Abstract
Daylight vision in most mammals is mediated predominantly by a middle/long wavelength-sensitive (M/LWS) pigment. Although spectral sensitivity and associated shifts in M/LWS are mainly determined by five critical sites, predicted phenotypic variation is rarely validated, and its ecological significance is unclear. We experimentally determine spectral tuning of M/LWS pigments and show that two highly divergent taxa, the gerbil and the elephant-shrew, have undergone independent dramatic blue-green shifts to 490 nm. By generating mutant proteins, we identify additional critical sites contributing to these shifts. Our results, which extend the known range of spectral tuning of vertebrate M/LWS, provide a compelling case of functional convergence, likely related to parallel adaptive shifts from nocturnal to brighter light conditions in similar habitats.
Keywords: opsin, middle/long wavelength-sensitive pigment, spectral tuning, functional convergence
Light sensitivity to daylight conditions in most mammals is conferred by a middle/long wavelength-sensitive (M/LWS) pigment, normally complemented by a short wavelength-sensitive visual pigment (SWS1), and rhodopsin (RH1) for dim-light vision, with the pigment opsin encoded by the genes OPN1MW/LW, OPN1SW, and RHO, respectively (1). While some lineages have lost the SWS1 opsin via relaxed selection linked to low-light niches (2), the M/LWS opsin is functional across mammals, with a few exceptions (3).
Evidence, to date, indicates that the spectral tuning of M/LWS pigments (499 nm to 571 nm) (4), measured by the wavelength of maximum sensitivity (λmax), is mainly controlled by amino acid identity at five critical sites (positions 180, 197, 277, 285, and 308) (5), although other sites (e.g., 213 and 294) may also affect spectral tuning via epistatic interactions (6). Predicted λmax values from known critical sites imply spectral shifts in M/LWS pigments during the diversification of mammals (7), yet these have rarely been validated (5), and their ecological significance is not known.
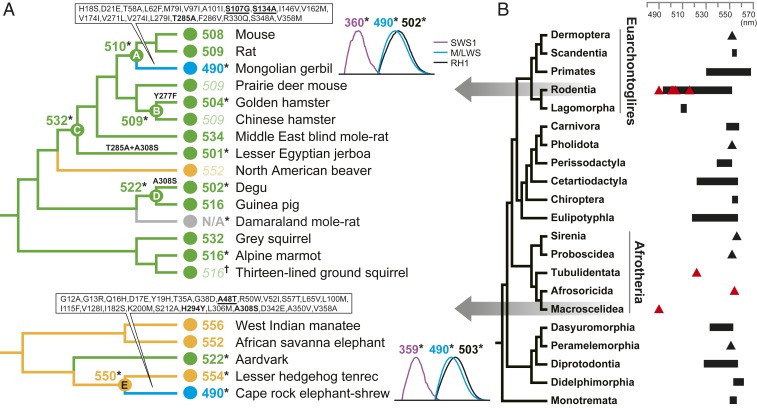
Here we determined the spectral phenotypes of M/LWS pigments in the highly divergent mammalian clades Rodentia (superorder Euarchontoglires) and Afrotheria, for which sequence-based predictions have suggested marked short-wavelength spectral shifts (7). Our results indicate that five rodents—the alpine marmot, degu, golden hamster (“hamster”), lesser Egyptian jerboa (“jerboa”), and Mongolian gerbil (“gerbil”)—are all sensitive to shorter wavelengths of light (516, 502, 504, 501, and 490 nm, respectively) than the predicted values (Dataset S1). We also found that the pigment of one species, the Damaraland mole-rat, showed no readable peak despite possessing an intact open reading frame, implying a potentially unstable pigment (Fig. 1).
Fig. 1.
Phenotypic evolution of M/LWS pigments in Rodentia and Afrotheria. (A) Values of λmax for M/LWS for extant and ancestral taxa. The values obtained in this study by either direct measurement (*) or estimation from critical sites (†). Published λmax values are based on either direct measurements or sequence-based predictions (Dataset S1). Across the two clades, branches and λmax values are colored by wavelength sensitivity, as yellow (550 nm to 556 nm), green (≤549 nm), and blue (490 nm). Gray indicates no value. Predicted values are shown in italics. For branches leading to gerbil and elephant-shrew, all amino acid substitutions are shown, with critical sites identified here underlined. Other focal branches show known critical substitutions only (bold) (7). For gerbil and elephant-shrew, recorded absorbance peaks (difference spectra) and λmax values for all three pigments are shown. (B) Phylogenetic tree showing position of the focal orders Rodentia and Macroscelidea (7). Published λmax values for each mammalian order are shown as either ranges (black bars) or single point values (black triangles). The λmax values measured in this study are indicated by red triangles (Dataset S1).
We also performed M/L opsin in vitro expression assays for representatives of three orders of Afrotheria: the aardvark (order Tubulidentata), Cape rock elephant-shrew (Macroscelidea), and lesser hedgehog tenrec (Afrosoricida), hereafter referred to as elephant-shrew and tenrec. For the aardvark and tenrec, the respective spectral peaks (522 and 554 nm) were similar to values predicted by sequences (7). In contrast, the elephant-shrew’s pigment had a λmax of 490 nm, showing a wide discrepancy (32 nm) with the predicted value (7) (Fig. 1). The highly divergent elephant-shrew and gerbil appear to have both undergone dramatic functionally convergent shifts (−60 and −20 nm) in M/L opsin sensitivity toward blue-green light, extending the lowest known limits for vertebrates (4). Both taxa occur in similar desert and/or grassland habitats (8, 9), and also show more daylight activity patterns than their respective closest relatives (e.g., tenrec or mouse/rat in Fig. 1) (10). Thus the observed spectral changes are likely to be associated with transitions from a nocturnal niche to one characterized by a wider range of light conditions.
To determine the molecular basis of spectral tuning, we performed in vitro expression of ancestral proteins (Fig. 1), and, using site-directed mutagenesis, generated mutants containing derived substitutions. For the degu, hamster, and jerboa, differences in spectral sensitivity between the expressed derived and respective ancestral proteins could be explained by observed substitutions at known critical sites (Y277F, T285A, and A308S). In contrast, the gerbil and elephant-shrew were each characterized by an amino acid substitution at a single critical site (T285A and A308S, respectively). Introducing these residues into the ancestral proteins did not recover the recorded λmax value of the wild types. We therefore searched the protein sequences of each taxon for other potentially important substitutions on the basis of changes in the presence of a hydroxyl group (11). For gerbil M/LWS pigment, we found that S107G and S134A gave rise to spectral shifts in λmax (−3 and −6 nm, respectively), and that the combination of S107G, S134A, and T285A recovered the λmax shift to 490 nm seen in the wild-type gerbil (Dataset S2).
We repeated this approach for the elephant-shrew, and found a critical site (position 48), with an A48T mutation producing a −9-nm λmax shift. This species also showed an amino acid substitution (H294Y) at a known critical site (6) that resulted in a −3-nm shift. When combined with the A308S critical replacement, these three substitutions caused a 44-nm λmax shift toward blue-green range, which is still 16 nm short of the observed λmax (Dataset S2). Our findings thus indicate that lineages of two distantly related superorders of mammals have undergone tuning to blue-green light via nonidentical molecular routes, and that shifts in the latter stem, in part, from unknown molecular mechanisms.
Both identified critical sites associated with large spectral shifts (i.e., S134A in gerbil and A48T in elephant-shrew) occur in exon 2. By comparing orthologous exonic sequence across vertebrates, we found the same S134A replacement in some ray-finned fishes, lobe-finned fishes, and jawless fishes (Dataset S3). Thus the mechanism underlying the −6-nm λmax shift in the gerbil may have evolved independently in multiple vertebrate lineages. Indeed, the S134A replacement may also explain tuning in the cave fish, where a reported shift from 564 nm to 558 nm could not be fully explained by the five critical sites (5). The second replacement, A48T, was only seen in hedgehog (Dataset S3).
While our results from the gerbil and elephant-shrew M/LWS reveal a possible link between molecular adaptations and ecological shifts, their daylight color vision is also mediated by SWS1, and possibly by RH1 pigment under mesopic conditions (1). To gain a more complete picture of visual phenotypes in these taxa, we also expressed these two pigments in vitro. We found that both of the SWS1 pigments are ultraviolet sensitive (λmax 360 and 359 nm, respectively) and that RH1 has near-identical sensitivities (502 and 503 nm) (Fig. 1). Moreover, neither SWS1 nor RH1 shows clear changes since the ancestral state (12, 13). We thus conclude that parallel transitions in color vision range of these two highly divergent mammals have been driven by functionally convergent changes in their M/LWS opsins, and may reflect ecological or temporal convergence on similar photic niches.
Materials and Methods
To interrogate reported shifts in M/LWS spectral sensitivity in rodents (Rodentia, Euarchontoglires) and some lineages of Afrotheria, we expressed pigments in vitro using published sequences (Dataset S1) and directly measured spectral sensitivity following previous experimental protocols (7). We confirmed extreme phenotypes by repeating these steps based on either newly generated sequences (Damaraland mole-rat and gerbil) or additional published sequence from elephant-shrew (Datasets S1 and S2). For the latter two taxa, we also determined the phenotypes for SWS1 and RH1 pigments.
To elucidate the molecular basis of phenotypic change in the Rodentia and Afrotheria clades, we generated variants of ancestral pigments (A to E in Fig. 1) in which we introduced one or more of the derived residues by PCR-based site-directed mutagenesis (7). We identified more critical sites and screened for these in other vertebrate species in National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/).
Data Availability.
All data discussed are available in Datasets S1–S3.
Supplementary Material
Acknowledgments
We thank R. Crouch (Medical University of South Carolina) and L. Neuhold (NIH) for 11-cis-retinal, and C. Faulkes for providing DNA. This study was funded by grants awarded to Y.L. by the National Natural Science Foundation of China (Grant 31601855) and Shaanxi Normal University, and European Research Council Starting Grant 310482 to S.J.R.
Footnotes
The authors declare no competing interest.
Data deposition: Data related to this work have been deposited in GenBank (accession nos. MT024768 and MT024769).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2002235117/-/DCSupplemental.
References
- 1.Davies W. I., Collin S. P., Hunt D. M., Molecular ecology and adaptation of visual photopigments in craniates. Mol. Ecol. 21, 3121–3158 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Emerling C. A., Huynh H. T., Nguyen M. A., Meredith R. W., Springer M. S., Spectral shifts of mammalian ultraviolet-sensitive pigments (short wavelength-sensitive opsin 1) are associated with eye length and photic niche evolution. Proc. Biol. Sci. 282, 20151817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Springer M. S., et al. , Inactivation of cone-specific phototransduction genes in rod monochromatic cetaceans. Front. Ecol. Evol. 4, 61 (2016). [Google Scholar]
- 4.Hart N. S., et al. , Visual opsin diversity in sharks and rays. Mol. Biol. Evol. 37, 811–827 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Yokoyama S., Radlwimmer F. B., The molecular genetics and evolution of red and green color vision in vertebrates. Genetics 158, 1697–1710 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsumoto Y., et al. , Evolutionary renovation of L/M opsin polymorphism confers a fruit discrimination advantage to ateline New World monkeys. Mol. Ecol. 23, 1799–1812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y., Chi H., Li L., Rossiter S. J., Zhang S., Molecular data support an early shift to an intermediate-light niche in the evolution of Mammals. Mol. Biol. Evol. 35, 1130–1134 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Smit H. A., Robinson T. J., Van Vuuren B. J., Coalescence methods reveal the impact of vicariance on the spatial genetic structure of Elephantulus edwardii (Afrotheria, Macroscelidea). Mol. Ecol. 16, 2680–2692 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., Liu W., Wang G., Wan X., Zhong W., Home-range sizes of social groups of Mongolian gerbils Meriones unguiculatus. J. Arid Environ. 75, 132–137 (2011). [Google Scholar]
- 10.Bennie J. J., Duffy J. P., Inger R., Gaston K. J., Biogeography of time partitioning in mammals. Proc. Natl. Acad. Sci. U.S.A. 111, 13727–13732 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekharan S., Katayama K., Kandori H., Morokuma K., Color vision: “OH-site” rule for seeing red and green. J. Am. Chem. Soc. 134, 10706–10712 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yokoyama S., Tada T., Liu Y., Faggionato D., Altun A., A simple method for studying the molecular mechanisms of ultraviolet and violet reception in vertebrates. BMC Evol. Biol. 16, 64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., et al. , Scotopic rod vision in tetrapods arose from multiple early adaptive shifts in the rate of retinal release. Proc. Natl. Acad. Sci. U.S.A. 116, 12627–12628 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed are available in Datasets S1–S3.