Abstract
The explosion of medical imaging data along with the advent of big data analytics has launched an exciting era for clinical research. One factor affecting the ability to aggregate large medical image collections for research is the lack of infrastructure for automated data annotation. Among all imaging modalities, annotation of magnetic resonance (MR) images is particularly challenging due to the non-standard labeling of MR image types. In this work, we aimed to train a deep neural network to annotate MR image sequence type for scans of brain tumor patients. We focused on the four most common MR sequence types within neuroimaging: T1-weighted (T1W), T1-weighted post-gadolinium contrast (T1Gd), T2-weighted (T2W), and T2-weighted fluid-attenuated inversion recovery (FLAIR). Our repository contains images acquired using a variety of pulse sequences, sequence parameters, field strengths, and scanner manufacturers. Image selection was agnostic to patient demographics, diagnosis, and the presence of tumor in the imaging field of view. We used a total of 14,400 two-dimensional images, each visualizing a different part of the brain. Data was split into train, validation, and test sets (9600, 2400, and 2400 images, respectively) and sets consisted of equal-sized groups of image types. Overall, the model reached an accuracy of 99% on the test set. Our results showed excellent performance of deep learning techniques in predicting sequence types for brain tumor MR images. We conclude deep learning models can serve as tools to support clinical research and facilitate efficient database management.
Keywords: Artificial intelligence, Deep learning, Magnetic resonance imaging, Sequence type, Automated annotation, Image database
Introduction
The use of diagnostic imaging has increased immensely in the past two decades. The volume of imaging services per Medicare beneficiary has been reported to move faster than the growth of all other services physicians provide [1]. Among all imaging modalities, the rapid increase in imaging utility is noticeable for magnetic resonance (MR) imaging. A report into the temporal patterns in diagnostic medical imaging calculated a three-fold increase in the rate of prescribed MR imaging services for Medicare beneficiaries over a 10-year period [2]. This rise in the quantity of data has been attributed to factors such as improved availability and quality of the technology leading to increased demand by patients and physicians [2].
Recent developments in imaging techniques have brought unparalleled advantages to clinical care. However, the speed of innovation has also introduced practical bottlenecks for data storage and image annotation. This is particularly the case for MR imaging databases due to varying sequence names across manufacturers and the lack of an annotation standard among different imaging centers—or even within an imaging center. Among MRI scanner manufacturers, the equivalent or similar sequence types can have wildly different names [3–5]. As new sequences are developed and introduced, this variation only increases. Further, within a given imaging center, new parameter combinations are frequently introduced as radiologists, MR physicists, and other imaging researchers explore adjustments to sequences and scan order to improve scan quality or reduce scan times. Often these customized sequences are named to distinguish one from another, however, the rationale behind these names is rarely communicated beyond the small group responsible for developing them. For multi-institutional data repositories, successful image annotation requires a designated, trained individual who focuses on the intrinsic image weighting and the characteristics of image content rather than manufacturer or institutional nomenclature. This lengthy and tedious manual process creates a bottleneck for aggregation of large image collections and impedes the path to research. An automated annotation system that can match the speed of image generation in the big data era is sorely needed.
Over that last two decades, our lab has focused on developing patient-specific mathematical models for brain tumor growth and response to therapy [6–17]. As such, we have built a large repository of curated brain tumor patient imaging and clinical data from over 20 institutions, a variety of MR imaging devices, and a diversity of imaging parameters. A major step in that curation is the annotation of an appropriate MR imaging type from these disparate sources. Proper image type identification guides the assignment of images for the segmentation of the abnormality seen on the image by our image analysis team. With over 70,000 images now in the database and that number increasing daily, it is clear that we have a unique resource for clinical investigation and a need to automate this process.
Harnessing the power of large training data, deep learning techniques have shown tremendous success in visual recognition tasks that include thousands of categories [18]. These techniques have also been utilized for classification of tumor types in various organs [19–21]. In this work, we aimed to use deep learning techniques to predict the sequence type of MR scans of brain tumor patients. We focused on the four most common sequence types of morphologic MR imaging in our database: T1-weighted (T1W), T1-weighted post-gadolinium contrast agent (T1Gd), T2-weighted (T2W), and T2 fluid-attenuated inversion recovery (FLAIR). To the best of our knowledge, no previous work has focused on automatic annotation of MR image sequence types. This form of classification can be highly useful for building large-scale imaging repositories that can receive submissions from heterogeneous data sources.
Materials
MR Sequence Types
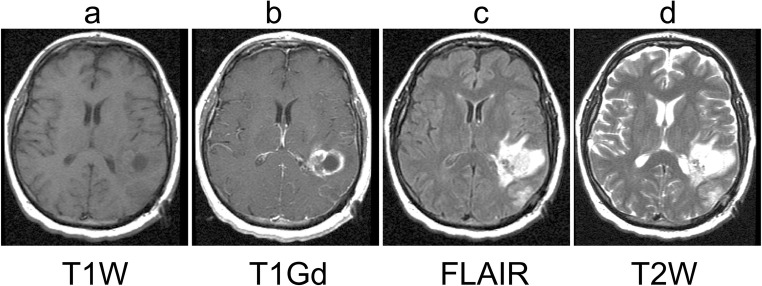
MR imaging is a powerful non-invasive imaging technique as it provides soft tissue contrast of brain structures. MR utilizes a powerful magnet and exploits the differing magnetic properties of water protons in various microenvironments within the tissue. Further, sequences and parameters can display the same organ with varying contrast [22]. The four most common morphologic MR image types ordered for brain tumor patients are T1-weighted (T1W), T1-weighted post-gadolinium contrast agent (T1Gd), T2-weighted (T2W), and T2 fluid-attenuated inversion recovery (FLAIR). Figure 1 shows examples of these sequence types for a brain tumor patient. On T1W images (Fig. 1a), normal gray matter is darker than normal white matter and the ventricles are black. Neoplasms are not particularly prominent on T1W images. However, gadolinium contrast agent (Gd) appears hyperintense on T1W images (Fig. 1b). Gd is given intravenously and crosses the broken-down blood-brain barrier (BBB) of gliomas and is thought to indicate the most aggressive tumor regions. On T2W images (Fig. 1d), normal gray matter is brighter than normal white matter and the ventricles are generally the brightest structure. Edema associated with leaky BBB appears hyperintense, which can sometimes appear with similar intensity to the ventricles. The FLAIR imaging sequence is generally a shorthand for a T2W-weighted image that inverts the intensity of the ventricles such that they appear dark, similar to their appearance on T1W images (Fig. 1c). The dark ventricles on FLAIR images can help to distinguish them from the hyperintense edema in adjacent periventricular parenchyma [23, 24].
Fig. 1.
Examples of the four MR sequence types included in this work: a T1-weighted (T1W), b T1-weighted post-gadolinium contrast (T1Gd), c T2-weighted fluid-attenuated inversion recovery (FLAIR), and d T2-weighted (T2W). Images were acquired from a brain tumor patient
Data
Our group has developed an unprecedented clinical research database of over 70,000 serially acquired MR studies of 2500+ unique patients with brain tumors of various grades, sizes, and at different locations in the brain. This IRB-approved repository contains pre- and post-treatment MR images acquired across 20 institutions, each using different scanner manufacturers, pulse sequences, sequence parameters, and field strengths. This repository contains patient information and therefore is subject to HIPPA regulations. Data may be available for sharing upon the request of qualified parties as long as patient privacy and intellectual property interests of our institution are not compromised. Typically, data access will occur through a collaboration and may require interested parties to obtain an affiliate appointment with our institution.
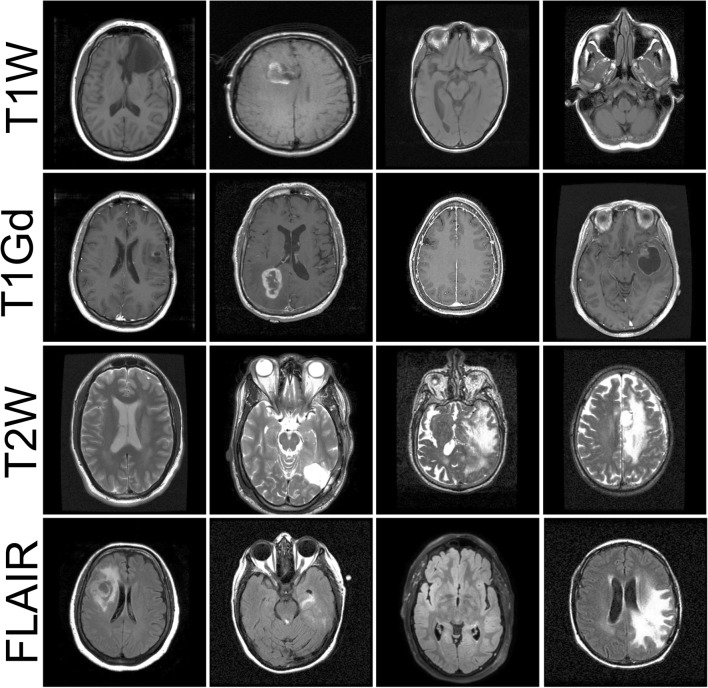
Several factors, including imaging protocol parameters/weights, were variable among the images in our database because of acquisition in various institutions and various devices. MR sequence types depend on the imaging modalities prescribed for treatment planning of a given patient. In this work, we focused on the most common sequence types in our repository including anatomical T1W, T1Gd, T2W, and FLAIR sequences. Reference annotation was created by image review by trained technicians with multiple years of segmentation experience. We randomly selected 1000 unique image series of each of the four sequence types. Image series were acquired from ~ 700 unique patients in various stages of treatment process. From each image series, we randomly selected four axial slices, excluding the first and last quarters of axial slices. This resulted in a total of 14,400 two-dimensional images. Figure 2 shows examples of images in our dataset. Note that the process of image selection was agnostic to the presence of tumor abnormality or surgical resection in the image field of view.
Fig. 2.
Examples of images in the training set. Images were acquired in multiple institutions using various devices. Several factors were variable among images including slice number, image contrast, levels of focus, head orientation, and presence of tumor in the imaging field of view
Preprocessing
We excluded the top and bottom portions of image series from the image collection to avoid regions without soft tissue (close to the skull) and within the neck that yield few visual indications of sequence type. Beyond variability in scanner manufacturer or location, there was immense heterogeneity in image field of view, signal to noise ratio, resolution, brightness, head orientation, and intensity range. To maintain the similarity of our data with future clinically collected data and to ensure the utility of the model in production, we performed minimal preprocessing on our image collection. Images were resized to a common image size of 128 × 128 pixels and dynamic range of intensities were scaled to a 0–1 range to improve convergence. No additional preprocessing was performed.
Training, Validation, and Test Sets
We divided 14,400 images into 9600 training, 2400 validation, and 2400 test samples. Each dataset contained equal numbers of each sequence type. To prevent model overfitting and improve learning, we used data augmentation at the training phase. Our strategy for data augmentation included introducing random rotation (range of ± 25°), width and height shift (ranging to 10% of total width/height), shear (range of 0.2), zoom (range of 0.2), horizontal flip, and filling points outside the boundaries of the input mode using nearest neighbor.
Methods
Network Architecture
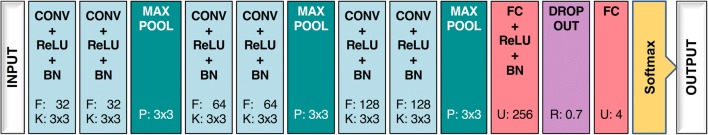
We used a variation of the Visual Geometry Group network (VGGNet) architecture [25]. Figure 3 presents the architecture of our network, which included 6 convolutional layers with 32, 64, and 128 filters for every 2 layers, kernel sizes of 3 × 3, and strides of 1 in all blocks. All convolutional layers were accompanied by a rectified linear unit (ReLU) activation function and batch normalization. Spatial pooling was carried out after two batch normalization layers using max pooling with a stride of 2 and pool size of 3 × 3. Convolutional layers were followed by two fully connected layers of 256 and 4 nodes. Dropout regularization with a probability of 0.7 was used after the first fully connected layer to avoid model overfitting. Finally, the outputs of the fully connected layers were converted into probability values using a softmax activation function.
Fig. 3.
Network architecture. Convolutional layers were all accompanied by ReLU activation and batch normalization (BN) layers. Numbers in each layer represent parameters of the layers: kernel (K) and filter size (F) for convolutional layers, pooling window size (P) for max pooling layers, number of units (U) in fully connected layers, and rate (R) in the dropout layer
Training Procedure
We used the Keras [26] package with TensorFlow [27] backend for our implementation and trained the network on a Nvidia TITAN V GPU. For learning the network weights, we optimized using the Adam [28] algorithm with a batch size of 32 and categorical cross-entropy as the loss function. The nonlinear rectified linear unit after every convolutional and fully connected layer ensured that the vanishing gradient problem during training would be avoided. Initial weights were selected at random from the Xavier normal initializer. Learning rate was initially set to 2e−4 with weight decay scheduled at the beginning of each epoch in which the learning rate was divided by the number of epochs at each scheduled weight update.
Metrics
We used maximum class probability as the prediction of the model for a given sample. To assess the performance of the model during training, we report the area under the receiver operating characteristic (ROC) curves for the validation set. To evaluate the performance of the model on previously unseen data, we report accuracy, sensitivity, and specificity of the model on the test set. Sensitivity of prediction per sequence type was defined as the percentage of correctly identified sequence-type images. Specificity for each sequence type was calculated as the percentage of cases of all other sequence types that were correctly identified. Overall sensitivity of the model for this classification task was calculated using the percentage of true positive and true negatives across the entire test set. All calculations were performed in python using Scikit-learn [29] and Scipy packages.
Results
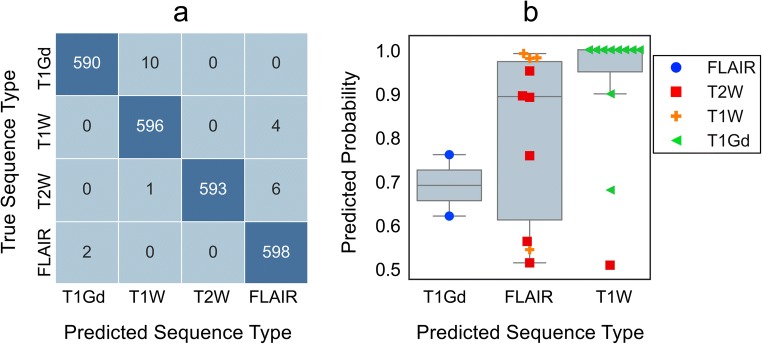
Sequence-specific and average area under the ROC curves on the validation set was > 0.99. Table 1 presents the overall and per-sequence performance of the model on the independent test set samples. Sequence-specific and average accuracy of the model was > 0.99 for the test set samples, similar to the result on validation set, suggesting the generalizability of the model on future and extended data. Among all sequence types, T1Gd achieved the lowest sensitivity (0.98). After reviewing misclassified T1Gd images, we noticed that four of 10 false-negative cases all had been mislabeled in the training dataset and actually belonged to the model-predicted sequence type. Figure 4 presents extended analysis of test set predictions. Figure 4a shows the confusion matrix associated with the predictions of the model, while Fig. 4b compares the distribution of assigned probabilities to misclassified images. T1W and T2W misclassified images were most probable to be labeled as FLAIR. The majority of T1W false positives were T1Gd images, indicating that whenever the gadolinium hyperintensity was not detected, T1Gd was most probably mistaken with T1W.
Table 1.
Model performance on previously unseen test set (N = 2400)
| Sequence type | Sensitivity | Specificity | Accuracy |
|---|---|---|---|
| T1W | 0.993 | 0.994 | 0.994 |
| T1Gd | 0.983 | 0.998 | 0.995 |
| T2W | 0.997 | 1.000 | 0.998 |
| FLAIR | 0.993 | 0.997 | 0.996 |
| All | 0.992 | 0.997 | 0.992 |
Fig. 4.
Detailed analysis of test set predictions. a Confusion matrix for test set predictions. T1Gd had the highest count of false negatives for which all false negative samples were predicted as T1W. b Comparison of probabilities assigned to misclassified test samples for different sequence types. Among all, probabilities assigned to misclassified T1W samples was high
Discussion
Previous works on classification and annotation of visual content using deep learning techniques have largely focused on detecting items of interest in real-world images (e.g., cats and dogs) [30–34]. Similar works within the biomedical imaging literature have assessed the utility of deep learning techniques in the classification of various diseases [22–24, 35, 36]. However, few studies have explored how these techniques can improve organization of multi-center biomedical image repositories. We found one study that demonstrated the utility of deep learning methods in automatic annotation of cardiac MR acquisition planes [37]. To the best of our knowledge, our study is the first of its kind that uses deep learning for annotation of MR sequence types.
Our result shows that convolutional neural network predictor of MR sequence type can achieve accuracy, sensitivity, and specificity of 99% in identification of sequence type on previously unseen MR images. This is a notable success given the two-fold variability in our training data: one with regard to image content (presence/absence of tumor, head position, slice number, treatment effects, etc.) and another as a result to diversity of imaging parameters (echo time, repeat time, field strength, etc.) across a spectrum of imaging sites. Our result suggests that the tedious, lengthy, and erroneous task of manual image labeling for multi-institutional medical image repositories can reliably be managed using automatic annotation systems using artificial intelligence.
Upon review of misclassified images, we found that in 4 cases, the model was correct while the original reference label was incorrect. Among others, T1Gd had the highest false negative rate which can be attributable to the lack of identifiable gadolinium enhancement on the slices (example in Fig. 5). Gadolinium enhancement is not necessarily present on all two-dimensional slices in a volumetric MR image, and may only be present within blood vessels and/or tissues with breakdown or absence of an intact blood-brain barrier (e.g., nasal mucosa). In addition, contrast-enhanced scans must be performed in relation to injection time of the contrast agent. Consequently, some scans may be acquired at sub-optimal times when Gd contrast has yet to reach the tumor region or has begun to clear from the tissue. For T1W misclassified cases, poor contrast between gray and white matter is a likely reason for confusion with FLAIR images. T2W images can be differentiated from FLAIR by the brightness of CSF (cerebrospinal fluid) within the ventricles and along the sulci (e.g., Sylvian fissure) and basal cisterns. Without proper identification of these CSF spaces, differentiating T2W from FLAIR can be challenging. Lateral ventricles are likely prioritized when visible (given their large size and distinctive morphology). The other aforementioned CSF spaces could be prioritized in the absence of visible lateral ventricles (e.g., 3rd and 4th ventricles, sulci, cisterns). We believe that further improvement of the model demands acknowledgment of these factors.
Fig. 5.
Examples of misclassified images in which a high probability was assigned to the wrong class. True and predicted labels are presented under each image. All predictions in this figure were based on a probability of > 0.75. Parts a and c show complete misses. In comparison, the image in part b shows no sign of contrast enhancement, which is the only difference between T1Gd and T1 sequence types. This can happen if image acquisition is performed too late in relation to injection time of contrast agent
We opted to use a 2D network structure for this work. Our results show that provided with a diverse set of training data, a 2D convolutional neural network can achieve high accuracy in annotation of MR sequence types. Selection of slices from the center of the image stacks increased the possibility of capturing images with gadolinium enhancement. While it can be argued that a 3D convolutional network is the more natural choice for detecting patterns given the volumetric nature of brain imaging data, we avoided using a 3D network for a few key reasons. First, MR imaging of the brain is typically performed in lower resolution in one of the three imaging dimensions (X-Y-Z). Slice thickness can differ substantially within and between examples of sequence types. Training a 3D network demands additional preprocessing to down-sample high-resolution series to match dimensionality of lower resolution images, leading to intensity interpolation and potentially loss of accuracy. Secondly, the use of 3D networks significantly increases memory requirement and computational cost during training as these networks have substantially more parameters compared to 2D nets. Memory resources must also be able to hold large 3D images during model training, frequently requiring multiple GPUs. Therefore, we utilized a 2D approach to enable a reasonable training time for our network and to work with our available resources.
Our current research focused on predicting individual 2D slices selected at random from volumetric image stacks. In practical application, we would annotate the image stack using the entire 3D volumetric image. In future work, we intend to bias our methods toward annotation of the image stack according to model predictions for a minority of important slices.
Conclusion
In this work, we demonstrated the utility of artificial intelligence in automated annotation of MR sequence types for multi-institutional image repositories. Our result showed that deep learning methods can successfully detect patterns related to MR sequence type even when trained on non-harmonized heterogeneous data collected from different institutions, each using different image acquisition parameters and scanner manufacturers. Future directions for this work will focus on extending the model to include additional sequence types and validating the model against normal and diseased brain MR imaging scans.
Funding Information
This study received support from the James S. McDonnell Foundation, the Ivy Foundation, the Mayo Clinic, the Zicarelli Foundation, and the NIH (R01 NS060752, R01 CA164371, U54 CA210180, U54 CA143970, U54 CA193489, U01 CA220378).
Footnotes
Statement of Impact
To the best of our knowledge, no previous work has focused on automatic annotation of MR image types. This form of classification can be highly useful for building large-scale imaging repositories that can receive submissions from heterogeneous data sources.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kristin R. Swanson and Leland S. Hu contributed equally to this work.
References
- 1.Commission, M. P. A. Book AD, others . Healthcare spending and the Medicare program. Washington, DC: MedPAC; 2012. [Google Scholar]
- 2.Smith-Bindman R, Miglioretti DL, Larson EB. Rising use of diagnostic medical imaging in a large integrated health system. Health Aff. 2008;27:1491–1502. doi: 10.1377/hlthaff.27.6.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nitz WR. MR imaging: Acronyms and clinical applications. Eur. Radiol. 1999;9:979–997. doi: 10.1007/s003300050780. [DOI] [PubMed] [Google Scholar]
- 4.MRI sequences acronyms. IMAIOS Available at: https://www.imaios.com/en/e-Courses/e-MRI/MRI-Sequences/Sequences-acronyms. (Accessed: 30th April 2019)
- 5.Enlarge, C. T. T. O. & Ge, G. GRE Acronyms.
- 6.Swanson KR, Rockne RC, Claridge J, Chaplain MA, Alvord EC, Anderson ARA. Quantifying the role of angiogenesis in malignant progression of gliomas: in silico modeling integrates imaging and histology. Cancer Res. 2011;71:7366–7375. doi: 10.1158/0008-5472.CAN-11-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson PR, Juliano J, Hawkins-Daarud A, Rockne RC, Swanson KR. Patient-specific mathematical neuro-oncology: Using a simple proliferation and invasion tumor model to inform clinical practice. Bull. Math. Biol. 2015;77:846–856. doi: 10.1007/s11538-015-0067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkins-Daarud A, Rockne R, Corwin D, Anderson ARA, Kinahan P, Swanson KR. In silico analysis suggests differential response to bevacizumab and radiation combination therapy in newly diagnosed glioblastoma. J. R. Soc. Interface. 2015;12:20150388. doi: 10.1098/rsif.2015.0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rockne RC. et al.: A patient-specific computational model of hypoxia-modulated radiation resistance in glioblastoma using 18F-FMISO-PET. J. R. Soc. Interface 12, 2015 [DOI] [PMC free article] [PubMed]
- 10.Alfonso JCL, Talkenberger K, Seifert M, Klink B, Hawkins-Daarud A, Swanson KR, Hatzikirou H, Deutsch A. The biology and mathematical modelling of glioma invasion: a review. J. R. Soc. Interface. 2017;14:20170490. doi: 10.1098/rsif.2017.0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rayfield CA, Grady F, de Leon G, Rockne R, Carrasco E, Jackson P, Vora M, Johnston SK, Hawkins-Daarud A, Clark-Swanson KR, Whitmire S, Gamez ME, Porter A, Hu L, Gonzalez-Cuyar L, Bendok B, Vora S, Swanson KR. Distinct phenotypic clusters of glioblastoma growth and response kinetics predict survival. JCO Clinical Cancer Informatics. 2018;2:1–14. doi: 10.1200/CCI.17.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randall EC, Emdal KB, Laramy JK, Kim M, Roos A, Calligaris D, Regan MS, Gupta SK, Mladek AC, Carlson BL, Johnson AJ, Lu FK, Xie XS, Joughin BA, Reddy RJ, Peng S, Abdelmoula WM, Jackson PR, Kolluri A, Kellersberger KA, Agar JN, Lauffenburger DA, Swanson KR, Tran NL, Elmquist WF, White FM, Sarkaria JN, Agar NYR. Integrated mapping of pharmacokinetics and pharmacodynamics in a patient-derived xenograft model of glioblastoma. Nat. Commun. 2018;9:4904. doi: 10.1038/s41467-018-07334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swanson KR, Rostomily RC, Alvord EC., Jr A mathematical modelling tool for predicting survival of individual patients following resection of glioblastoma: A proof of principle. Br. J. Cancer. 2008;98:113–119. doi: 10.1038/sj.bjc.6604125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Massey SC, Rockne RC, Hawkins-Daarud A, Gallaher J, Anderson ARA, Canoll P, Swanson KR. Simulating PDGF-Driven Glioma Growth and Invasion in an Anatomically Accurate Brain Domain. Bull. Math. Biol. 2018;80:1292–1309. doi: 10.1007/s11538-017-0312-3. [DOI] [PubMed] [Google Scholar]
- 15.Johnston SK. et al.: ENvironmental Dynamics Underlying Responsive Extreme Survivors (ENDURES) of Glioblastoma: a Multi-disciplinary Team-based. Multifactorial Analytical Approach. bioRxiv 461236, 2018. doi:10.1101/461236 [DOI] [PMC free article] [PubMed]
- 16.Barnholtz-Sloan JS, Swanson KR: Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci. Transl. Med, 2019 [DOI] [PMC free article] [PubMed]
- 17.Massey SC et al.: Image-based metric of invasiveness predicts response to adjuvant temozolomide for primary glioblastoma. bioRxiv 509281, 2019. doi:10.1101/509281 [DOI] [PMC free article] [PubMed]
- 18.Brown RW et al.: Magnetic Resonance Imaging: Physical Principles and Sequence Design. (John Wiley & Sons, 2014
- 19.Cha S. Neuroimaging in neuro-oncology. Neurotherapeutics. 2009;6:465–477. doi: 10.1016/j.nurt.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Armstrong TS, Cohen MZ, Weinberg J, Gilbert MR. Imaging techniques in neuro-oncology. Semin. Oncol. Nurs. 2004;20:231–239. doi: 10.1016/S0749-2081(04)00087-7. [DOI] [PubMed] [Google Scholar]
- 21.Krizhevsky A, Sutskever I, Hinton GE: ImageNet classification with deep convolutional neural networks. in Advances in Neural Information Processing Systems 25 (eds. Pereira, F., Burges, C. J. C., Bottou, L. & Weinberger, K. Q.) 1097–1105 (Curran Associates, Inc., 2012
- 22.Mohsen H, El-Dahshan E-SA, El-Horbaty E-SM, Salem A-BM. Classification using deep learning neural networks for brain tumors. Future Computing and Informatics Journal. 2018;3:68–71. doi: 10.1016/j.fcij.2017.12.001. [DOI] [Google Scholar]
- 23.Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. Journal of Medical Imaging. 2016;3:034501. doi: 10.1117/1.JMI.3.3.034501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y-JY-J, Hua K-L, Hsu C-H, Cheng W-H, Hidayati SC: Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets and Therapy, 2015. 10.2147/ott.s80733 [DOI] [PMC free article] [PubMed]
- 25.Simonyan K, Zisserman A: Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv [cs.CV], 2014
- 26.Chollet F, Others. Keras. 2015
- 27.Abadi, M. et al.: Tensorflow: A system for large-scale machine learning. in 12th ${USENIX} Symposium on Operating Systems Design and Implementation ({OSDI}$ 16) 265–283, 2016
- 28.Kingma DP, Adam BJ: A method for stochastic optimization. arXiv [cs.LG], 2014
- 29.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 30.Gong, Y., Jia, Y., Leung, T., Toshev, A. & Ioffe, S. Deep convolutional ranking for multilabel image annotation. arXiv [cs.CV], 2013
- 31.Wu, F., Wang Z., Zhang Z., Yang Y., Luo J., Zhu W., Zhuang Y. Weakly Semi-Supervised Deep Learning for Multi-Label Image Annotation. IEEE Transactions on Big Data 1, 109–122 (2015), 109, 122.
- 32.Wu J, Yu Y, Huang C, Yu K: Deep multiple instance learning for image classification and auto-annotation. in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 3460–3469, 2015
- 33.Murthy VN, Maji S, Manmatha R: Automatic Image Annotation Using Deep Learning Representations. in Proceedings of the 5th ACM on International Conference on Multimedia Retrieval 603–606 (ACM), 2015.
- 34.Ojha U, Adhikari U, Singh DK: Image annotation using deep learning: A review. 2017 International Conference on Intelligent Computing and Control (I2C2), 2017. 10.1109/i2c2.2017.8321819
- 35.Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, Lin MD, Duncan JS, Weinreb JC, Chapiro J, Letzen B. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur. Radiol. 2019;29:3338–3347. doi: 10.1007/s00330-019-06205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hermessi, H., Mourali, O. & Zagrouba, E. Deep feature learning for soft tissue sarcoma classification in MR images via transfer learning. Expert Systems with Applications 120, (2019), 116, 127.
- 37.Margeta J, Criminisi A, Cabrera Lozoya R, Lee DC, Ayache N. Fine-tuned convolutional neural nets for cardiac MRI acquisition plane recognition. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization. 2017;5:339–349. [Google Scholar]