Abstract
Collecting and curating large medical-image datasets for deep neural network (DNN) algorithm development is typically difficult and resource-intensive. While transfer learning (TL) decreases reliance on large data collections, current TL implementations are tailored to two-dimensional (2D) datasets, limiting applicability to volumetric imaging (e.g., computed tomography). Targeting performance enhancement of a DNN algorithm based on a small image dataset, we assessed incremental impact of 3D-to-2D projection methods, one supporting novel data augmentation (DA); photometric grayscale-to-color conversion (GCC); and/or TL on training of an algorithm from a small coronary computed tomography angiography (CCTA) dataset (200 examinations, 50% with atherosclerosis and 50% atherosclerosis-free) producing 245 diseased and 1127 normal coronary arteries/branches. Volumetric CCTA data was converted to a 2D format creating both an Aggregate Projection View (APV) and a Mosaic Projection View (MPV), supporting DA per vessel; both grayscale and color-mapped versions of each view were also obtained. Training was performed both without and with TL, and algorithm performance of all permutations was compared using area under the receiver operating characteristics curve. Without TL, APV performance was 0.74 and 0.87 on grayscale and color images, respectively, compared to 0.90 and 0.87 for MPV. With TL, APV performance was 0.78 and 0.88 on grayscale and color images, respectively, compared with 0.93 and 0.91 for MPV. In conclusion, TL enhances performance of a DNN algorithm from a small volumetric dataset after proposed 3D-to-2D reformatting, but additive gain is achieved with application of either GCC to APV or the proposed novel MPV technique for DA.
Keywords: Deep neural network, Data augmentation, Photometric conversion, Transfer learning, Coronary artery computed tomography angiography, Artificial intelligence, Medical imaging
Background
The frequent shortage of large datasets in medical imaging often restricts training of a deep neural network (DNN) for tasks, including image-data classification, image segmentation, or disease localization. Compared with available large general-image datasets, such as ImageNet [1], medical-image datasets are relatively small. When a dataset is limited in size, data augmentation (DA) techniques are often used to improve training; commonly applied techniques include random rotations, horizontal and vertical flips, random crops, and small multi-directional translations [2]. In addition, improved performance of a DNN can be achieved by enhancements in image representation, such as grayscale-to-color map conversion (GCC) [3]. Lastly, transfer learning (TL) has also been shown to improve algorithm performance in datasets with limited size [4]; TL is a deep learning method leveraging pre-trained weighted models from large datasets to accomplish an identified task by fine-tuning the final layers of a DNN while the weights of the initial layers are held stable [5]. In medical imaging, TL is easily applicable to 2-dimensional (2D) images due to an abundance of suitable pre-trained models. However, this is not the case for 3D images, as is commonly found in advanced forms of imaging (e.g., computed tomography angiography (CTA)). DNNs have been successfully trained on multi-angle 2D projections of 3D objects in order to overcome this limitation, and there is evidence that the characteristics of a 3D shape can be recognized with greater accuracy from a collection of independent 2D projections [6, 7].
Although the aforementioned techniques have been individually studied outside of the medical-imaging domain, their combined effect on DNN algorithm performance for medical images is not known and was the focus of this work; a 3D-to-2D representation method enabling both TL and other options for image DA is proposed. The purpose of this study was to evaluate the incremental impact of 3D-to-2D reformation methods along with novel image DA, photometric conversion (i.e., GCC), and/or TL on training of an algorithm from a small coronary CTA (CCTA) dataset. This was in preparation for a larger clinical study of the feasibility of use of a DNN algorithm to support augmented intelligence in CCTA screening for atherosclerosis absence (versus any degree of disease) [8], thereby facilitating prompt and safe emergency room discharge of a chest-pain patient to home [9].
Methods
Image-Data Description
All CCTA image-dataset utilization was retrospective, performed locally under Institutional Review Board approval (including HIPAA compliance) with the waiver of patient consent.
The image dataset was derived from 200 randomly selected CCTA examinations (male/female 108/92; age mean/standard deviation 50.6/12.1 years), with 100 demonstrating atherosclerosis and 100 atherosclerosis-free based on review by an investigator-expert (RDW with 33-year experience in cardiac imaging and ACC/AHA level III CCT certification). All imaging (March 2013 to July 2018) was standard-of-care, clinically indicated by chest pain without known coronary artery disease [10, 11] and was performed used multi-detector CT systems (Siemens Healthineers, Erlangen, Germany).
Image-Data Processing
A custom graphical user interface (GUI) was developed using Windows-based MeVisLab 2.8 [12] for detecting and localizing atherosclerosis (e.g., calcification and stenosis) in the coronary artery system (Fig. 1). Through GUI integration, coronary artery/branch-courses were initially delineated automatically utilizing a work-in-progress version of commercial capabilities of the CT cardio-vascular engine [13] with proprietary centerline methodology [14, 15]. Next, the GUI allowed the investigator-expert to manually demarcate the extent of atherosclerosis per affected artery/branch (“diseased”; n = 245) in the 100 atherosclerotic cases. While any unlabeled artery/branch was considered atherosclerosis-negative, negative arteries/branches in atherosclerotic cases were excluded from further analysis due to potential similarities with diseased arteries/branches. On the other hand, all negative arteries/branches in the 100 atherosclerosis-free cases (“normal”, n = 1127) were included.
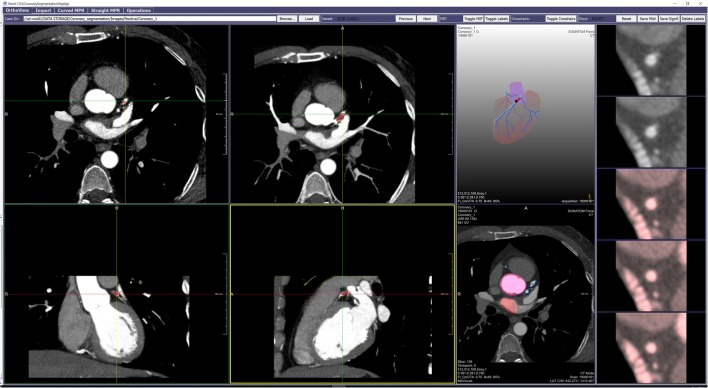
Fig. 1.
GUI for segmentation of the coronary artery system. It includes capabilities for production of the following: (1) multiple orthogonal or oblique multi-planar reformatted or thin-maximum intensity projection 2D images (left sided 2 × 2 panel); (2) a stacked short-axis image series of a coronary artery [right edge strip], with manually applied tinting (red) reflecting local presence of atherosclerosis; and (3) centerline-dependent rotatable coronary artery 3D “branching tree” display (upper, between 2 × 2 panel and right edge strip), with artery enhancement (light-blue) indicating manual selection of artery-of-interest, and ball marker (dark-blue) and segment overlay (red) indicating specific level of manually demarcated atherosclerotic plaque
3D-to-2D Projection Reformatting
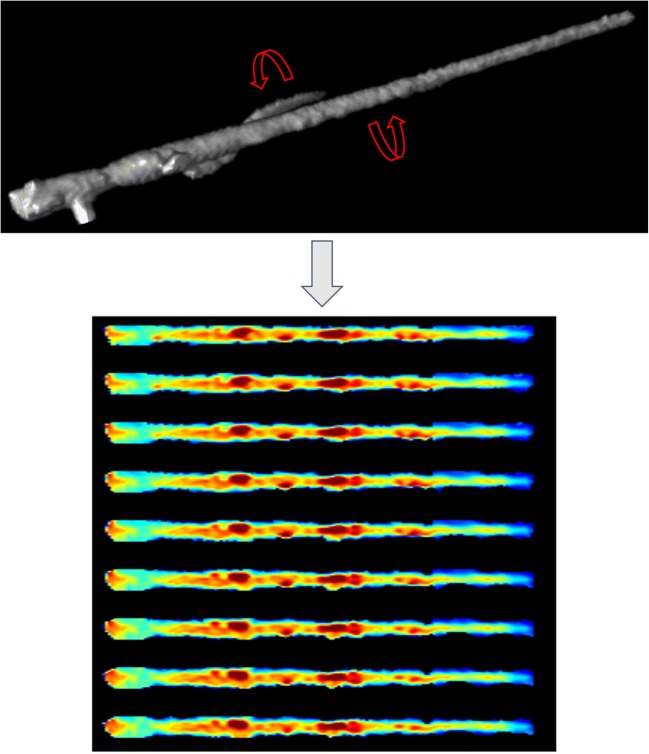
Supporting the 3D-to-2D projection transformation, circumferentially arranged straightened-Multiplanar Reformatted (MPR) displays of each coronary artery/branch, all longitudinally co-registered to the shared centerline, underwent surface-illumination processing to produce a cylindrical-appearing volume image (Fig. 2). During subsequent rotation of each volume image, to keep image resolution as close as possible to original image dimensions used for training, unique ray-traced (RT) projections were created every 10° (0–180°), producing 18 distinctive 2D RT representations per artery/branch [16]; when all 18 were combined by averaging the overlapping intensities, an “Aggregate Projected View” (APV) of the original centerline-extracted arteries/branches was produced.
Fig. 2.
Circumferentially arranged straightened-MPR displays of a diseased coronary artery/branch, all longitudinally co-registered to the shared centerline and then surface-illumination processed, produced a “stretched-appearing” volume image (top). Rotation of each volume image with creation of unique ray-traced (RT) projections every 10° (only 9 shown), produced multiple RT representations per artery/branch (bottom)
For DNN algorithm development, the image data was subdivided according to a 3:1:1 ratio for training, validation, and testing [17]. For the APV evaluation 142, 50, and 53 diseased arteries/branches, as well as 657, 225, and 245 normal arteries/branches, were placed into training, validation, and test sets respectively (Table 1A). However, in order to provide balance in the training and validation sets, similar numbers of normal arteries/branches (142 and 50, respectively) were randomly selected to match those of diseased arteries/branches [Table 1B].
Table 1.
A relative shortage of diseased artery/branch APV representations was demonstrated when a 3:1:1 image-dataset distribution was used for training:validation:testing, leading to an undesirable imbalance for training and testing (1A). When 1:1 diseased:normal dataset balancing was applied for training and validation, low case volumes (e.g., only 142:142) was expected to limit training performance (1B). To increase diseased artery/branch representations for training, while maintaining diseased:normal dataset balance, a novel DA method was developed for dataset enlargement through creation of additional depictions of the same arteries/branches (i.e., MPV); by this “mosaicking” DA method alone, the 142 original diseased APVs identified for training were amplified to 710 diseased MPV representations, approximating the 657 non-permuted normal MPVs (1C)
| 1A | 3:1:1 Distributed | |||
| Training | Validation | Testing | Total | |
| Diseased APV | 142 | 50 | 53 | 245 |
| Normal APV | 657 | 225 | 245 | 1127 |
| 1B | 3:1:1 Distributed+1:1 balanced | |||
| Training | Validation | Testing | Total | |
| Diseased APV | 142 | 50 | 53 | 245 |
| Normal APV | 142 | 50 | 245 | 437 |
| 1C | 3:1:1 Distributed+1:1 balanced+data-augmented for training | |||
| Training | Validation | Testing | Total | |
| Diseased MPV | 710 | 50 | 53 | 813 |
| Normal MPV | 657 | 50 | 245 | 952 |
Image-Data Augmentation
Recognizing that aggregating 18 distinctive 2D RT representations per coronary artery/branch as a single APV is associated with information loss due to averaging, a mosaic alternative was proposed. By this option, rather than averaging the intensities of all 18 distinctive RT representations, a mosaic display of the same 18 views as a 2 × 9 image matrix [18] created a “Mosaic Projected View” (MPV); this enabled a novel image DA technique in which the views of the diseased arteries/branches could be randomly permuted to increase representations. Specifically for this work, the 18 views of each artery/branch were randomly re-ordered five times for the purposes of generating 710 diseased images for more robust learning while maintaining diseased:normal balance in training (Table 1C); no such DA was performed on the validation and test sets.
Photometric Grayscale-to-Color Conversion
To date, most available pre-trained models, such as ImageNet, Pascal-VOC, and Microsoft COCO, have been trained on colored image datasets. However, the aforementioned 2D projection views generated for each artery/branch were represented as a single-channel grayscale image. Therefore, the effect of GCC on improved visualization was also investigated. This process involved transforming a 12-bit grayscale image to a 24-bit RGB image (8-bit for each of the color channels) [3, 19].
Transfer Learning
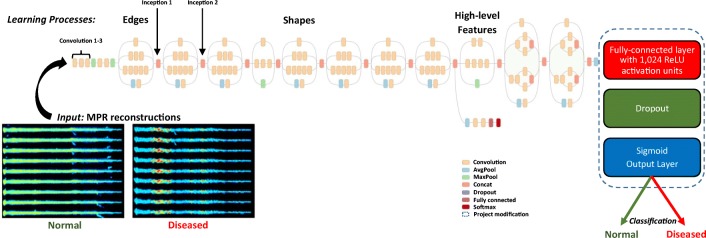
TL is a common approach utilized to improve DNN algorithm performance on a small dataset. While Inception-V3 [20] was used as the base DNN, in order to refine the model for the CCTA dataset, the final layers of the DNN were replaced by a fully connected layer of 1024 nodes in a ReLU activation unit [21], followed by a Sigmoid output function for binary classification as normal vs. diseased (Fig. 3). The weights of the model were initialized with training weights based on the ImageNet dataset.
Fig. 3.
DNN algorithm training with Inception-V3. The additional fully connected ReLU layer and Sigmoid output layer are added at the end of the DNN as shown (right). A sample input of DA coronary artery representations is also shown (left)
All training was performed using Keras [22] with TensorFlow-1.8 [23]. The initial learning rate was 0.001 on a stochastic gradient descent optimizer [24] with batch size of 8; re-training was terminated after 120 epochs. Traditional DA routines (e.g., random rotation, horizontal and vertical flipping, random crops, and translation) were performed for each case [2]. During the training/validation process, algorithm performance (monitoring binary cross-entropy) on the validation set was observed per epoch with preservation of the model with highest accuracy to that point; if the validation accuracy increased in subsequent epochs, the model was updated.
Evaluation of the Proposed Methods
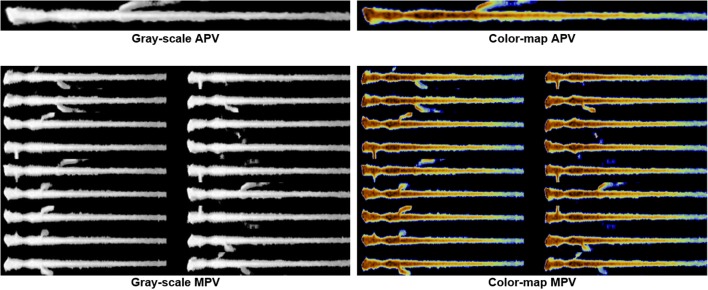
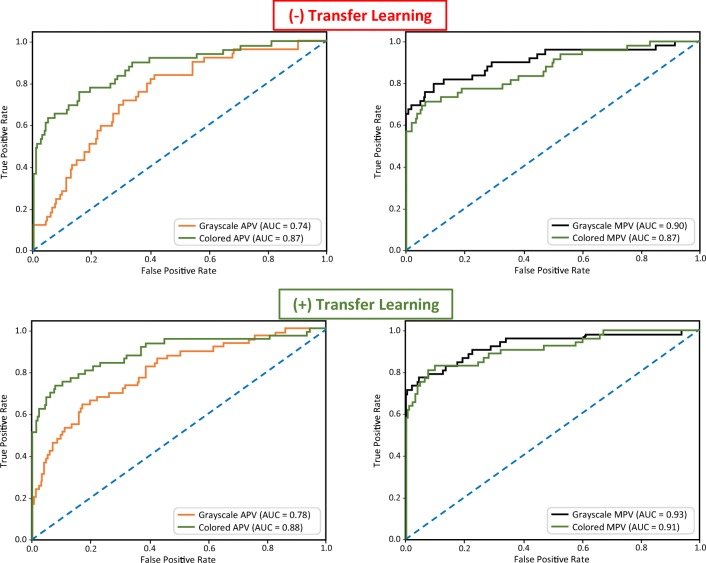
In order to assess incremental impact of the following proposed methods: (1) DA (by MPV “mosaicking”), (2) photometric conversion (by GCC), and (3) TL (by initialization with pre-trained weights), eight models reflecting the possible combinations of aforementioned variables (i.e., APV vs. MPV gray-scale, APV vs MPV colored, either without/with TL) underwent assessment (Fig. 4). Classification accuracies were based on the area under the receiver operating characteristics curve (AUC) evaluation.
Fig. 4.
To assess incremental impact of the following proposed methods on DNN algorithm performance: (1) DA (by MPV “mosaicking”), (2) photometric conversion (by GCC), and (3) TL (by initialization with pre-trained weights), eight models reflecting the possible combinations of aforementioned variables were developed; they included APV vs MPV in gray-scale (left), as well as APV vs MPV in color-map (right), both without and with application of TL
Results
DA based on mosaicked 3D-to-2D transformation (by MPV technique) alone showed the highest DNN algorithm performance improvement (Fig. 5). MPV results either equaled (AUC = 0.87), but more often exceeded (MPV AUC = 0.90–0.93 > APV AUC = 0.74–0.88), algorithm performance by non-mosaicked training (i.e., APV only). Next, color-mapping from GCC alone was superior to corresponding gray-scale comparisons only for APV (both color APV AUC = 0.87–0.88 > gray APV AUC = 0.74–0.78), while being inferior for MPV (both color MPV AUC = 0.87–0.91 < gray MPV AUC = 0.90–0.93). Last, TL using pre-trained model weights on ImageNet alone was found to be consistently beneficial (with pre-trained weights AUC = 0.78–0.93 > random/un-trained weights AUC = 0.74–0.90). When TL was combined with the proposed novel MPV-based DA method, the greatest amplification of algorithm performance was achieved (with both DA and TL AUC = 0.93).
Fig. 5.
Changing AUC based on applications of proposed DA, GCC, and/or TL methods is shown. The increase in performance from MPV-based DA (right) over APV use alone (left), as well as from the utilization of pre-trained model weights (TL with ImageNet) (bottom), compared to random-weight initialization of training (i.e., no TL) (top), demonstrate both the individual and additive value of the proposed novel DA and TL methods towards yield in DNN algorithm classification
Discussion
With the goal of strengthening the development of a DNN algorithm for binary classification supporting augmented intelligence in CCTA screening for atherosclerosis absence versus any degree of disease [8] via improved image dataset size and balance, this work was focused on a strategy potentially incorporating the following: (1) 3D-to-2D projection reformatting methods supporting novel DA (by MPV mosaicking); (2) photometric conversion for enhanced image representation (by GCC); and (3) TL initialized by pre-trained weighting (by ImageNet), rather than by random-weighting. The results indicate that while a positive impact from TL can be achieved on volumetric datasets using the proposed 3D-to-2D reformatting, greater gain in algorithm performance is realized with application of either color-mapping (by GCC) to APV or novel DA (by MPV technique).
Most pre-trained imaging-focused DNNs require 2D images, while advanced medical images (e.g., CCTA) are 3D in nature. Thus, methods to generate 2D images from 3D counterparts have become vital. This work demonstrated that multiple mosaicked projected views of a single MPR-based coronary artery/branch representation (i.e., MPV) can help train a classifier using Inception-V3 after TL initialization (e.g., with ImageNet weights), thereby facilitating the achievement of high DNN algorithm performance of AUC = 0.93 based on a small image dataset from only 200 examinations.
Since many of the available deep learning models were designed for colored images, it is possible that GCC may be beneficial in some cases in medical imaging. However, the opposite results revealed on MPV suggests that the color-mapping technique may only be advantageous when detection of subtle changes in density are needed, such as in the case of APV which averages the densities of all projections whereas MPV displays the projections side-by-side in a mosaic configuration.
Recently, recurrent convolution neural networks were used to characterize coronary stenosis and plaque with accuracies of 0.80 and 0.77, respectively; it produced encouraging “proof of concept” for artificial intelligence-based coronary analysis [25]. In the current technical development, higher algorithm performance was achieved with a 3D-to-2D image conversion technique combined with novel DA and TL.
Nevertheless, there are technical limitations to the proposed CCTA evaluation. First, a 2D, rather than 3D, approach to the representation of CCTA image data is used. However, this is justifiable because of the following: (1) ,most curated/labeled medical-imaging datasets available are relatively small, compared with publicly available datasets, and need amplification; (2) it is impractical to have radiologists labor over the annotation of large volumes of images, compounded further by the 3D nature of CCTA data; (3) CCTA data access and image-annotation expertise are currently limited to our site and their cardiac radiologist workforce; and (4) there has been previously reported success with use of transfer learning for 2D views of 3D image datasets. Second, there is reliance on vessel auto-segmentation which proved imperfect, with infrequent incorrect extraction of arteries/branches due to unpredictable centerline deviation, with no easy option for manual rectification. Last, observed occasional incorrect displacements of coronary calcifications from the peri-centric field of view during segmentation may have contributed to falsely atherosclerosis-free algorithmic interpretations. However, both issues would have impacted global algorithm performance but should not have impacted comparisons as no selective modification was made to the training sets other than the augmentations described earlier.
Conclusion
This report corroborates the use of 3D-to-2D reformatting techniques along with novel DA (by MPV technique) and TL-combined strategy to overcome recognized image-data limitations common in medical imaging. The mosaicking method for DA, as well as the GCC strategy on APV, described in this report could alone have important applications in other areas of medical imaging [18, 26], but further investigation is needed to determine whether these techniques can also be successfully utilized in other clinical scenarios including different imaging modalities and regions of the body. Nonetheless, this report addresses practical opportunities for managing the anticipated ongoing issues related to shortages of curated image-data for DNN algorithm training.
Funding Information
This project was partially funded by:
1. Donation from the Edward J. DeBartolo, Jr. Family.
2. Master Research Agreement with Siemens Healthineers.
Compliance with Ethical Standards
All CCTA image-dataset utilization was retrospective, performed locally under Institutional Review Board approval (including HIPAA compliance) with the waiver of patient consent.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Krizhevsky A, Sutskever I, Hinton GE. Proceedings of the 25th international conference on neural information processing systems 1. 2012. ImageNet classification with deep convolutional neural networks; pp. 1097–1105. [Google Scholar]
- 2.Perez L, Wang J: The effectiveness of data augmentation in image classification using deep learning. CoRR abs, arXiv:1712.04621, 2017.
- 3.Prasad S, Kumar P, Sinha KP: Grayscale to color map transformation for efficient image analysis on low processing devices. In: El-Alfy ES., Thampi S., Takagi H., Piramuthu S., Hanne T. (eds) Advances in Intelligent Informatics. Advances in Intelligent Systems and Computing, vol 320. Springer, cham, 2015, 10.1007/978-3-319-11218-3_2
- 4.Ng HW, Nguyen VD, Vonikakis V, Winkler S: Deep learning for emotion recognition on small datasets using transfer learning. Proceedings of ACM International conference on multimodal interaction, 443–449, 2015.
- 5.Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35:1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su H, Maji S, Kalogerakis E, Learned-Miller E: Multi-view convolutional neural networks for 3D shape recognition. IEEE International Conference on Computer Vision (ICCV), 945–953, 2015.
- 7.Kanezaki A, Matsushita Y, Nishida Y: RotationNet: joint object categorization and pose estimation using multi views from unsupervised viewpoints. International Conference on Computer Vision and Pattern Recognition (CVPR), arXiv:1603.06208, 2018.
- 8.White RD, Erdal BS, Bigelow MT, Demirer M, Galizia MS, Gupta V, Carpenter JL, Candemir S, Dikici E, O'Donnell TP, Halabi AH, Prevedello LM et al. Augmented intelligence to facilitate exclusion of coronary atherosclerosis on CCTA during emergency department chest-pain presentations: algorithm development. Radiology, Cardiothoracic Imaging (Submitted).
- 9.Litt HI, Gatsonis C, Snyder B, Singh H, Miller CD, Entrikin DW, Leaming JM, Gavin LJ, Pacella CB, Hollander JE. CT angiography for safe discharge of patients with possible acute coronary syndromes. N Engl J Med. 2012;366:1393–1403. doi: 10.1056/NEJMoa1201163. [DOI] [PubMed] [Google Scholar]
- 10.Ghoshhajra BB, Engel LC, Major GP, Goehler A, Techasith T, Verdini D, Do S, Liu B, Li X, Sala M, Kim MS, Blankstein R, Prakash P, Sidhu MS, Corsini E, Banerji D, Wu D, Abbara S, Truong Q, Brady TJ, Hoffmann U, Kalra M. Evolution of coronary computed tomography radiation dose reduction at a tertiary referral center. Am J Med. 2012;125:764–772. doi: 10.1016/j.amjmed.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lubbers MM, Dedic A, Kurata A, Dijkshoorn M, Schaap J, Lammers J, Lamfers EJ, Rensing BJ, Braam RL, Nathoe HM, Post JC, Rood PP, Schultz CJ, Moelker A, Ouhlous M, van Dalen BM, Boersma E, Nieman K. Round-the-clock performance of coronary CT angiography for suspected acute coronary syndrome: results from the BEACON trial. Eur Radiol. 2018;28:2169–2175. doi: 10.1007/s00330-017-5082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demirer M, Candemir S, Bigelow M, Yu SM, Gupta V, Prevedello LM et al.: A user interface for optimizing radiologist engagement in image-data curation for artificial intelligence. Radiology, 2019 (In Press) [DOI] [PMC free article] [PubMed]
- 13.Siemens Healthineers, Erlangen, Germany. Available at https://www.healthcare.siemens.com/computed-tomography/clinical-imaging-solutions/ct-cardio-vascular-engine Accessed 14 April 2018
- 14.Siemens Research. Princeton, USA. Available at http://www.usa.siemens.com/en/about_us/research/research_dev/imaging_visualization/imaging_visualization_publications_2009.htm. Accessed 3 Jan 2019
- 15.Zheng Y, Tek H, Funka-Lea G. Robust and accurate coronary artery centerline extraction in CTA by combining model-driven and data-driven approaches. Med Image Comput Comput Assist Interv. 2013;16:74–81. doi: 10.1007/978-3-642-40760-4_10. [DOI] [PubMed] [Google Scholar]
- 16.Dalrymple NC, Prasad SR, Freckleton MW, Chintapalli KN. Informatics in radiology (infoRAD): Introduction to the language of three-dimensional imaging with multidetector CT. RadioGraphics. 2005;25:1409–1428. doi: 10.1148/rg.255055044. [DOI] [PubMed] [Google Scholar]
- 17.Dobbin KK, Simon RM: Optimally splitting cases for training and testing high dimensional classifiers. BMC Med Genomics 4(31), 2011. 10.1186/1755-8794-4-31 [DOI] [PMC free article] [PubMed]
- 18.ArcMap: Understanding the mosaicking rules for a mosaic dataset. ESRI. Available at http://desktop.arcgis.com/en/arcmap/latest/manage-data/raster-and-images/understanding-the-mosaicking-rules-for-a-mosaic-dataset.htm. Accessed 18 November 2018.
- 19.Zickler T, Mallick SP, Kriegman DJ, Belhumeur PN. Color subspaces as photometric invariants. International Journal of Computer Vision. 2008;79:13–30. doi: 10.1007/s11263-007-0087-3. [DOI] [Google Scholar]
- 20.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z: Rethinking the inception architecture for computer vision. Proceedings of the IEEE conference on computer vision and pattern recognition. 2818–2826. arXiv:1512.00567, 2016.
- 21.Dahl GE, Sainath TN, Hinton GE: Improving deep neural networks for LVCSR using rectified linear units and dropout. IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 8609–8613, 2013.
- 22.Chollet F: User experience design for APIs. The Keras Blog; Available at https://blog.keras.io/user-experience-design-for-apis.html. Accessed 18 November 2018.
- 23.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, Kudlur M, Levenberg J, Monga R, Moore S, Murray DG, Steiner B, Tucker P, Vasudevan V, Warden P, Wicke M, Yu Y, Zheng X: TensorFlow: a system for large-scale machine learning. USENIX symposium on operating systems design and implementation 16:265–283, 2016. arXiv:1605.08695
- 24.Bottou L: Large-scale machine learning with stochastic gradient descent. Proceedings of COMPSTAT 177–186, 2010.
- 25.Zreik M, van Hamersvelt RW, Wolterink JM, Leiner T, Viergever MA, Isgum I: Automatic detection and characterization of coronary artery plaque and stenosis using a recurrent convolutional neural network in coronary CT angiography. Available at https://openreview.net/forum?id=BJenxxhof. arXiv:1804.04360v1. Accessed 18 November 2018.
- 26.Loewke KE, Camarillo DB, Jobst CA, Salisbury JK. Real-time image mosaicing for medical applications. Stud Health Technol Inform. 2007;125:304–309. [PubMed] [Google Scholar]