Abstract
Peripheral blood smear analysis plays a vital role in diagnosing many diseases including cancer. Leukemia is a type of cancer which begins in bone marrow and results in increased number of white blood cells in peripheral blood. Unusual variations in appearance of white blood cells indicate leukemia. In this paper, an automated method for detection of leukemia using image processing approach is proposed. In the present study, 1159 images of different brightness levels and color shades were acquired from Leishman stained peripheral blood smears. SVM classifier was used for classification of white blood cells into normal and abnormal, and also for detection of leukemic WBCs from the abnormal class. Classification of the normal white blood cells into five sub-types was performed using NN classifier. Overall classification accuracy of 98.8% was obtained using the combination of NN and SVM.
Keywords: Peripheral blood smear analysis, Leukemia, Computer aided detection, Classification
Introduction
Peripheral blood smear (PBS) analysis is a gold standard technique used for diagnosis of many diseases including cancer. Diagnosis in laboratories involves various steps which are carried out by experts. There are three types of blood cells namely red blood cells (RBCs), platelets, and white blood cells (WBCs). WBCs are of five types namely lymphocytes, monocytes, neutrophils, eosinophils, and basophils. Change in count and/or morphological changes of WBCs indicate abnormal health status and help in diagnosing many diseases. Leukemia is a type of cancer which begins in bone marrow and results in increased number of WBCs in peripheral blood. Leukemia represents about 3.5% of all new cancer cases in the USA with an estimation of 60,300 cases in the year 2018 [1] and 399,967 people living with or in remission from leukemia in the USA in 2019 [2]. In the year 2017, the prevalence of leukemia was about 2.5% worldwide as per World Cancer Research Fund International [3]. Leukemic cells grow in uncontrolled manner and may not mature like normal cells or may not attain maturity level. Because of immaturity, these cells are unable to function properly like normal cells. The non-specific nature of leukemic cells, often leads to wrong diagnosis. Therefore, careful observation of stained blood smear or bone marrow aspiration must be considered for detection and diagnosis of leukemia effectively. Methods such as fluorescence in situ hybridization, immunophenotyping, cytogenic analysis, and cytochemistry are being used to detect leukemia. These methods are expensive and are not available in many medical centers. Also, it is time consuming and requires manual interpretation.The result of manual evaluation depends on skill and experience of the pathologist. The subjectivity in laboratory results can also be due to lengthy procedure and massive amount of samples [4, 5]. Therefore, there is a need for cost-effective, automatic, and robust technique to detect leukemia. Automatic microscopic evaluation of blood samples helps pathologists to speed-up and enhance the accuracy of the evaluation process [6]. Also, computer-aided diagnosis system provides objective results [7].
Many automated methods have been proposed to detect leukemia using image processing approach. However, automated WBC segmentation still remains a challenging task due to its complex biological nature, inconsistent staining procedure, and illumination source [8, 9]. Foran et al. [10] used a combination of nonparametric clustering method and CIE LUV color space representation for segmentation of WBCs, thereby reporting a correct classification rate around 83%. In order to detect acute myeloid leukemia (AML), Agaian et al. [11] employed a combination of k-means clustering and CIE LAB color space representation for detection of nuclei. The study reported extraction of shape, color, and texture features which were used to train a SVM classifier which provided classification accuracy around 98%. Moradi et al. [12] used k-means clustering and SVM for detection and classification of acute lymphoblastic leukemia (ALL). They reported an average classification accuracy around 97%. Neoh et al. [13] presented a decision support system for detection of leukemia using marker-controlled watershed segmentation for detection of WBCs and a clustering algorithm with stimulating discriminant measures for segmentation of WBCs into nucleus and cytoplasm. Several classifiers namely multi-layer perceptron (MLP), SVM and ensembles with diverse weighting combination methods were evaluated for classification of WBCs into normal and leukemic WBCs and they achieved an average accuracy of 96.7%. An automated system to diagnose acute leukemia was proposed by Reta et al. [14] by using a combination of CIE LAB color space representation and k-means clustering for segmentation of WBCs. Further, overlapped cells were separated using linear interpolation method which yielded an approximate segmentation accuracy of 95%. A binary classifier with majority vote criteria was considered for classification of WBCs into normal and leukemic type which provided an overall classification accuracy of 95%. A similar study of detection and classification of acute leukemia using k-means clustering was reported in [15, 16] wherein, neural network (NN) classifier was employed by Negm et al. [15] which yielded classification accuracy around 97%. Patel et al. [16] used histogram equalization and Zack’s thresholding along with k-means clustering for extraction of WBCs. SVM classifier was trained for detection of leukemic WBCs which resulted in average detection accuracy around 94%. Belacel et al. [17] proposed a method for classification of sub-types of leukemia using supervised machine learning method by considering 108 images for training and 83 images for testing. They obtained an average correct classification rate of 96.4%.
In order to design a decision support system for diagnosis of ALL using microscopic images, Zeinab et al. [18] used exponential intuitionistic fuzzy divergence and Zack’s thresholding methods for detection of nuclei and entire region of WBCs, marker-controlled watershed segmentation for separation of overlapped cells, and ensemble classifier for detection of leukemic WBCs. The combination of methods used resulted in classification accuracy around 97%. A similar study was reported by Srisukkham et al. [19]. Marker-controlled watershed segmentation was used for detection of WBCs and stimulating discriminant measure for segmentation of WBCs into nucleus and cytoplasm. Bare-bone particle swarm optimization technique was utilized for selection of discriminant features. SVM and NN classifiers were evaluated using the discriminant features, thereby achieving classification accuracy around 96% using NN. Mohapatra et al. [20] also proposed a method for detection of ALL using PBS images. Shadowed C-means clustering was used for segmentation of WBCs. Several classifiers namely Naive Bayes (NB), K Nearest Neighbor (KNN), MLP, SVM, and ensemble of classifiers were evaluated for detection of ALL. They reported an average classifier sensitivity around 90% using ensemble of classifiers.
Fatichah [21] employed fuzzy morphology-based segmentation method and fuzzy decision tree-based classifier for classification of sub-types of leukemia, thereby achieving an average classification accuracy around 84%. Jair et al. [22] proposed a method for classification of acute leukemia. Markov random field-based method and k-means algorithm were a part of WBC segmentation and particle swarm optimization technique was used for selection of best classifier. They obtained an average classification accuracy of 97.68%.
Azevedo et al. [23] used a combination of wavelet transform, fuzzy 2-partition entropy, genetic algorithm, and morphological operations for segmentation of WBCs considering images of chronic lymphoblastic leukemia (CLL) cases, thereby reporting an overall Dice score of 0.89.
Classification of WBCs into normal and abnormal was presented by Osowski et al. [24] using bone marrow images. Classification mean error rate of 19.5% was achieved using genetic algorithm-based SVM classifier. Jie et al. [25] employed k-means clustering, hidden-Markov random field, expectation maximization algorithm for segmentation of WBCs considering 61 bone marrow aspirate images including normal WBCs and blasts, thereby achieving average segmentation accuracy between 96 and 98%.
Nikitaev et al. [26] reported classification of WBCs into normal, lymphoblast, myeloblast, and monoblast using wavelet analysis which resulted in recognition rate of 82%. Jyoti et al. [27] proposed a method for classification of WBCs into ALL, AML, and normal cells using 331 features extracted from 420 microscopic images. Histogram equalization, filtering, Otsu’s thresholding, and morphological operations were used for segmentation of WBCs. Genetic algorithm was used for feature selection. The selected features were trained using MLP for classification which resulted in classification accuracy around 97%. Summary of the prior art is listed in Table 1. The percentage accuracy given in the table is for detection of leukemia by various research groups.
Table 1.
Summary of prior art
| Author, year | Method | No. of images | Accuracy (%) |
|---|---|---|---|
| Foran et al. [10], 2000 | Nonparametric clustering, Mahalanobis distance | 261 | 83 |
| Belacel et al. [17], 2001 | PROAFTN | 191 | 96 |
| Jair et al. [22], 2012 | Ensemble particle swarm model selection | 633 | 94 |
| Mohapatra et al. [20], 2014 | c-means clustering, NB, KNN, MLP, SVM, and ensemble classifier | − | 90 |
| Agaian et al. [11], 2014 | k-means clustering, SVM | 80 | 98 |
| Fatichah [21], 2015 | Fuzzy morphology, decision tree | 120 | 84 |
| Moradi et al. [12], 2015 | k-means, SVM | 312 | 96 |
| Neoh et al. [13], 2015 | Watershed algorithm, clustering method, MLP, SVM, ensemble classifier | 180 | 96 |
| Reta et al. [14], 2015 | k-means, binary classifier | 633 | 95 |
| Patel et al. [16], 2015 | k-means, SVM | 180 | 94 |
| Nikitaev et al. [26], 2016 | Wavelet analysis | 1289 | 82 |
| Negm et al. [15], 2017 | k-means, NN | 624 | 97 |
| Srisukkham et al. [19], 2017 | Marker controlled watershed, SVM, NN | 180 | 93 |
| Jyoti et al. [27], 2017 | Otsu’s thresholding, MLP | 420 | 97 |
| Zeinab et al. [18], 2018 | Marker-controlled watershed, ensemble classifier | 368 | 97 |
Although significant contributions have been made by previous researchers, there exists a need for optimal solution to handle variations present in microscopic images. Also, there is a need for an automated system which can reduce the work load of pathologists by achieving 100% confidence factor in classifying WBCs into normal and abnormal. In the proposed study though the aim is to detect leukemia, the method also classifies the WBCs into normal and abnormal, thus eliminating the need for review of normal cases by pathologists.
The contributions of this paper are summarized as follows.
Automated threshold calculation for detection of nuclei which is robust to brightness and color variations present in PBS images.
Automated cropping of original images using location of the nuclei.
Automated accurate detection of WBCs from the cropped images.
Classification of WBCs into normal and abnormal.
Detection of leukemic WBCs from the abnormal class.
Materials and Methods
This section is divided into four sections namely data collection, detection of WBCs, feature extraction, and classification. The “Data Collection” section describes about data collection. The details of the proposed method for WBC detection are described in the “Segmentation of WBC” section. The details of feature extraction and classification are given in the “Feature Extraction” and “Classification” sections respectively.
Data Collection
Images were acquired using OLYMPUS CX51 microscope under × 100 magnification with 1600 × 1200 resolution from hematology laboratory in Kasturba Medical College (KMC) hospital, Manipal, India. We collected 1159 PBS images which consists of 170 lymphocytes, 109 monocytes, 295 neutrophils, 156 eosinophils, 81 basophils, and 607 abnormal WBCs. Abnormal WBCs include reactive lymphocytes, degenerated cells, myelocytes, and leukemic WBCs which are called as blasts. The subset of abnormal WBCs consists of 483 leukemic and 124 nonleukemic WBCs. For classification of the WBCs, 80% of each type was considered for training and the remaining 20% was considered for testing.
We have acquired images from archive data from KMC which is a teaching hospital for which written consent was taken to be used for research purposes.
To develop an algorithm which is robust to variations that generally occur in microscopic images, we have purposefully collected images from different laboratories of the hospital. Sample preparation was carried out by different people with different levels of expertise. A few sample images of the dataset are shown in Fig. 1. It can be observed from the figure that the dataset consists of images of different brightness levels and color shades.
Fig. 1.
Sample images of the dataset
Segmentation of WBC
In this section, the method used for WBC detection is described. Detection of region of nuclei was considered as first step in the proposed method. Information of the nuclei was used for detection of WBCs. Thresholding, morphological operations, and area filter were used for detection of nuclei. Active contours method was considered for detection of WBCs. The block diagram of the proposed method is shown in Fig. 2. The input image is an original image of size 120 × 1600. It consists of RBCs, platelets, and WBCs as shown in the figure.
Fig. 2.
Block diagram of WBC detection
Detection of Nuclei
The block diagram representation of nuclei detection method is shown in Fig. 2a–d. Input image is a color image. G component of original image was used for further processing. It can be observed from Fig. 2a that the region of nuclei appears dark compared with other regions in the input image. To address the problem of brightness and color shade variations, a new approach was considered to compute an appropriate threshold value for detection of region of nuclei. Rosin’s thresholding method [28] and histogram-based thresholding method which was applied on the resultant images of Rosin’s method were used for selection of optimal threshold value. The combination of these two methods provides the threshold value based on the image brightness level.
Rosin’s method is based on the intensity histogram of the input image. In the proposed method, histogram of bin size 150 was considered because the region of nuclei appears dark with the intensity value below 150 as shown in Fig 3a. The maximum peak max-peak was obtained for the gray level equal to 145 which is defined as P1 and the region corresponding to P1 is shown in Fig. 3a. This method draws a straight line from maximum peak position P1 to the last bin P2 which is 150 in the proposed method. It computes the perpendicular distance from the straight line to the gray levels between P1 and P2. The gray level value which corresponds to maximum perpendicular distance is considered as threshold value thresh. The threshold value varied between 145 to 149 for the images in the dataset. The result of Rosin’s thresholding is a grayscale image obtained by replacing the pixel values above the threshold value thresh to 255 as shown in Fig. 3b. The results of Rosin’s thresholding method were approximate regions of nuclei.
Fig. 3.
Image with different regions for threshold selection. a Regions with different pixel values. b Thresholded image
To obtain an accurate threshold value, the histogram of the image obtained using Rosin’s thresholding method was considered with number of bins equal to thresh. Maximum pixel count pixelCount1 and corresponding gray level grayLevel1 were computed from the histogram but by excluding gray level whose value was equal to thresh. The grayLevel1 was considered as threshold value which was used to detect region of nuclei. The grayLevel1 was applied on original G component image IG to obtain region of nuclei. The threshold value varied between 56 to 85 for the images in the dataset. The algorithm steps for selection of suitable threshold value are as follows.
Apply Rosin’s thresholding method on G component IG of original image with bin size of 150 to obtain a threshold value thresh
Obtain Ithresh by replacing pixel values >= thresh to 255 (Fig. 2b)
Obtain pixel count pixelCount and corresponding gray level grayLevel of Ithresh with number of bins = thresh
Eliminate pixelCount and grayLevel corresponding to gray level =thresh to obtain pixelCount1 and grayLevel1
Determine the gray level grayLevel1 with maximum pixelCount1
Select the grayLevel1 as threshold value to obtain binary image Ibw (Fig. 2c). If grayLevel1 < 50, use grayLevel1 = 70 as threshold value to obtain Ibw
Post-processing was considered in the proposed method to obtain the accurate region of nuclei. This includes area filling, area filtering, and morphological closing. Area filter was used to remove platelets and other unwanted regions in the binary image. The objects with area below 5000 pixels were removed from the binary image. The closing operation was considered based on the areas of nuclei. The areas of nuclei between 6000 pixels and 25000 pixels were subjected to closing operation using disk-shaped structuring element of radius 14 and the areas above 25000 pixels were closed using disk-shaped structuring element of radius 8. Area-based morphological closing operation was considered to selectively apply closing of lobulated nuclei.
Detection of WBC
Detection of WBC is a challenging task due to its cytoplasm region which appears different for different types of WBCs. Also, the variation can be observed in shape and size of WBCs. Active contours method which is robust to any shape was utilized for detection of WBCs in the proposed method. The block diagram of WBC detection is shown in Fig. 2e–g. Active contours work on the basis of energy minimization technique. It is a flexible curve which can adapt to the required object boundaries. It is an iterative process where user has to specify an initial contour and number of iterations [29]. Michael et al. [30] introduced active contours algorithm with three energy functions as given in Eq. (1).
| 1 |
where, v(s) = (x(s),y(s)) is the contour of an object, Eint is internal energy, Eimg is image force, and Econ is external constraint force.
Internal energy is sum of elasticity and stiffness, defined such that it is minimum at object boundary. It helps in smoothing the curve. Image energy is calculated from image pixel values, so that it takes small values at the boundaries. The third term in Eq. (1) is a constraint which controls external force and also helps in placing the contour near the desired local minimum. The shape and size of WBCs vary depending on their type. Therefore, we used active contours to detect and extract WBCs which is robust to shape and size variations.
Several pre-processing steps such as cropping, color-transfer method, and background removal were used before applying the active contours. Cropping the original images to obtain sub-images was considered to obtain images with single WBC which helped to overcome the problem of overlapping cells and also active contour method converges in less number of iterations with images of small size. Automated image cropping was carried out using the location of nuclei in the images. Color transfer method as explained by Congcong et al. [31] was used for the purpose of color normalization. This method transforms the color characteristics of the input image to that of color characteristics of the considered template image. Hence, use of this method eliminates the color variations present in the dataset. Background removal was considered to effectively use the energy minimization technique of active contours so that it converges at the exact boundary of WBCs. The pre-processing steps of the proposed method are as follows.
Read original image IRGB
Read binary representation of detected nuclei Ibw
- Repeat the steps (a)–(d) for every nuclei present in the image
- (a) Determine the corners of the bounding box of the nucleus
- (b) Subtract a constant value 110 from the upper corners to obtain upper
- (c) Add a constant value 110 to the lower corners to obtain lower
- (d) Crop the image IRGB and Ibw using coordinate points upper and lower to obtain IcroppedRGB and Icroppedbw respectively
Apply color transfer method on IcroppedRGB to obtain Itrans
Convert Itrans to grayscale image to obtain Igray
Replace the pixel values of Igray above 218 by ‘zero’ (Fig. 2e)
It can be observed from Fig. 2e that pre-processing eliminated the background without loss of boundary of WBCs.
Mask for active contours was generated from the nucleus. Geometrical features namely area and circularity of the nuclei were computed. These nuclei were approximately classified as segmented nucleus, small or large nucleus. Convex hull area of nucleus was dilated depending on approximate classification of nucleus as depicted in Fig. 2f. Disk-shaped structuring elements of sizes between 20 and 45 were used to obtain appropriate masks to initiate the active contours. The mask generation steps are shown in Fig. 4. Image (a) in the figure is binary representation of the nucleus, image (b) is the rotated nucleus. image (c) represents addition of the nucleus and rotated nucleus, and image (d) is the dilated nucleus. The steps for generation of mask are as follows.
Read binary representation of cropped nucleus Icroppedbw (Fig. 4a)
Obtain area and circularity of the nucleus
Rotate Icroppedbw to 900 to obtain I90 (Fig. 4b)
Add Icroppedbw and I90 to obtain Iadd (Fig. 4c)
- Dilate nuclei (Iadd) using disk shaped structuring element to obtain Imask (Fig. 4d)
- For area < 60000 and circularity > 0.8, radius of the disk = 20
- For area < 50000 and circularity < 0.7, radius of the disk = 30
- For other cases, radius of the disk = 45
Use Imask as mask on Igray to initiate the contours (Fig. 2g)
Fig. 4.
Generation of mask for active contours. a Nucleus. b Rotated nucleus. c Added nucleus. d Dilated nucleus
The combination of dilated convex hull of nucleus and active contours detects WBCs correctly irrespective of lighting conditions of images. The rotation and addition of nucleus as mentioned in step 3 helped in proper detection of WBC in case nucleus is not centered at the cytoplasm region.
Feature Extraction
Feature extraction plays an important role in classification of WBCs. Features from the nuclei, its cytoplasm, and WBCs were extracted. Features namely size, shape, color, and texture vary depending on the type of WBCs. Segmentation of WBC into nucleus and cytoplasm is shown in Fig. 5. It can be observed from the figure that nucleus of neutrophil and eosinophil is lobulated. Color and texture variations can be observed in the cytoplasm region. Also, texture variation can be observed in the basophil and abnormal WBCs. Hence, shape, color, and texture features were considered for the classification in the present study.
Fig. 5.
Segmentation of WBC into nucleus and cytoplasm; L, lymphocyte; M, monocyte; N, neutrophil; E, eosinophil; Ab, abnormal WBC; B, basophil
Shape Features
Shape features such as area, perimeter, circularity, convexity, solidity, and ratio of nucleus area to RBC area (Nucleus-RBC ratio) were extracted from the nuclei of WBCs as given in Table 2. Also, area of WBCs and area of nucleus to area of WBC ratio (NC ratio) were computed. All these shape features were extracted from binary representation of the nuclei and WBCs.
Table 2.
Shape features used in the present study
| Feature | Relation |
|---|---|
| Area | Total number of pixels |
| Perimeter | Path length that surrounds area |
| Circularity | Perimeter2 / 4*pi*area |
| Convexity | Perimeter of convexhull / perimeter of nucleus |
| Solidity | Area of nucleus / area of convexhull |
| Nucleus-RBC ratio | Area of nucleus / area of RBC |
| NC Ratio | Area of nucleus / area of WBC |
In addition to all these shape features, number of dents were computed from the nuclei using FFT approach [32, 33] to differentiate between degenerated WBCs and blasts. The method detects number of lobes which can be used to differentiate types of WBCs [34].
Color Features
Cytoplasm of WBCs varies in color depending on the type as shown in Fig. 5. Also nuclei of abnormal WBCs show color variations. Hence, mean and variance of R, G, and B components of RGB image; H, S, and V components of HSV color space representation; and L, A, and B components of CIE LAB color space representation were extracted from the nuclei and the cytoplasm of WBCs.
Texture Features
It can be observed from Fig. 5 that the texture variation can be observed in both nucleus and cytoplasm. Hence, texture features from both the nucleus and cytoplasm of WBCs were considered in the present study. Histogram-based features, Spatial Gray Level Dependence Matrix (SGLDM) features, Statistical Feature Matrix (SFM) features, and Hu moments of order 3 were extracted. The considered various texture features are given in Table 3.
Table 3.
Texture features used in the present study
| Type | Features |
|---|---|
| Histogram based | Mean, variance, skewness, kurtosis, energy |
| SGLDM | Angular second moment, contrast, correlation, variance, homogeneity, sum average, sum variance, sum entropy, entropy, difference variance, difference entropy, information measures of correlation |
| SFM | Coarseness, contrast, periodicity, roughness |
| Hu moments | 7 central moments |
Classification
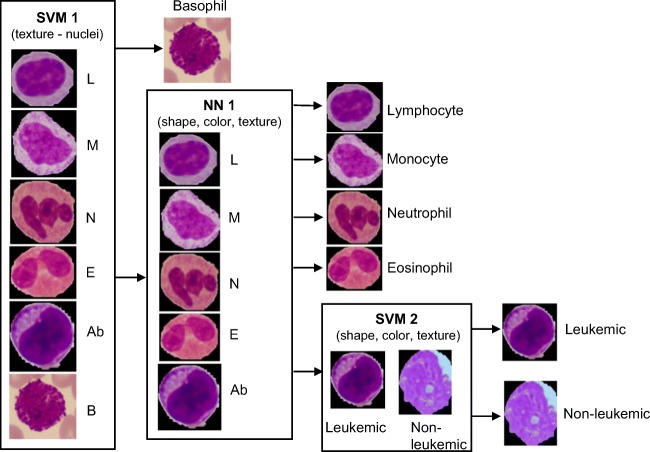
In this phase, main aim of the proposed system was to discriminate leukemic WBCs. Also, classification of the normal WBCs into its sub-types was attempted in the present study. A combination of SVM and NN classifiers were used for classification of the WBCs. The classification steps are shown in Fig. 6. Classification of WBCs is a 3-step process in the proposed method. Basophils consist of numerous dark granules on the nucleus as shown in Fig. 6.
Fig. 6.
Classification steps of the proposed method
Nucleus of basophil is hard to segment out from the region of cytoplasm. Hence, basophils were detected using texture features of nuclei derived from SGLDM. In the second step, NN was used to classify the WBCs into lymphocyte, monocytes, neutrophils, eosinophils, and abnormal WBCs. The abnormal class consists of blasts, reactive lymphocytes, degenerated cells, and myelocytes. In the third step, the features extracted from that of nuclei and cytoplasm were used to train the SVM classifier to detect blasts.
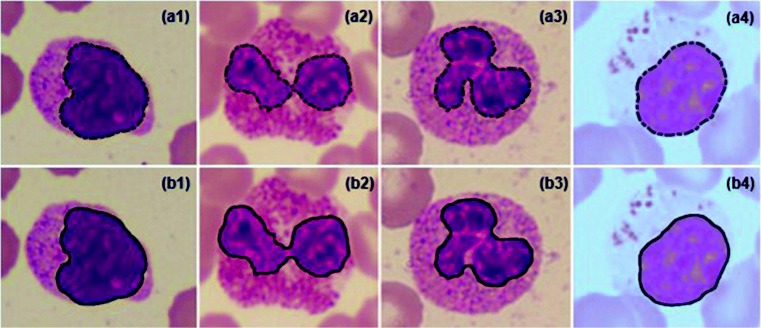
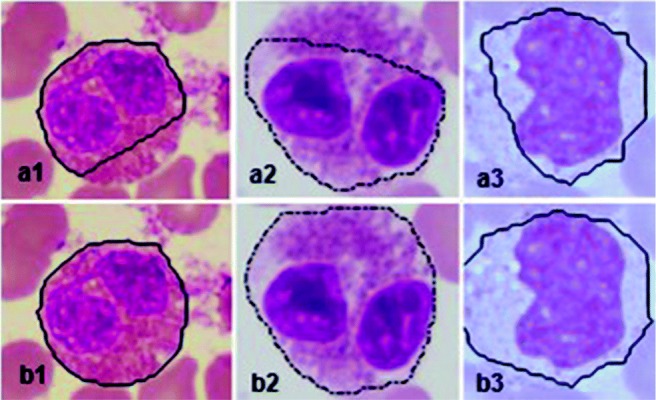
The texture features of the degenerated WBCs overlap with that of blasts in many cases. Hence, elimination of degenerated WBCs was considered before detection of the blasts from the abnormal class. Removal of degenerated WBCs was based on the number of dents and circularity of nuclei. The sample images of degenerated WBCs and blasts are shown in Fig. 7. It can be observed from the figure that nuclei of blasts are more circular compared with the degenerated WBCs. Also, boundaries of nuclei of degenerated WBCs are not continuous and several dents can be observed. Cytoplasm region of the degenerated WBC is so pale that it is indistinguishable from the background as shown in the figure. In the proposed method, WBCs with circularity value less than 0.65 and number of dents more than 3 were considered as degenerated WBCs. This step was considered to avoid the degenerated WBCs getting identified as blasts.
Fig. 7.
Sample images of degenerated WBCs and blasts; (a1–a4) degenerated WBCs, (b1–b4) blasts
In the proposed classification, SVM with polynomial kernel of order 3 and box constraint of 1.2 was used for detection of basophils. Polynomial kernel of order 2 and box constraint of 0.3 was used for detection of blasts. To test the performance of the SVM classifier, “hold-out validation” with 80% of the data was considered for training and the rest of the dataset was considered for testing.
Scaled conjugate gradient back-propagation method was used to train NN with hidden layer size of 7. The network was trained using 80% of the data and the rest of the dataset was used for testing the network.
Results and Discussion
This section provides the results of the proposed method. Results of segmentation of WBCs are given in the “Segmentation Results” section. The classification results are given in the “Results of Classification” section.
Segmentation Results
Segmentation in the proposed method is a two-step process. The first corresponds to the detection of nuclei and the second step corresponds to the detection of WBCs. The “Results of Nuclei Detection” and “Results of WBC Detection” sections describe the results of nuclei detection and WBC detection respectively. Performance of the proposed segmentation method was carried out by computing the performance measures namely Dice score, accuracy, precision, and recall rates. The mathematical formulation for the performance measures is given in Table 4.
Table 4.
Mathematical relations for the performance measures
| Performance measure | Relation |
|---|---|
| Dice score | 2(SG) / (S+G) |
| Accuracy | (TP + TN)/ (TP + TN + FP + FN) |
| Precision rate | TP / (TP + FP) |
| Recall rate | TP / (TP + FN) |
“S” in Table 4 is segmented region using the proposed method, “G” is the region obtained from the ground truth, TP is true positive, TN is true negative, FP is false positive, and FN is false negative.
Results of Nuclei Detection
Performance of the proposed nuclei detection method was evaluated by comparing the results of the proposed method with ground truth obtained from an expert. A few sample images showing nuclei detection results of the proposed method along with expert annotated images are shown in Fig. 8. It can be observed from the figure that results of the proposed method match well with the expert annotated images for all the images, but a small variation can be observed in case of lobulated nucleus (Fig. 8(b2) and (b3)). This is due to the morphological closing operation used in the proposed method.
Fig. 8.
Results of nuclei detection method; images (a1)–(a4): expert annotated images, images (b1)–(b4): results of the proposed method
The mean and standard deviation (STD) values of the performance measures are tabulated in Table 5. The overall accuracy of the nuclei detection method is 99%. This is due to the optimal threshold value computed in the proposed method. We computed the appropriate threshold value by eliminating the background using Rosin’s threshold method and using the histogram-based thresholding method on the background removed image. The combination of Rosin’s method and histogram-based thresholding resulted in an optimal value which can be used for detection of region of nuclei with brightness and color shade variations. The proposed method is robust to manage the variations present in microscopic images.
Table 5.
Performance measures of segmentation of WBCs
| Measures | Nuclei detection | WBC detection | ||
|---|---|---|---|---|
| Mean | STD | Mean | STD | |
| Dice | 0.97 | 0.03 | 0.98 | 0.03 |
| Accuracy | 0.99 | 0.0025 | 0.99 | 0.002 |
| Precision | 0.97 | 0.04 | 0.98 | 0.03 |
| Recall | 0.97 | 0.04 | 0.98 | 0.04 |
Results of WBC Detection
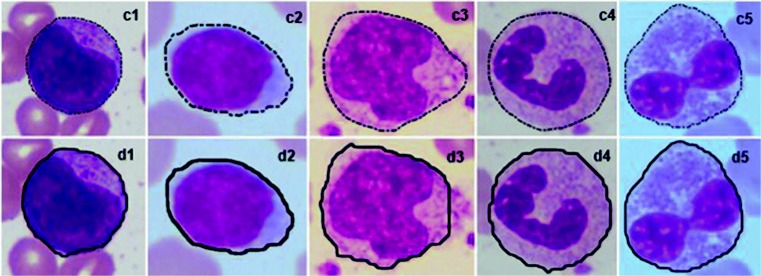
The performance of the proposed WBC detection method was evaluated by comparing the results with expert annotated images. A few sample images are shown in Fig. 9. It can be observed from the figure that results of the proposed method for detection of WBCs match well with the ground truth images. This could be due to the adaptive approach considered for the design of mask. We used dilated nuclei as mask to initiate the contours. Also, we rotated the nucleus and added the rotated nucleus to original nucleus before applying dilation. This helped to detect WBCs even when the nucleus is not located at the center of cytoplasm.
Fig. 9.
Results of WBC detection method; images –(c5): expert annotated images, images (d1)–(d5): results of the proposed method
The effect of mask used in the proposed method is shown Fig. 10. Dilated nucleus without rotation results in under-detection of the WBC whereas use of rotation and addition operations results in appropriate detection of the WBC. The proposed method of mask generation overcomes the problem of under-detection. The results of the proposed WBC detection method were evaluated by computing performance measures as listed in Table 5. The overall Dice score of 0.98 was obtained for WBC detection. This indicates that the proposed method can efficiently detect WBCs in PBS images. Cropping the images to obtain sub-images was performed to avoid touching and overlapping of WBCs during the application of active contours. Also, cropping the image reduced the time requirement of active contours which is an iterative technique. Dilation of the mask based on area and circularity of the nucleus was considered in the proposed method. Use of variable mask size reduced the number of iterations by using small mask size for small WBCs such as lymphocytes which converges in less number of iterations. Color-transfer method was employed to eliminate the color variations present in the dataset. Thus, the combination of pre-processing and active contours detected WBCs even in presence of shape, size, and color variations in the images.
Fig. 10.
Effect of mask used in active contours; image a1–a3: result of active contours without rotation and addition of nucleus, b1–b3: result of active contours with rotation and addition
Results of Classification
The main aim of classification was to detect leukemic WBCs in PBS images. Also, classification of the normal WBCs into its sub-types namely lymphocytes, monocytes, neutrophils, eosinophils, and basophils was considered in the proposed method. A combination of SVM and NN classifiers was used to achieve the goal of classification. The results of the classifier were evaluated by computing accuracy, sensitivity, and specificity. A step-by-step classification was employed which is as shown in Fig. 6.
In the first step of classification, basophils were detected using the texture features extracted from the nuclei. The texture features were derived from SGLDM. Basophils are less common in peripheral smear and the nucleus is completely covered by dark granules. The proposed nuclei detection method detected entire region of basophil. This is due to the fact that it is hard to distinguish between nucleus and cytoplasm in case of basophils. Hence, detection of basophils was considered as a first step and eliminated the basophils in further processing. Result of “SVM 1” classifier is listed in Table 6. It can be observed from the table that training accuracy of 100% was obtained and almost equal accuracy was obtained for testing with one basophil misclassified into the “others” class. This resulted in overall F1-score of 98.5%. Also, positive predictive value (PPV) and negative predictive value (NPV) of 0.996 and 1 were obtained for detection of basophils.
Table 6.
Performance of “SVM 1” for basophil detection
| Training | Testing | |||
|---|---|---|---|---|
| Basophil | Others | Basophil | Others | |
| Basophil | 65 | 0 | 15 | 0 |
| Others | 0 | 1070 | 1 | 267 |
In the second step, classification of WBCs into lymphocyte, monocytes, neutrophils, eosinophils, and abnormal WBCs was considered using the NN classifier. The training and testing performances in terms of accuracy, sensitivity, and specificity are listed in Table 7.
Table 7.
Training and testing performance of “NN” classifier
| Performance measures | Training | Testing |
|---|---|---|
| Accuracy (%) | 99.6 | 97.8 |
| Sensitivity (%) | 99.6 | 98.6 |
| Specificity (%) | 99.3 | 97.2 |
In the second step, overall accuracy of 99.5% was obtained. The average values of performance of the NN classifier in terms of accuracy, sensitivity, specificity, and F1-score for the types of WBCs are listed in Table 8. Overall accuracy of 99.5% was obtained. It can be observed from the table that detection of abnormal WBCs is 100% accurate. Such a classifier which can classify WBCs into normal and abnormal with 100% accuracy helps to reduce the workload of pathologists. Abnormal cases can be sent for expert opinion for further tests and hence need not wait for long time. An automated system which classifies WBCs into normal and abnormal accurately can be used for practical application so that normal cases are not misled and abnormal cases are not missed for further evaluation.
Table 8.
Performance of “NN 1” classifier
| WBC type | Accuracy (%) | Sensitivity (%) | Specificity (%) | F1-score (%) |
|---|---|---|---|---|
| Lymphocyte | 100 | 100 | 100 | 100 |
| Monocyte | 99.8 | 100 | 99.8 | 99.1 |
| Neutrophil | 99.5 | 98.6 | 98 | 99 |
| Eosinophil | 99.7 | 98.7 | 99.8 | 98.7 |
| Abnormal | 100 | 100 | 100 | 100 |
Classification of normal WBCs into its sub-types helps in counting number of each type of WBC which can be used for diagnosing count-related diseases such as neutropenia, eosinophila, and basophilia.
The abnormal class consists of leukemic WBCs, reactive lymphocytes, degenerated cells, and myelocytes. The third step consists of detection of leukemic WBCs (blasts) from the abnormal class. The degenerated WBCs were eliminated at this stage to bring down the misclassification rate. Out of 90 degenerated WBCs, 84 were accurately detected using the shape features and they were excluded from the abnormal class for further processing. This step was performed after the classification of WBCs into normal and abnormal because, overlap of a few shape and texture features can be observed between degenerated WBCs and monocytes. The shape and size of nuclei of these two types of WBCs are similar.
The leukemic WBCs appear large and they consist of nucleoli which are circular opening in region of nucleus. Shape features of nuclei, NC ratio, texture features of both nuclei and cytoplasm, and color features of WBC and area of WBCs were considered to train “SVM 2.” The results of leukemic WBC detection are given in Table 9. This classifier is used to identify leukemic WBCs from the cells classified as abnormal. We tried two approaches for identification of leukemic WBCs. In the first approach, we aimed at high accuracy and obtained an average accuracy of 98% with specificity around 94%. In the second approach, we aimed for high specificity and we achieved 100% specificity so as to identify leukemic WBCs accurately. Sensitivity of 69% was a trade-off to achieve 100% specificity. Such a classifier which can identify the leukemic WBCs with 100% specificity would help in decision making. Hence, this classifier has been specifically designed for detection of leukemic WBCs. Experts need not look into the detected leukemic cases, which can be directly sent for further confirmation test such as bone marrow biopsy. This reduces the burden of pathologists and also helps in providing quick results.
Table 9.
Overall performance of “SVM 2” for detection of leukemic WBCs
| Performance measure | Value (%) |
|---|---|
| Accuracy | 92.8 |
| Sensitivity | 69 |
| Specificity | 100 |
| F1-score | 89 |
The overall accuracy of the proposed step-by-step classifier is compared with the other existing methods which aimed at designing a decision support system to detect leukemia from PBS images. The comparison is given in Table 10. It can be observed from the table that although the dataset consists of different types of abnormal WBCs and images of brightness and color variations, the proposed method provides high accuracy. The dataset used in the current study comprises both myeloblasts and lymphoblasts. Most of the existing methods have been proposed by considering only lymphoblasts. An automated method which is designed by considering only a specific type of abnormal WBCs may not be a practically useful application.
Table 10.
Comparison of the proposed method with the existing methods
| Dataset | Overall accuracy | |
|---|---|---|
| Moradi et al. [12] | ALL sub-types, reactive WBCs, atypical lymphocyte, normal WBCs (312 images) | 97% |
| Neoh et al. [13] | Lymphocytes, lymphoblasts (180 images) | 96.7% |
| Srisukkham et al. [19] | Lymphocytes, lymphoblasts (180 images) | 96% |
| Zeinab et al. [18] | Normal WBCs and lymphoblats (368) images | 98% |
| Proposed Method | Normal WBCs, reactive WBCs, myelocytes, degenerated WBCs, blasts (1159 images) | 98.85% |
Accurate classification of WBCs into normal and abnormal will reduce the workload of pathologists. This is because, experts need not look into the normal cases. This also helps speeding up the evaluation process for abnormal cases. Another importance of the proposed method is detection of blasts with 100% specificity. This again helps in reducing the number of cases of manual evaluation. The detected leukemia cases can be directly sent for further tests. Manual evaluation is required only for the abnormal cases. Reduced number of cases indirectly reduces the manual errors. Hence, the proposed method can effectively bring down the manual errors and reduces the workload of pathologists.
Though the proposed segmentation method is robust to brightness and color variations, it is not tested for different staining methods. Hence, the future scope of the segmentation method is to test the method with images of different stains. Overall accuracy of the proposed classifier provides the promising result for classification of WBCs into normal and abnormal, and also for classification of the normal WBCs into its sub-types, but the accuracy of detection of blasts from the abnormal class needs further improvement.
Conclusion
In this paper, a method for detection of leukemia using peripheral blood smear images was presented. A robust method for computing accurate threshold value for detection of region of nuclei was introduced. Also, a novel approach for generating mask for detection of WBC from the cropped sub-images was described. Average Dice score of 0.97 and 0.98 was obtained for detection of nuclei and WBCs respectively. Average accuracy of detection of nuclei and WBCs was around 99%.
Shape, color, and texture features were extracted from the nuclei and cytoplasm of WBCs. A combination of SVM and NN classifier was used to classify the WBCs into five sub-types and abnormal WBCs. Accuracy of 100% was obtained for classification of WBCs into normal and abnormal using SVM classifier. We obtained average accuracy of 99.5% for classification of normal WBCs into five sub-types using NN classifier. SVM classifier was used for detection of blasts form the abnormal class which provided an accuracy around 93% and specificity of 100%. The proposed method provides the promising results for classification of WBCs into normal and abnormal which can effectively bring down the workload of pathologists. This is because, experts need not look into the normal cases. Further, the method needs improvement for detection of blasts from the abnormal class.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Roopa B. Hegde, Email: roopabhegde@gmail.com
Keerthana Prasad, Email: keerthana.prasad@manipal.edu.
Harishchandra Hebbar, Email: harish.hebbar@manipal.edu.
Brij Mohan Kumar Singh, Email: brij.singh@manipal.edu.
I Sandhya, Email: drsandhyai@yahoo.co.in.
References
- 1.Cancer Stat Facts: Leukemia, https://seer.cancer.gov/statfacts/html/leuks.html, Accessed on 15-05-2018
- 2.Facts and Statistics, https://www.lls.org/http%3A/llsorg.prod.acquia-sites.com/facts-and-statistics/facts-and-statistics-overview/facts-and-statistics, Accessed on 16-07-2019
- 3.https://www.wcrf.org/int/cancer-facts-figures/worldwide-data, Accessed on 15-05-2018
- 4.Irshad H, Veillard A, Roux L, Racoceanu D. Methods for nuclei detection, segmentation, and classification in digital histopathology: a review - current status and future potential. IEEE Rev Biomedic Eng. 2014;7:97–114. doi: 10.1109/RBME.2013.2295804. [DOI] [PubMed] [Google Scholar]
- 5.Xing F, Yang L. Robust Nucleus/Cell detection and segmentation in digital pathology and microscopy images: a comprehensive review. IEEE Trans Biomed Eng. 2016;9:234–263. doi: 10.1109/RBME.2016.2515127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen P, Park DS, Yong Y, Min YH. Leukocyte image segmentation by visual attention and extreme learning machine. Neural Comput Appl. 2012;21(6):1217–1227. doi: 10.1007/s00521-011-0522-9. [DOI] [Google Scholar]
- 7.do Nascimento MZ, Martins AS, Tosta TAA, Neves LA. Lymphoma images analysis using morphological and non-morphological descriptors for classification. Comput Methods Programs Biomed. 2018;163:65–77. doi: 10.1016/j.cmpb.2018.05.035. [DOI] [PubMed] [Google Scholar]
- 8.Feilong C, Miaomiao C, Jianjun C, Jianwei Z, Zhenghua Z. A novel segmentation algorithm for nucleus in white blood cells based on low-rank representation. Neural Comput Appl. 2017;28(1):503–511. [Google Scholar]
- 9.Ananthi VP, Balasubramaniam P. A new thresholding technique based on fuzzy set as an application to leukocyte nucleus segmentation. Comput Methods Programs Biomed. 2016;134:165–177. doi: 10.1016/j.cmpb.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Foran DJ, Comaniciu D, Meer P, Goodell LA. Computer-assisted discrimination among malignant lymphomas and leukemia using immunophenotyping, intelligent image repositories, and telemicroscopy. IEEE Trans Inf Technol Biomed. 2000;4(4):265–273. doi: 10.1109/4233.897058. [DOI] [PubMed] [Google Scholar]
- 11.Agaian S, Madhukar M, Chronopoulos AT. Automated screening system for acute myelogenous leukemia detection in blood microscopic images. IEEE Systems Journal. 2014;8(3):995–1004. doi: 10.1109/JSYST.2014.2308452. [DOI] [Google Scholar]
- 12.Amin MM, Kermani S, Talebi A, Oghli MG. Recognition of acute lymphoblastic leukemia cells in microscopic images using means clustering and support vector machine classifier. J Med Signals Sens. 2015;5(1):49–58. doi: 10.4103/2228-7477.150428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neoh SC, Srisukkham W, Zhang L, Todryk S, Greystoke B, Lim CP, Hossain MA, Aslam N (2015) An Intelligent Decision Support System for Leukaemia Diagnosis Using Microscopic Blood Images. Scientific Reports, 10.1038/srep14938 [DOI] [PMC free article] [PubMed]
- 14.Reta C, Altamirano L, Gonzalez JA, Hernandez RD, Peregrina H, Olmos I, Alonso JE, Lobato R. Segmentation and classification of bone marrow cells images using contextual information for medical diagnosis of acute Leukemias. PLoS One. 2015;10(6):e0130805. doi: 10.1371/journal.pone.0130805,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negm AS, Hassan OA, Kandil AH. A decision support system for Acute Leukaemia classification based on digital microscopic images. Alex Eng J. 2018;57(4):2319–2332. doi: 10.1016/j.aej.2017.08.025. [DOI] [Google Scholar]
- 16.Patel N, Mishra A. Automated leukaemia detection using microscopic images. Procedia computer science. Second International Symposium on Computer Vision and the Internet (VisionNet’15) 2015;58:635–642. [Google Scholar]
- 17.Belacel N, Vincke Ph, Scheiff JM, Boulassel MR. Acute leukemia diagnosis aid using multicriteria fuzzy assignment methodology. Comput Methods Programs Biomed. 2001;64(2):145–151. doi: 10.1016/S0169-2607(00)00100-0. [DOI] [PubMed] [Google Scholar]
- 18.Zeinab M, Habibollah D, Sadegh HM. An automatic and robust decision support system for accurate acute leukemia diagnosis from blood microscopic images. J Digit Imaging. 2018;31:702–717. doi: 10.1007/s10278-018-0074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srisukkham W, Zhang L, Neoh SC, Todryk S, Lim CP. Intelligent leukemia diagnosis with bare-bones PSO based feature optimization. Appl Soft Comput. 2017;56:405–419. doi: 10.1016/j.asoc.2017.03.024. [DOI] [Google Scholar]
- 20.Subrajeet M, Dipti P, Sanghamitra S. An ensemble classifier system for early diagnosis of acute lymphoblastic leukemia in blood microscopic images. Neural Comput Appl. 2014;24(7):1887–1904. [Google Scholar]
- 21.Fatichah C, Tangel ML, Yan F, Betancourt JP, Widyanto MR, Dong F, Hirota K. Fuzzy feature representation for white blood cell differential counting in acute leukemia diagnosis. Int J Control Autom Syst. 2015;13(3):742–752. doi: 10.1007/s12555-012-0393-6. [DOI] [Google Scholar]
- 22.Escalante HJ, Montes-y-Gómez M, González JA, Gómez-Gil P, Altamirano L, Reyes CA, Reta C, Rosales A. Acute leukemia classification by ensemble particle swarm model selection. Artif Intell Med. 2012;55:163–175. doi: 10.1016/j.artmed.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Azevedo Tosta TA, Faria PR, Batista VR, Neves LA, do Nascimento MZ. Using wavelet sub-band and fuzzy 2-partition entropy to segment chronic lymphocytic leukemia images. Appl Soft Comput. 2018;64:49–58. doi: 10.1016/j.asoc.2017.11.039. [DOI] [Google Scholar]
- 24.Osowski S, Siroic R, Markiewicz T, Siwek K. Application of support vector machine and genetic algorithm for improved blood cell recognition. IEEE T Instrum Meas. 2009;58(7):2159–2168. doi: 10.1109/TIM.2008.2006726. [DOI] [Google Scholar]
- 25.Su J, Liu S, Song J. A segmentation method based on HMRF for the aided diagnosis of acute myeloid leukemia. Comput Meth Prog Bio. 2017;152:115–123. doi: 10.1016/j.cmpb.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Nikitaev VG, Nagornov OV, Pronichev AN, Polyakov EV, Dmitrieva V. The blood smear image processing for the acute leukemia diagnostics. Inter J Bio Biomedic Eng. 2016;10:109–114. [Google Scholar]
- 27.Rawat J, Singh A, Bhadauria HS, Virmani J, Devgun JS. Computer assisted classification framework for prediction of acute lymphoblastic and acute myeloblastic leukemia. Biocybern Biomed Eng. 2017;37:637–654. doi: 10.1016/j.bbe.2017.07.003. [DOI] [Google Scholar]
- 28.Rosin PL. Unimodal thresholding. Pattern Recognit. 2001;34:2083–2096. doi: 10.1016/S0031-3203(00)00136-9. [DOI] [Google Scholar]
- 29.Bakos M Active contours and their utilization at image segmentation. In: Proc 5th Slovakian-Hungarian Joint Symposium on Applied Machine Intelligence and Informatics, Poprad, Slovakia, 2007, pp 313–317
- 30.Kass M, Witkin A, Terzopoulos D. Snakes: Active contour models. Int J Comput Vision. 1988;1(4):321–331. doi: 10.1007/BF00133570. [DOI] [Google Scholar]
- 31.Zhang C, Xiao X, Li X, Chen Y-J, Zhen W, Chang J, Zheng C, Liu Z. White blood cell segmentation by Color-Space-Based K-Means clustering. Sensors (Basel) 2014;14(9):16128–16147. doi: 10.3390/s140916128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson SG Notes on FFT-based Differentiation, MIT Applied Mathematics, 1-11 (2011). http://math.mit.edu/stevenj/fft-deriv.pdf
- 33.Matthew Sottile: Finding dents in a blobby shape, 2013, https://syntacticsalt.com/2013/01/11/finding-dents-in-an-blobby-shape/
- 34.Roopa B. Hegde, keerthana prasad, harishchandra hebbar, brij mohan kumar singh: Development of a robust algorithm for detection of nuclei and classification of white blood cells in peripheral blood smear images. J Med Syst. 2018;42(6):110. doi: 10.1007/s10916-018-0962-1. [DOI] [PubMed] [Google Scholar]