cell Res. Ther. 2017;8:76. doi: 10.1186/s13287-017-0518-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Falke LL, Gholizadeh S, Goldschmeding R, Kok RJ, Nguyen TQ. Diverse origins of the myofibroblast-implications for kidney fibrosis. Nat. Rev. Nephrol. 2015;11:233–244. doi: 10.1038/nrneph.2014.246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.