Abstract
Epidermal growth factor receptor (EGFR) amplification and EGFR variant III (EGFRvIII, deletion of exons 2–7) are of clinical interest for glioblastoma. The aim was to develop a digital PCR (dPCR)-based method using locked nucleic acid (LNA)-based hydrolysis probes, allowing the simultaneous detection of the EGFR amplification and EGFRvIII variant. Sixty-two patients were included. An exploratory cohort (n = 19) was used to develop the dPCR assay using three selected amplicons within the EGFR gene, targeting intron 1 (EGFR1), junction of exon 3 and intron 3 (EGFR2) and intron 22 (EGFR3). The copy number of EGFR was estimated by the relative quantification of EGFR1, EGFR2 and EGFR3 amplicon droplets compared to the droplets of a reference gene. EGFRvIII was identified by comparing the copy number of the EGFR2 amplicon to either the EGFR1 or EGFR3 amplicon. dPCR results were compared to fluorescence in situ hybridization (FISH) and next-generation sequencing for amplification; and to RT-PCR-based method for EGFRvIII. The dPCR assay was then tested in a validation cohort (n = 43). A total of 8/19 EGFR-amplified and 5/19 EGFRvIII-positive tumors were identified in the exploratory cohort. Compared to FISH, the EGFR3 dPCR assay detected all EGFR-amplified tumors (8/8, 100%) and had the highest concordance with the copy number estimation by NGS. The concordance between RT-PCR and dPCR was also 100% for detecting EGFRvIII using an absolute difference of 10.8 for the copy number between EGFR2 and EGFR3 probes. In the validation cohort, the sensitivity and specificity of dPCR using EGFR3 probes were 100% for the EGFR amplification detection compared to FISH (19/19). EGFRvIII was detected by dPCR in 8 EGFR-amplified patients and confirmed by RT-PCR. Compared to FISH, the EGFR2/EGFR3 dPCR assay was estimated with a one-half cost value. These results highlight that dPCR allowed the simultaneous detection of EGFR amplification and EGFRvIII for glioblastoma.
Keywords: Glioblastoma, Digital PCR, EGFR amplification, EGFRvIII variant, Cost-effectiveness
Introduction
Glioblastoma is the most frequent primary brain tumor in adults, with 125,000 to 150,000 new cases per year worldwide [1]. Despite extensive treatment based on surgery, radiotherapy and chemotherapy combination, recurrence remains the rule with a median overall survival of less than 18–24 months [2]. Diagnosis is commonly based on histopathological examination and characterization of isocitrate dehydrogenase (IDH)1/2 mutations [3]. Recent advances also highlighted a key role of other molecular alterations, such as those located on the epidermal growth factor receptor (EGFR) gene, which is altered in approximately 57% of cases [4]. EGFR amplification and EGFR variant III (EGFRvIII), which is characterized by the deletion of exons 2–7, are the two most frequent EGFR alterations in glioblastoma observed in 40–50% and 10% of patients, respectively [4–7]. Interestingly, it has been reported that the presence of EGFRvIII is associated with EGFR gene amplification in most cases [8]. In this context, specific treatments that directly target the EGFR pathway or activate the immune response against EGFRvIII have been recently developed using either as a single therapy or in combination with standard treatment [9–12]. Antibody-drug conjugates targeting EGFR may improve survival at the time of recurrence in EGFR-amplified glioblastoma [13]. In addition, identification of EGFR amplification associated with either a telomerase reverse transcriptase promoter (TERTp) mutation or chromosomal alterations (chromosome 7 gain and chromosome 10 loss) in diffuse or anaplastic astrocytoma has led to a reclassification proposal of grade II-III 1p19q non-codeleted gliomas into glioblastoma-like tumors [14, 15]. Taken together, these data support that the detection of EGFR alterations may be considered relevant in patients treated for glioblastoma.
Until now, EGFR alterations have been detected by separate methods. Indeed, fluorescence in situ hybridization (FISH) is the gold standard for the detection of EGFR amplification, and the use of other methods, such as genomic hybridization (array CGH) or next-generation sequencing (NGS), has also been reported. On the other hand, the detection of the EGFRvIII variant, leading to an abnormal expression of ARNm, is performed commonly using RT-PCR-based methods [9].
The development of a specific molecular method allowing the simultaneous detection of EGFR alterations may be of interest in glioblastoma. Targeted copy number variation (CNV) detection by digital PCR (dPCR) using locked nucleic acid (LNA)-based hydrolysis probes has recently been shown to be efficient in genetic diseases [16]. LNA-hydrolysis probes are very short nucleotides, and repeated sequences across the human genome may be incorporated in a dPCR amplicon. Gene copy number estimation is then based on the ratio of detected LNA probes between a gene of interest and a reference gene.
In this context, we aimed to develop a novel dPCR assay using LNA-hydrolysis probes located within and outside the region spanning from exon 2 to exon 7 to allow the simultaneous detection of EGFR amplification and EGFRvIII variant. First, we used an exploratory cohort of patients with glioblastoma to develop a dPCR assay in comparison to FISH for EGFR amplification and to an RT-PCR-based method for EGFRvIII. In the second step, we tested the ability of our dPCR assay to simultaneously detect these two EGFR alterations in an independent validation cohort of patients with glioblastoma.
Patients and methods
Patients and tumor samples
The present study is ancillary to the ongoing prospective GLIOPLAK trial (registered in ClinicalTrials.gov, NCT02617745), which is investigating predictive markers of chemo-induced toxicities. A total of 62 patients were recruited from November 2015 to November 2017. Eligible patients were at least 18 years old and had a newly diagnosed and histologically confirmed supratentorial glioblastoma, according to the 2016 WHO classification [3]. Patients received concomitant radiotherapy with temozolomide followed by sequential temozolomide treatment [2]. Tumor samples were obtained during surgery (biopsy, gross-total or partial resection) and processed for routine histopathology, immunohistochemistry and molecular biology experiments. Tumor DNA was extracted from formalin-fixed paraffin-embedded FFPE samples using the Maxwell 16 FFPE Plus LEV DNA Purification® Kit on a Maxwell 16 Instrument® (Promega®, Fitchburg, Wisconsin, United States). IDH1/2 mutations within exon 4 were analyzed using the ABI PRIM SNaPshot® Multiplex Kit (ThermoFisher Scientific®, Waltham, Massachusetts, USA); MGMT promoter (MGMTp) methylation was analyzed with the pyrosequencing method (therascreen MGMT Pyro®, Qiagen®, ThermoFisher Scientific®).
For the purpose of the present study, the population was divided into two groups: an exploratory cohort, which included the first 19 patients, and a validation cohort, which was based on the next 43 consecutive patients. The exploratory cohort was used to develop the dPCR assay by selecting amplicons and allowing the simultaneous detection of EGFR amplification and EGFRvIII, according to the standard methods of FISH or NGS and RT-PCR-based methods, respectively. In the second step, we used an independent validation cohort to evaluate the ability of the dPCR assay to detect both EGFR alterations.
Development of dPCR assay
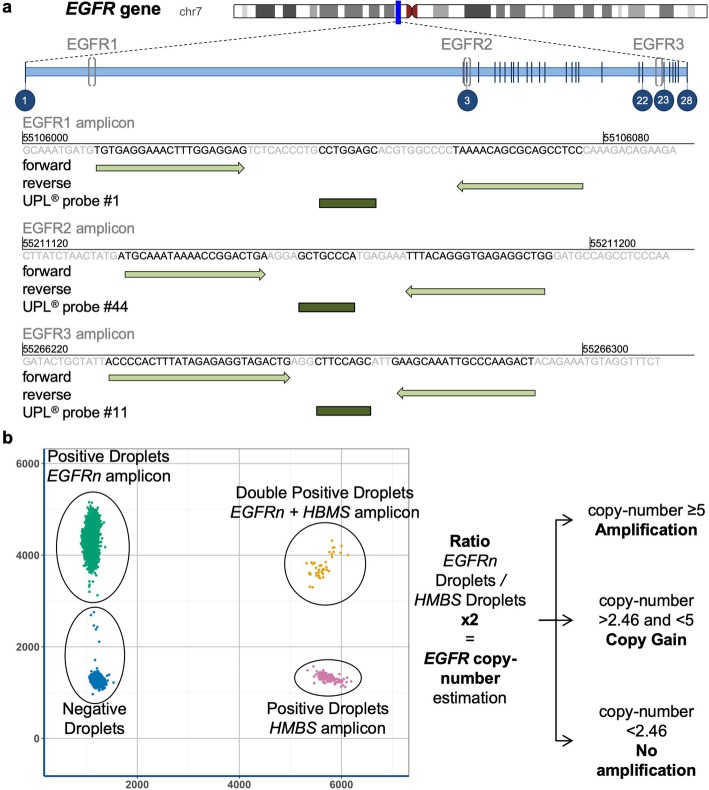
According to recently published methods of dPCR using universal LNA-hydrolysis probes from the 96 Universal Probe Library® (UPL, Sigma-Aldrich®, St. Louis, Missouri, USA), three dPCR assays were performed for each tumor sample [16]. These assays used a duplex PCR: one PCR amplicon within the EGFR gene and one reference PCR amplicon located in the hydroxymethylbilane synthase (HMBS) gene, a housekeeping gene located in 11q23. Three different amplicons of the EGFR gene were designed: the EGFR1 amplicon located within intron 1 with UPL® probe #1 (reference: 04684974001), the EGFR2 amplicon located between exon 3 and intron 3 with UPL® probe #44 (reference: 04688040001) and the EGFR3 amplicon located within intron 22 with UPL® probe #11 (reference: 04685105001) (Fig. 1a). The reference HMBS amplicon is located in intron 1 using the forward primer (5′-GGGACAGTGTACCCAAGGTC-3′), the reverse primer (5′-CTGAGGTAAACGGATCTGACG-3′) and a custom trichloro-phenylcarboxyfluorescein oligonucleotide (VIC)-labeled probe (5′-CCAAGAGGCTGAGCAGGACT-3′, ThermoFisher Scientific®). dPCR experiments were performed using a Qx200® droplet digital PCR (ddPCR) System (Biorad®, Hercules, California, USA). ddPCR was run in a final volume of 22 μL with tumor DNA, 10 μl ddPCR Supermix for probes (no dUTP), primers for the EGFR- and HMBS-targeted amplicons (0.9 μM), 6-carboxyfluorescein (FAM)-labeled LNA-based hydrolysis probe for the EGFR-targeted sequence (0.18 μM), and VIC-labeled probe for the HMBS amplicon (0.18 μM). Thermal cycling was performed, according to the manufacturer’s instructions: 10 min at 95 °C; then 40 cycles at 94 °C for 30 s and 56 °C for 1 min; and a final step of 10 min at 98 °C. The software QuantaSoft® was used for the interpretation of the profiles. The EGFR copy number was determined by calculating the ratio of EGFR FAM-labeled droplets over the HMBS VIC-labeled droplets multiplied by the number of HMBS copies (× 2 in the human genome) (Fig. 1b). CNV assessment was then based on comparisons to the EGFR copy number estimation from FFPE control tissues (normal copy number, 1.66–2.46). According to molecular analyses from INTELLANCE trials, glioblastoma was considered to be EGFR-amplified if the copy number was greater than or equal to 5 [7]. For EGFRvIII identification, we hypothesized that the copy number estimated by the EGFR2 amplicon would be lower than the copy number estimated by EGFR1/EGFR3 for the same sample. The optimal amount of DNA for one dPCR experiment was set at 30 ng.
Fig. 1.
Design of the dPCR assay using LNA-hydrolysis probes for detecting the EGFR amplification and EGFRvIII variant. a Three amplicons were designed within the EGFR gene from Universal Probe Library® (Sigma-Aldrich). EGFR1, EGFR2 and EGFR3 are located within three different regions in the gene. EGFR2 is inserted into the deleted region of the EGFRvIII variant (deletion of exons 2–7). b Two-dimensional cluster plot representing the 6-carboxyfluorescein (FAM)-labeled LNA-based hydrolysis probe for the EGFR-targeted sequence (EGFR1, EGFR2 or EGFR3) against the trichloro-phenylcarboxyfluorescein oligonucleotide (VIC)-labeled hydrolysis probe for the HMBS amplicon. Droplets are grouped as clusters: FAM/VIC-negative (double-negative droplets, blue), FAM-positive/VIC-negative (green), FAM-negative/VIC-positive (pink), and FAM/VIC-positive (double-positive droplets, orange). The EGFR copy number was determined by calculating the ratio of EGFR FAM-labeled droplets over the HMBS VIC-labeled droplets multiplied by the number of HMBS copies (× 2 in the human genome)
EGFR amplification detection by fluorescence in situ hybridization
FISH was performed on FFPE tumor samples. After the selection of an area containing more than 70% tumor cells, 4-μm sections were deparaffinized, dehydrated with ethanol and pretreated with Vysis Paraffin Pretreatment IV (Abbott®, Illinois, USA). Hybridization was performed using the EGFR/CEP7 FISH Probe Kit® (Abbott®), according to the manufacturer’s instructions. The probes used covers 303 kb located in the 7p11 region, which contains the EGFR gene. The reference probe was located on the centromere of chromosome 7. The post-hybridization step was performed with a Wash Buffer Kit (Abbott®). Samples were considered EGFR-amplified when the quantity of fluorescence of the EGFR-targeting probe (red fluorescence) was greater than twofold per nucleus than the number of centromere-targeting probes (green fluorescence) and only when at least 15% of tumor cell nuclei were EGFR-amplified [7].
NGS experiments and EGFRvIII detection
An Ion Torrent Personal Genome Machine (PGM, Life Technologies®, Carlsbad, California, United States of America) was used for EGFR somatic point mutations and EGFR amplification detection. Tumor DNA was sequenced using a custom EGFR-targeted panel dedicated to highly recurrent altered region of the gene (Additional File: Table S1) [4]. Amplified libraries (Ion AmpliSeq® Library Kit 2.0) were submitted to emulsion PCR using the Ion OneTouch® 200 Template Kit (Life Technologies®) with the Ion OneTouch® System (Life Technologies®). Data analysis was performed using Torrent Suite version 5.4 software (ThermoFisher Scientific®). Reads were mapped to the human hg19 reference genome. Copy-number was estimated using the ONCOCNV algorithm compared to control DNA from healthy subjects [17].
EGFRvIII identification was performed using ligation-dependent reverse transcription polymerase chain reaction (LD-RT-PCR), which allows the detection of fusion transcripts and exon skipping [18–20]. RNA was extracted from FFPE tumor samples using the Maxwell® 16 LEV RNA FFPE Purification Kit (reference AS1260, Promega®) and following manufacturer instructions. RNA was converted into complementary DNA (cDNA) using reverse transcription probes located on the end of EGFR exon 1 and the start of EGFR exon 8; the cDNA was then hybridized. In the case of EGFRvIII (deletion of exons 2–7), by adding DNA ligase, a covalent link between the two probes was formed, allowing PCR amplification and subsequent identification by NGS on MiSeq® (Illumina®, San Diego, California, USA).
Cost-effectiveness estimation
An exploratory cost-effectiveness study was conducted to compare the cost of EGFR amplification detection with dPCR with the reference method (FISH). Total costs per patient included reagent costs and medical/technician times.
Standard protocol approvals, registrations and patient consent
Informed written consent to participate in the study was obtained from all patients. The French National Committee for the Protection of Persons approved the study (RCB ID 2015-A00377–42).
Statistical analyses
In the exploratory cohort, to assess the equivalence of the EGFR copy number estimation between NGS and dPCR, a correlation matrix plot was performed. The sensitivity, specificity and positive and negative predictive values were calculated to assess the diagnostic performance. The gold standard was the FISH results. The copy number difference and its threshold between the EGFR2 assay and the two other techniques to predict the EGFRvIII variant were determined using receiver operating characteristic (ROC) curves. In the validation cohort, the sensitivity and specificity of the dPCR assay for detecting EGFR amplification was compared to those of FISH. All dPCR and FISH analyses were carried out in a double-blind manner. Statistical analyses and figures were performed using R software (R version 3.5.1, 2018, Vienna, Austria) [21].
Results
Baseline characteristics
The characteristics of all patients are detailed in Table 1. Among them, 59 (95%) had wild-type IDH (IDH-wt) glioblastoma, and 3 had IDH-mutated glioblastoma. Overall, EGFR amplification was identified in 27 tumors (44%) using FISH; a total of 8 (42%) and 19 (44%) were in the exploratory and validation cohorts, respectively. In the group of patients with an IDH-wt glioblastoma, three had a rare histological subtype (1 with giant cell glioblastoma and 2 with gliosarcoma), and none of the three tumors had EGFR amplification.
Table 1.
Clinical and Tumor characteristics
| Characteristics | Entire cohort N = 62 |
Exploratory cohort N = 19 |
Validation cohort N = 43 |
|
|---|---|---|---|---|
| Age (years), mean [min. – max.] | 56.9 [21–76] | 55.5 [28–76] | 57.7 [21–72] | |
| Sex | Female | 28 (45%) | 7 (37%) | 21 (49%) |
| Male | 34 (55%) | 12 (63%) | 22 (51%) | |
| Surgery | Biopsy | 25 (40%) | 8 (42%) | 17 (40%) |
| Resection | 37 (60%) | 11 (58%) | 26 (60%) | |
| Glioblastoma IDH wild type | 59 (95%) | 18 (95%) | 41 (95%) | |
| Giant cell glioblastoma | 1 (2%) | 1 (5%) | 0 | |
| Gliosarcoma | 2 (3%) | 0 | 2 (5%) | |
| Glioblastoma IDH mutant | 3 (5%) | 1 (5%) | 2 (5%) | |
| EGFR amplification by FISH | 27 (43%) | 8 (42%) | 19 (44%) | |
| MGMTp methylation | Non-methylated | 37 (60%) | 13 (68%) | 24 (56%) |
| Methylated | 17 (27%) | 6 (32%) | 11 (26%) | |
| Unknown | 8 (13%) | 0 | 8 (18%) | |
| TERTp mutation | C228T | 41 (66%) | 14 (74%) | 27 (63%) |
| C250T | 14 (23%) | 4 (21%) | 10 (23%) | |
| Wild-type | 7 (11%) | 1 (5%) | 6 (14%) |
Development of a dPCR assay for detecting EGFR alterations in the exploratory cohort
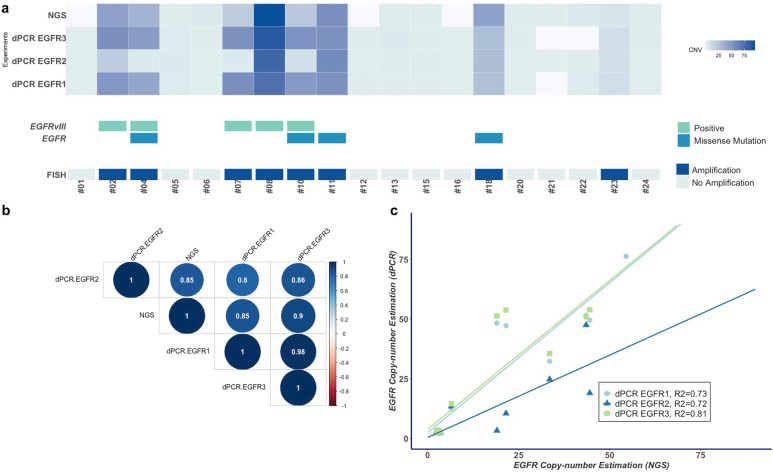
Among the 19 patients in the exploratory cohort, EGFR amplification was identified in eight (8/19, 42%) patients using FISH. EGFR1 and EGFR3 assays strictly correlated with FISH results, making it possible to distinguish all EGFR-amplified (8/8) and EGFR-non-amplified glioblastoma (11/11), with sensitivity, specificity, positive and negative predictive values of 100%. Overall, the mean copy number estimation by dPCR was 25 (range 2–76) using EGFR1 and 29.4 (range 2–98) using EGFR3. Using a threshold of copy number amplification greater than or equal to 5, the mean EGFR1 copy number amplification was 47.5 (range 12.3–76.3), and the mean EGFR3 copy number amplification was 56.1 (14.5–98.3) (Fig. 2a). Using FISH, seven out of the 8 EGFR-amplified glioblastomas contained at least 90% cell nuclei with EGFR amplification. Interestingly, the remaining EGFR-amplified glioblastoma patient (patient #23) had 30% amplified cell nuclei and concordant copy number estimation by dPCR of 12.3 for EGFR1 and 14.5 for EGFR3. The diagnostic performance of EGFR2 was lower with one discordant case; the EGFR was not amplified by dPCR but was amplified using FISH (sensitivity of 87.5% and specificity of 100%).
Fig. 2.
Concordance between the results of the dPCR assay, FISH, next-generation sequencing (NGS) and LD-RT-PCR for the detection of the EGFR amplification and EGFRvIII variant in the exploratory cohort (n = 19). a Heatmap of EGFR copy number estimated by NGS and the three dPCR assays. Each column represents a tumor sample (n = 19). The blue gradient represents the estimated value of the EGFR copy number. There is a strong agreement between the EGFR1 and EGFR2 dPCR assays and NGS. The absence of results using the NGS experiment is indicated by the light gray color. Below the heatmap, the presence of EGFR mutations and EGFRvIII variant as well as the results of FISH are presented. The presence of somatic mutations was detected by the EGFR-targeted NGS panel, and the presence of the EGFRvIII variant was detected by LD-RT-PCR. Patient #08 harbors both the EGFR amplification and EGFRvIII variant with tumor heterogeneity regarding the copy number estimation by dPCR (EGFR1 63, EGFR2 70 and EGFR3 91). b Correlation matrix plot of EGFR copy number estimation using three dPCR assays (EGFR1, EGFR2 and EGFR3) and NGS (n = 16). The dPCR EGFR3 assay results have the highest correlation with the NGS results. On the other hand, the dPCR EGFR2 assay results have the poorest correlation, mainly due to its underestimation of the EGFR CNV in the case of EGFRvIII-positive glioblastoma. c Linear regression curves representing EGFR copy number values estimated by NGS (x-axis) and the copy number estimated by the three dPCR assays (y-axis). As expected with the results of the matrix correlation plot, the estimation using the dPCR EGFR3 assay was confirmed to have the best correlation to the NGS estimation
NGS experiments were performed in 16 out of the 19 tumors, and three patients had a tumor DNA quantity that was too low to be analyzed. The 8/16 patients with an EGFR amplification detected by NGS were the same as those identified with dPCR assay or FISH. Notably, the correlation coefficient between the copy number calculated by dPCR assays (EGFR1, EGFR2 and EGFR3) and by NGS was higher than 0.8 (Fig. 2b). The copy number estimated by the EGFR3 assay had the highest correlation coefficient with NGS values (correlation coefficient of 0.9 and R-squared of 0.81) (Fig. 2c). The copy number estimation by the EGFR2 assay had the lowest correlation with the EGFR1/EGFR3 dPCR assays and with NGS. Moreover, the mean copy number by the EGFR2 assay was significantly lower than that estimated by the EGFR3 assay (18.8 vs 29.4, P = 0.023). Five EGFR-amplified glioblastomas had a lower estimated copy number by EGFR2 than those by EGFR1/EGFR3 assays; in one patient (patient #07), an EGFR2 copy number was estimated as 3.3, which was below the established cutoff.
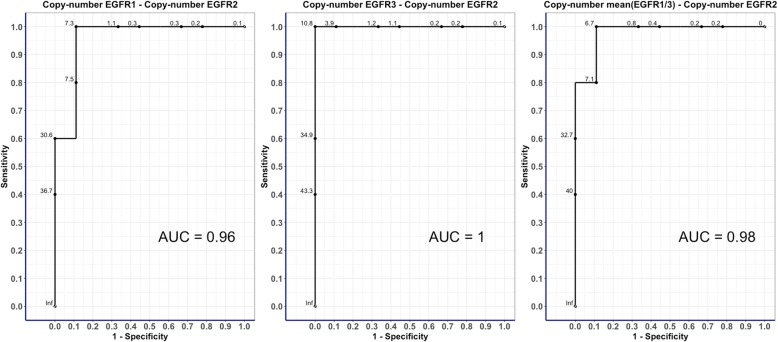
A total of 19 patients were tested for the EGFRvIII variant. Among them, five patients were positive for an EGFRvIII variant using LD-RT-PCR. The dPCR assays detected a total of 5/19 patients with the EGFRvIII variant, all of which were identical to those detected by the LD-RT-PCR method (Fig. 2a and Additional File: Fig. S2). We observed that the most predictive copy number differences between dPCR assays for detecting the EGFRvIII variant was between the EGFR3 and EGFR2 assays, rather than between the EGFR1 and EGFR2 assays, with a copy number absolute difference of 10.8 (AUC 1) (Fig. 3).
Fig. 3.
ROC curves of the copy number differences between the three dPCR assays for the prediction of the EGFRvIII variant. The three ROC curves represent the identification of the best diagnostic test to identify EGFRvIII using the absolute copy number differences between EGFR2 and the other dPCR assays, namely, EGFR1, EGFR3 and mean (EGFR1 + EGFR3). The best predictive test was selected using the highest AUC (difference of EGFR3 and EGFR2) and the threshold of the copy number difference that maximizes the sensitivity and specificity (10.8)
Taken together, using these results, the EGFR3 assay and the EGFR2/EGFR3 assay were selected to detect EGFR-amplified glioblastoma and to identify the EGFRvIII variant in the validation cohort (Fig. 4).
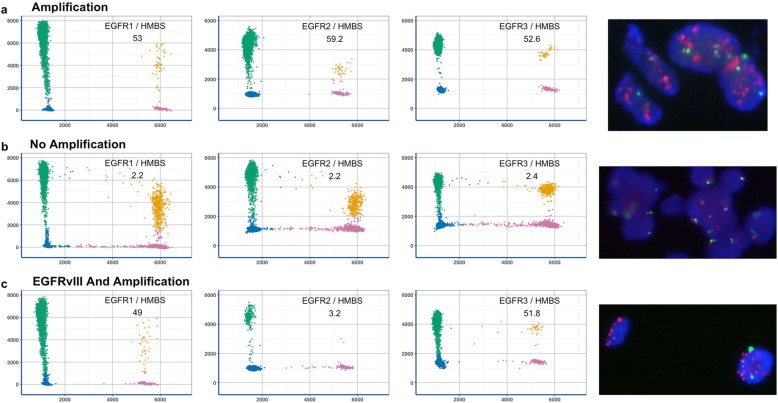
Fig. 4.
Three illustrative sample tumor examples using three dPCR assays and FISH. a Tumor with EGFR amplification. b Tumor without EGFR amplification. c Tumor with EGFR amplification and concomitant EGFRvIII variant. Corresponding FISH images are shown at the end of the line on the right
Results of the dPCR assays in the validation cohort
A total of 43 patients were included in the validation cohort. Among them, 19/43 (44%) were EGFR-amplified IDH-wt glioblastomas using FISH (19/43, 44%). Using the dPCR assay, based on the EGFR3 assay, the same 19 patients with an EGFR amplification were identified, leading to a sensitivity, specificity, positive and negative predictive values of 100%. The mean estimated copy number was 56.9 (range 13.6–196.5), and the median was 48.7. The EGFR3 assay allowed for the identification of EGFR copy gain in 16 EGFR-non-amplified glioblastomas with a mean copy number of 3.3 (range 2.6–3.9). A single patient harbored an EGFR amplification in 5% of tumor cell nuclei (Additional File: Fig. S3), and this was considered non-amplified both by FISH and by dPCR. Among the 19 EGFR-amplified glioblastomas, EGFRvIII was identified in 8 patients by dPCR EGFR2/EGFR3 assay, and all were confirmed using LD-RT-PCR.
Interestingly, two EGFR-amplified glioblastomas, identified by dPCR and confirmed by FISH, had very low amount of DNA (2 ng and 6 ng). One tumor had concomitant EGFRvIII variant confirmed by LD-RT-PCR, highlighting the value of the dPCR-based method for glioblastoma samples with small amount of DNA.
Cost estimation of dPCR
The estimated cost for one patient (CNV detected in EGFR1, EGFR2 and EGFR3) was 43% lower using dPCR than FISH (60.88€: 30.33€ for reagents and 30.55€ for working time) for dPCR and 106.01€ for FISH) (Additional File: Table S4). The total cost of dPCR decreased to 50.77€ when only EGFR2 and EGFR3 assays were used. Moreover, dPCR assays with EGFR2/EGFR3 detect both the EGFR amplification and EGFRvIII variant, whereas the FISH assay can only identify an EGFR amplification.
Discussion
This study shows that the specific dPCR assay using LNA-hydrolysis probes from UPL® is a reliable and simple method to simultaneously detect an EGFR amplification and EGFRvIII variant, and this can be used in clinical practice in glioblastoma. Indeed, using an experimental design based on two independent cohorts, we showed that the dPCR assay was better than standard methods and was able to detect the main somatic EGFR alterations in DNA extracted from FFPE tumor samples with a diagnostic performance of 100%.
The current molecular findings in our work were similar to those previously published using larger cohorts of glioblastoma patients eligible for Stupp treatment [22, 23]. Indeed, the overall proportion of EGFR-amplified tumors is similar to that reported in the TCGA (43%) [4], especially when accounting for the criteria recently suggested by French et al. to classify tumors as EGFR amplified (EGFR copy number higher than 5 and more than 50% of the nuclei were amplified) [7]. Moreover, it has also been recently confirmed that the proportion of patients with EGFR-amplified glioblastoma using NGS, FFPE-based or CGH-array techniques is between 35 and 45% [24, 25]. One of the major strengths of our study is that, in addition to the qualitative assessment of EGFR amplification, the dPCR assay using LNA-hydrolysis probes also provides a reliable quantitative copy number estimation compared to NGS. We also confirmed the high number of EGFR copy number amplicons in glioblastoma, including 13% of tumors (n = 8) with greater than 50 copy gains. Although the therapeutic impact of high EGFR-amplified tumors remains to be evaluated, our results clearly showed that our dPCR assay may be used to screen the EGFR copy number for decision making, particularly in further studies focusing on therapies targeting this molecular alteration.
One of the other benefits of the dPCR assay developed in our work is its potential economic cost compared to FISH. For one patient, the cost decreases from 40 to 50% when using only the EGFR2/EGFR3 assay. Moreover, in contrast to FISH, dPCR allows the simultaneous detection of the EGFRvIII variant, which has been shown to be a potential therapeutic target [26]. Lower copy number values observed between EGFR2 and EGFR3 amplicons are very likely explained by the presence of the EGFRvIII variant. The EGFR3 amplicon is not located in a specific gene region affected by recurrent splicing variants or deletions but is located between exon 25 and the C-terminal region. The qualitative discrepancy between EGFR1 and EGFR2 amplicons for detecting the gene copy number from the same tumor is probably due to breakpoint variability of the EGFRvIII variant. The EGFR1 amplicon is located at the start of intron 1 in an area containing various breakpoints for the EGFRvIII splicing variant [27]. The EGFR1 amplicon may therefore match with tumor DNA if the breakpoint is closer to exon 2 but may mismatch with the tumor DNA if the breakpoint is closer to exon 1. As shown in our results, the value of using EGFR2 resides in its location within the spliced area, regardless of the breakpoint. Therefore, the comparison of the copy number estimation using EGFR2 and EGFR3 assays should be a more sensitive method than dPCR using an amplicon located at the end of exon 1 [27, 28].
NGS-based CNV identification using panels dedicated to glioblastoma has been demonstrated to be as sensitive as FISH or CGH array [25, 29]. In our exploratory cohort, the diagnostic performance for the detection of EGFR amplification was 100% when comparing dPCR and EGFR-targeted NGS. The major advantage of NGS resides in the fact that a single assay may detect somatic point mutations and multiple CNVs. However, the cost of a single NGS assay remains high, which currently hampers its routine use. Moreover, it has also been reported that multiple CNVs may be easily detected with the LNA-probe hydrolysis dPCR method without any proportional cost increase, for example, the concomitant detection of other amplicons located on MET, PDGFRA, KIT, AKT1 or CDKN2A homozygous deletion [30]. Moreover, the quantity of tumor DNA necessary is lower for dPCR than for NGS, making this technique more suitable for small tumor fragments, including those derived from biopsies or fragments containing low amounts of tumor DNA, notably in the case of tumor necrosis. Detection of CNV by NGS requires the comparison between patient-matched and unmatched normal tissue. In the specific situation of EGFR amplification detection in glioblastoma sample, the ideal comparison tissue should have been healthy brain tissue, which is virtually impossible to obtain in daily practice [31, 32]. The major advantage of dPCR use is that there is no need for healthy brain tissue since HMBS reference gene is not altered in tumor samples.
At last, our results are based on experiments using LNA-probes provided by Roche®. The experimental procedure is not restricted to specific manufacturer’s probes and could easily be used with other manufacturers’ LNA-probes provided that these probes are designed to be used at a hybridization temperature of 56 °C.
In conclusion, our results highlight that the dPCR assay using LNA-hydrolysis probes allowed the simultaneous detection of the EGFR amplification and EGFRvIII variant and may be used routinely in patients treated for glioblastoma.
Supplementary information
Additional file 1: Supplementary Table. Details of EGFR amplicons by NGS.
Additional file 2: Supplementary Data. Results of LD-RT-PCR for EGFRvIII detection.
Additional file 4: Supplementary Material: cost evaluation of FISH method.
Abbreviations
- aCGH
Array-comparative genomic hybridization
- CNV
Copy number variation
- dPCR
Digital PCR
- ddPCR
Droplet digital PCR
- EGFR
Epidermal Growth Factor Receptor
- EGFRvIII
EGFR variant III
- FISH
Fluorescence in situ hybridization
- HMBS
Hydroxymethylbilane synthase
- LD-RT-PCR
Ligation-dependent reverse transcription polymerase chain reaction
- LNA
Locked nucleic acid
- IDH
Isocitrate dehydrogenase
- MGMT
O-6-methylguanine-DNA methyltransferase
- NGS
Next-generation sequencing
- ROC
Recursive operating curve
- TERT
Telomerase reverse transcriptase
- UPL
Universal Probe Library®
- wt
Wild-type
Authors’ contributions
MF, FC and NSV concepted the study, organized the technical support and analyzed the data. MF, NSV, FC, FM, AL and FDF wrote the manuscript; MF, DR, CA, IT, OL, FDF, FC enrolled patients; FM, PR, CB, AD, LB, MV, PJV contributed to experiment performance for FISH and LD-RT-PCR. KC and PC designed dPCR experiments and developed dPCR-based method to detect somatic CNA. All authors contributed to and approved the final manuscript.
Funding
This project was supported by the Institute of Research and Innovation in Biomedicine of Normandy (IRIB, Rouen, Normandie, France) and by Cancer Center Henri Becquerel.
Availability of data and materials
Deidentified data are available on request.
Ethics approval and consent to participate
An informed written consent to participate to the study was obtained from all patients and the French National Committee for the Protection of Persons approved the study (RCB ID 2015- A00377–42).
Consent for publication
Not applicable.
Competing interests
Maxime Fontanilles: Reports Honoria from Bristol-Myers Squibb® and Congress Fee from La Roche-Hauffman®.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Florian Clatot and Nasrin Sarafan-Vasseur contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40478-020-00917-6.
References
- 1.Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro-oncology. 2017;19:v1–v88. doi: 10.1093/neuonc/nox158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 4.Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quan AL, Barnett GH, Lee S-Y, Vogelbaum MA, Toms SA, Staugaitis SM, et al. Epidermal growth factor receptor amplification does not have prognostic significance in patients with glioblastoma multiforme. Int J Radiat Oncol Biol Phys. 2005;63:695–703. doi: 10.1016/j.ijrobp.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 6.Lassman AB, Aldape KD, Ansell PJ, Bain E, Curran WJ, Eoli M, et al. Epidermal growth factor receptor (EGFR) amplification rates observed in screening patients for randomized trials in glioblastoma. J Neuro-Oncol. 2019;144:205–210. doi: 10.1007/s11060-019-03222-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.French PJ, Eoli M, Sepulveda JM, de Heer I, Kros JM, Walenkamp A, et al (2019) Defining EGFR amplification status for clinical trial inclusion. Neuro-oncology 21(10):1263-1272. [DOI] [PMC free article] [PubMed]
- 8.An Z, Aksoy O, Zheng T, Fan Q-W, Weiss WA. Epidermal growth factor receptor and EGFRvIII in glioblastoma: signaling pathways and targeted therapies. Oncogene. 2018;37:1561–1575. doi: 10.1038/s41388-017-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weller M, Butowski N, Tran DD, Recht LD, Lim M, Hirte H, et al. ACT IV trial investigators. Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017;18:1373–1385. doi: 10.1016/S1470-2045(17)30517-X. [DOI] [PubMed] [Google Scholar]
- 10.Uhm JH, Ballman KV, Wu W, Giannini C, Krauss JC, Buckner JC, et al. Phase II evaluation of gefitinib in patients with newly diagnosed grade 4 astrocytoma: Mayo/north central Cancer treatment group study N0074. Int J Radiat Oncol Biol Phys. 2011;80:347–353. doi: 10.1016/j.ijrobp.2010.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen PY, Chang SM, Lamborn KR, Kuhn JG, Norden AD, Cloughesy TF, et al. Phase I/II study of erlotinib and temsirolimus for patients with recurrent malignant gliomas: north American brain tumor consortium trial 04-02. Neuro-oncology. 2014;16:567–578. doi: 10.1093/neuonc/not247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sepúlveda-Sánchez JM, Vaz MÁ, Balañá C, Gil-Gil M, Reynés G, Gallego Ó, et al. Phase II trial of dacomitinib, a pan-human EGFR tyrosine kinase inhibitor, in recurrent glioblastoma patients with EGFR amplification. Neuro-oncology. 2017;19:1522–1531. doi: 10.1093/neuonc/nox105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Bent M, Eoli M, Sepulveda JM, Smits M, Walenkamp A, Frenel J-S, et al (2019) INTELLANCE 2/EORTC 1410 randomized phase II study of Depatux-M alone and with temozolomide vs temozolomide or lomustine in recurrent EGFRamplified glioblastoma. Neuro-oncology. In press. [DOI] [PMC free article] [PubMed]
- 14.Brat DJ, Aldape K, Colman H, Holland EC, Louis DN, Jenkins RB, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018;136:805–810. doi: 10.1007/s00401-018-1913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stichel D, Ebrahimi A, Reuss D, Schrimpf D, Ono T, Shirahata M, et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018;136:793–803. doi: 10.1007/s00401-018-1905-0. [DOI] [PubMed] [Google Scholar]
- 16.Cassinari K, Quenez O, Joly-Hélas G, Beaussire L, Le Meur N, Castelain M, et al. A simple, universal, and cost-efficient digital PCR method for the targeted analysis of copy number variations. Clin Chem. 2019;65:1153–1160. doi: 10.1373/clinchem.2019.304246. [DOI] [PubMed] [Google Scholar]
- 17.Boeva V, Popova T, Lienard M, Toffoli S, Kamal M, Le Tourneau C, et al. Multi-factor data normalization enables the detection of copy number aberrations in amplicon sequencing data. Bioinformatics. 2014;30:3443–3450. doi: 10.1093/bioinformatics/btu436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bobée V, Ruminy P, Marchand V, Viailly P-J, Abdel Sater A, Veresezan L, et al. Determination of molecular subtypes of diffuse large B-cell lymphoma using a reverse transcriptase multiplex ligation-dependent probe amplification classifier: a CALYM study. J Mol Diagn. 2017;19:892–904. doi: 10.1016/j.jmoldx.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Piton N, Ruminy P, Gravet C, Marchand V, Colasse É, Lamy A, et al. Ligation-dependent RT-PCR: a new specific and low-cost technique to detect ALK, ROS, and RET rearrangements in lung adenocarcinoma. Lab Investig. 2018;98:371–379. doi: 10.1038/labinvest.2017.124. [DOI] [PubMed] [Google Scholar]
- 20.Drieux F, Ruminy P, Abdel-Sater A, Lemonnier F, Viailly P-J, Fataccioli V, et al (2019) Defining the signatures of peripheral T-cell lymphoma, with a targeted 20-markers gene expression profiling assay (RT-MLPA). Haematologica. In press. [DOI] [PMC free article] [PubMed]
- 21.Chiu A, Ayub M, Dive C, Brady G, Miller CJ. Twoddpcr: an R/bioconductor package and shiny app for droplet digital PCR analysis. Bioinformatics. 2017;33:2743–2745. doi: 10.1093/bioinformatics/btx308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramkissoon SH, Bi WL, Schumacher SE, Ramkissoon LA, Haidar S, Knoff D, et al. Clinical implementation of integrated whole-genome copy number and mutation profiling for glioblastoma. Neuro-oncology. 2015;17:1344–1355. doi: 10.1093/neuonc/nov015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNulty SN, Cottrell CE, Vigh-Conrad KA, Carter JH, Heusel JW, Ansstas G, et al. Beyond sequence variation: assessment of copy number variation in adult glioblastoma through targeted tumor somatic profiling. Hum Pathol. 2019;86:170–181. doi: 10.1016/j.humpath.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Sharifi Z, Abdulkarim B, Meehan B, Rak J, Daniel P, Schmitt J, et al. Mechanisms and antitumor activity of a binary EGFR/DNA-targeting strategy overcomes resistance of Glioblastoma stem cells to Temozolomide. Clin Cancer Res. 2019;25:7594–7608. doi: 10.1158/1078-0432.CCR-19-0955. [DOI] [PubMed] [Google Scholar]
- 27.Koga T, Li B, Figueroa JM, Ren B, Chen CC, Carter BS, et al. Mapping of genomic EGFRvIII deletions in glioblastoma: insight into rearrangement mechanisms and biomarker development. Neuro-oncology. 2018;20:1310–1320. doi: 10.1093/neuonc/noy058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxena D, Sheikh S, Kao G, Binder ZA, Alonso-Basanta M, O’Rourke DM, et al. Rapid and ultrasensitive digital PCR (dPCR) profiling of EGFRvIII in tumor cells and tissues. Neurooncol Adv. 2019;1:vdz030. doi: 10.1093/noajnl/vdz030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zacher A, Kaulich K, Stepanow S, Wolter M, Köhrer K, Felsberg J, et al. Molecular diagnostics of Gliomas using next generation sequencing of a Glioma-tailored gene panel. Brain Pathol. 2017;27:146–159. doi: 10.1111/bpa.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller ML, Tome-Garcia J, Waluszko A, Sidorenko T, Kumar C, Ye F, et al. Practical Bioinformatic DNA-sequencing pipeline for detecting oncogene amplification and EGFRvIII mutational status in clinical Glioblastoma samples. J Mol Diagn. 2019;21:514–524. doi: 10.1016/j.jmoldx.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuśmirek W, Szmurło A, Wiewiórka M, Nowak R, Gambin T (2019) Comparison of kNN and k-means optimization methods of reference set selection for improved CNV callers performance. BMC Bioinformatics [cited 2020 Feb 26];20. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6537193/ [DOI] [PMC free article] [PubMed]
- 32.Grasso C, Butler T, Rhodes K, Quist M, Neff TL, Moore S, et al. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn. 2015;17:53–63. doi: 10.1016/j.jmoldx.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table. Details of EGFR amplicons by NGS.
Additional file 2: Supplementary Data. Results of LD-RT-PCR for EGFRvIII detection.
Additional file 4: Supplementary Material: cost evaluation of FISH method.
Data Availability Statement
Deidentified data are available on request.