Significance
Progress in studying the androgen receptor (AR), the primary drug target in prostate cancer, has been hampered by challenges in expressing and purifying active multidomain AR for use in cell-free biochemical reconstitution assays. Here we successfully express full-length and truncated AR variants and demonstrate that the oncogenic ETS protein ERG, responsible for half of all prostate cancers, enhances the ability of AR to bind DNA through direct interaction with AR. In addition to providing a biochemical system to evaluate AR activity on different DNA templates, our findings provide insight into why ERG-positive prostate cancers have an expanded AR cistrome.
Keywords: prostate cancer, cistrome, antiandrogen
Abstract
The androgen receptor (AR) is a type I nuclear hormone receptor and the primary drug target in prostate cancer due to its role as a lineage survival factor in prostate luminal epithelium. In prostate cancer, the AR cistrome is reprogrammed relative to normal prostate epithelium and particularly in cancers driven by oncogenic ETS fusion genes. The molecular basis for this change has remained elusive. Using purified proteins, we report a minimal cell-free system that demonstrates interdomain cooperativity between the ligand (LBD) and DNA binding domains (DBD) of AR, and its autoinhibition by the N terminus of AR. Furthermore, we identify ERG as a cofactor that activates AR’s ability to bind DNA in both high and lower affinity contexts through direct interaction within a newly identified AR-interacting motif (AIM) in the ETS domain, independent of ERG’s own DNA binding ability. Finally, we present evidence that this interaction is conserved among ETS factors whose expression is altered in prostate cancer. Our work highlights, at a biochemical level, how tumor-initiating ETS translocations result in reprogramming of the AR cistrome.
The androgen receptor (AR) is a nuclear hormone receptor transcription factor critical for development and maintenance of normal prostate tissue, and a driver of castration-resistant prostate cancer (CRPC) (1). AR-targeted therapies are primarily used to treat CRPC, the most widely used being the androgen synthesis inhibitor abiraterone (2) and the antiandrogen, enzalutamide (ENZ) (3). However, half of patients treated with antiandrogens develop resistance through genomic amplification of AR (4). A major challenge in the design of better AR targeting drugs is the limited molecular understanding of: how AR binds DNA in the presence of androgen and ENZ, how the different domains of AR communicate allosterically, and how AR is regulated by protein cofactors. This problem is further exacerbated by the lack of a biochemical system to study AR DNA binding using active, multidomain purified proteins.
AR is composed of a N-terminal transactivation domain (NTD), a DNA binding domain (DBD), and a ligand binding domain (LBD) that binds androgens (5). In the absence androgens, AR is cytoplasmic and bound in a transcriptionally inactive conformation by molecular chaperones (6). Upon dihydrotestosterone (DHT) binding, AR is released and shuttled to the nucleus where it dimerizes and specifically regulates transcription of genes bearing partially palindromic hexameric sequences known as androgen response elements (AREs) (7, 8). Regulation of AR activity involves interaction with a diverse range of protein coactivators and corepressors (9). The best characterized of these is the hydrophobic interaction between a LXXLL or related motif within the protein cofactor with the Activation Function 2 (AF2) domain in the LBD of AR (10). AR is also regulated by an intramolecular interaction between the FXXLF motif in the NTD of AR and AF2 when AR is not in proximity to DNA (7, 8, 11, 12).
The AR cistrome in prostate cancers (PCas) is distinct from that in normal prostate cells, indicative of a shift in the repertoire of AR target genes (13, 14). Translocations of ETS family transcription factors are the most prevalent genetic lesions in prostate cancer (4, 15–17) and, in the case of the common TMPRSS2-ERG fusion, results in an expanded AR cistrome. In these instances, ERG has been postulated to be both AR activating (18–21) and repressive (22). These seemingly contradictory findings suggest that ERG modulation of AR activity is more complex than previously appreciated, an unsurprising notion given the vast heterogeneity of the AR transcriptional landscape and the degeneracy within the ARE consensus sequence (23–25). Furthermore, physical interaction of ERG with AR has been documented by mass spectrometry (26) and coimmunoprecipitation (22).
These findings raise the possibility that ERG, through direct binding to AR, may modulate its DNA binding activity. To address this question, we developed a minimal system using recombinantly purified AR and ERG to reconstitute AR binding to different DNA templates. We find that AR is autoinhibited by its NTD and that ERG directly stimulates the ability of AR to bind DNA through an AR coactivator motif present in the ETS domain of ERG. Our work uncovers a biochemical mechanism by which ERG expression alters AR binding and, consequently, the AR cistrome in prostate cells. These findings with ERG also appear relevant with other ETS factors whose expression is altered in PCa such as ETV1.
Results
Intramolecular Regulation of AR Binding to DNA.
To determine how the various domains of AR modulate its ability to bind DNA, we first purified recombinant DHT-bound AR variants with the N terminus intact (FL) or deleted (ΔNTD AR) using a bacterial expression system (Fig. 1A and SI Appendix, Fig. S1A). To control for potential ligand dissociation in the absence of the AR NTD (5, 27), we used supraphysiological concentrations of DHT in our assays. We then presented AR with a range of synthetic ARE containing double-stranded DNA (dsDNA) templates and measured binding via fluorescence polarization. DNA templates included a minimal ARE of 18 base pairs (bps) and two 43-bp dsDNAs with an internal ARE separated by a 6-bp insertion from either ETS consensus sites or scrambled (Scr) sequences on the 5′ and 3′ ends.
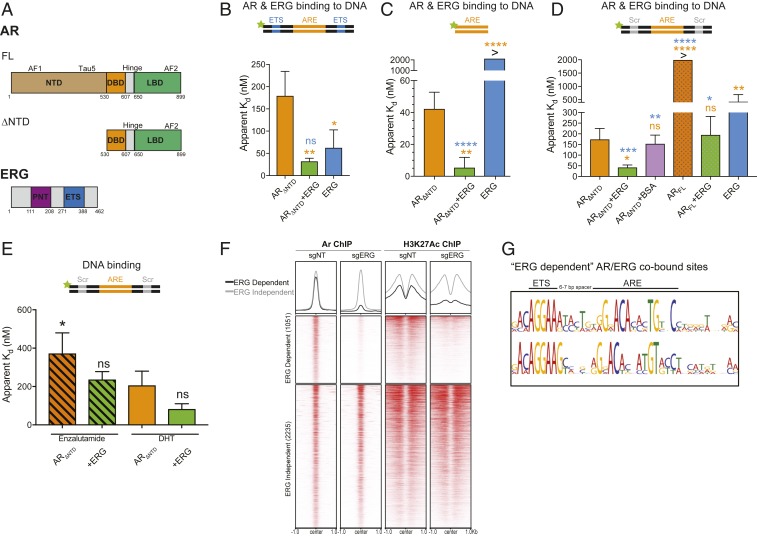
Fig. 1.
ERG directly stimulates AR independent of its DNA binding activity. (A) Domain structures of AR (FL and ∆NTD) and ERG. (B–E) AR binding to various 5′ fluorescein-labeled ARE dsDNA in the presence or absence of ERG was evaluated by fluorescence polarization. Data shown as mean ± SD (n = 3 technical replicates; one-way ANOVA, with orange and blue asterisks comparing to either AR and ERG, respectively). (B) ARΔNTD binding to ARE dsDNA containing an ETS consensus site. (C) ARΔNTD DNA binding on a minimal ARE sequence. (D) ARΔNTD and ARFL binding to ARE dsDNA bearing a scrambled (Scr) ETS site. (E) ARE/Scr DNA binding of antagonist (enzalutamide)- and agonist (DHT)-bound ARΔNTD. (F) ChIP-seq profile and heatmap derived from previously published Pten−/−;Rosa26-ERG mouse prostate organoids with ERG intact (sgNT) or depleted by CRISPR/Cas9-mediated knockdown (sgERG) (19). AR/ERG cobound peaks are separated into ERG dependent (AR binding is diminished by ERG knockout) and ERG independent (AR binding unchanged). (G) Top motif spacing of ERG/AR codependent peaks using SPAMO shows a 6- to 7-bp insertion is consistently observed between an ETS consensus motif and AREs. ****P ≤ 0.0001; ***P ≤ 0.001; **P ≤0.01; *P ≤ 0.05; ns, not significant, P > 0.05.
In all three dsDNAs tested, we observed ΔNTD AR bound to DNA at ∼100-nM affinity (Fig. 1 B–D), a high-affinity interaction similar to that reported for the closely related estrogen receptor and more distantly related type 2 nuclear hormone receptors (28–30), all of which lack the ∼50-kDa NTD. However, when the NTD was present, DNA binding by FL AR was impaired greater than 10-fold (Fig. 1C and SI Appendix, Fig. S1 B–D), consistent with earlier work using two-hybrid systems or fluorescence resonance energy transfer (FRET) to demonstrate an autoinhibitory interaction between the NTD and LBD (7, 27). Finally, to test whether a competitive antagonist of the LBD impacts the ability of AR to bind DNA, we expressed and purified ΔNTD AR in the presence of the antiandrogen enzalutamide (SI Appendix, Fig. S1E). Consistent with chromatin immunoprecipitation (ChIP) assays documenting that ENZ displaces AR binding from chromatin in cells (3, 31), we found that ENZ reduced DNA binding twofold relative to DHT in our minimal reconstitution system (Fig. 1E). This difference is comparable to the magnitude of DNA binding inhibition of the estrogen receptor induced by the antiestrogen fulvestrant under similar conditions using purified proteins (32). Taken together, these findings establish that both the NTD and the LBD allosterically modulate the ability of AR to bind DNA.
ERG Stimulates AR DNA Binding.
To test whether ERG influences the ability of AR to bind dsDNA, we generated purified full-length ERG, containing an N-terminal PNT transactivation domain and the DNA binding ETS domain (Fig. 1A). We observed cooperative binding between ERG and ΔNTD AR on ARE-ETS dsDNA, with a threefold increase in DNA binding in the presence of ERG (Fig. 1B). To discriminate between a purely additive effect of two transcription factors binding to DNA versus a cooperative event, we measured the ability of AR to bind to the two dsDNA templates that lacked ETS sites (ARE only; ARE-Scr). As expected, ERG bound both templates poorly (Fig. 1 C and D). However, ERG stimulated ΔNTD AR DNA binding on both, with AR plus ERG consistently exhibiting a threefold higher DNA binding affinity than AR alone on these DNA duplexes. This effect is specific to ERG as bovine serum albumin (BSA) failed to stimulate AR (Fig. 1C). We also observed in a system where AR levels were kept constant, ERG significantly enhanced DNA binding to the ARE-Scr (SI Appendix, Fig. S1F).
Importantly, ERG also partly relieved the autoinhibition of the NTD seen with FL AR, with dissociation constants comparable to that seen with ΔNTD AR (Fig. 1D and SI Appendix, Fig. S1 B–D). ERG also diminished the inhibitory effects of ENZ, with binding affinities resembling that seen with agonist-bound AR (Fig. 1E), consistent with recent observations that ERG overexpression confers relative resistance to AR-targeted therapies (19). Interestingly, ERG also enhanced AR binding to lower affinity ARE half sites (SI Appendix, Fig. S1G), which have been reported to comprise the vast majority of AR binding sites in PCa cells (23, 24, 33).
To determine whether AR binding to DNA is influenced by ERG in cells (in vivo), we examined AR and H3K27Ac ChIP-sequencing (ChIP-seq) data from isogenic mouse organoid lines derived from Pten−/−;Rosa26 ERG mice that underwent ERG deletion through CRISPR/Cas9-mediated gene editing (19). Among 3,286 enhancer sites cobound by AR and ERG, ∼30% lost AR binding upon ERG silencing. These “ERG-dependent” AR/ERG genes concomitantly lost the H3K27Ac mark of active enhancers (exemplified by multiple sites at the Plau gene locus), while “ERG-independent” AR/ERG cobound sites, such as in the Tmprss2 locus, maintained AR and H3K27Ac binding independent of ERG status (Fig. 1F, SI Appendix, Fig. S1H and Datasets S1 and S2). Transcriptome alterations by RNA-seq (19) mirrored the changes in ChIP-seq patterns (SI Appendix, Fig. S1I and Datasets S1 and S2), with selective up-regulation of prooncogenic transcriptional programs, including enhanced vascularization and cellular motility as evidenced by gene set enrichment analysis (GSEA) (SI Appendix, Fig. S1J). Motif analysis using MEME showed AR/ERG cobound sites contained both ERG and AR motifs regardless of whether AR binding was ERG dependent. We next queried the spacing between AR and ERG peaks and found that ERG-dependent AR binding sites were uniquely demarcated by 6 to 7 base pairs between ETS and ARE motifs (Fig. 1G). In contrast, no enriched spacing was detectable between ETS and ARE motifs within ERG-independent AR sites. These data suggest that in vivo, ERG can optimally interact with AR and modulate its activity when in close proximity.
Of note, the synthetic ARE/ETS duplex DNA described above for the fluorescence polarization assays (Fig. 1B and SI Appendix, Fig. S1B) contains a 6-bp insertion similar to that observed within ERG-dependent AR binding sites in vivo. Interestingly, this corresponds to approximately half a helical turn of DNA, which could favorably accommodate a direct interaction between AR and ERG. If this model is correct, then increased spacing between ARE and ETS sites would be predicted to interfere with this direct interaction, potentially diminishing the stimulatory effects of ERG on AR activity. Indeed, when the spacing between ARE and ETS sites was increased to 18 bp, no cooperativity between AR and ERG was detected (SI Appendix, Fig. S1K). In contrast, maximal enhancement of DNA binding between AR and ERG was observed with smaller spacing and when no ETS sequence was present (minimal ARE).
AR and ERG Interact through the ERG ETS Domain.
To evaluate the interaction between AR and ERG using an orthogonal method, we developed a qualitative DNA mobility shift assay with which we compared the effects of AR, ERG, or both proteins on unlabeled ARE/Scr dsDNA. AR alone shifted the entire population of dsDNA and produced a doublet, while ERG alone exhibited a mixture of free DNA and a high molecular weight smear, consistent with low-affinity binding to the scrambled ETS sequence. When AR and ERG were combined, the AR doublet mostly disappeared and a larger distinct species unique to the AR/ERG interaction formed (Fig. 2A).
Fig. 2.
AR and ERG interact through an AIM in the ERG ETS domain. (A) DNA gel shift of unlabeled ARE/Scr DNA in the presence of indicated concentrations of ARΔNTD and ERG. Gel (10% TBE PAGE) is stained with Sybr Gold. (B) Representative Sypro-stained SDS/PAGE gels of AR, ERG, and AR/ERG BM(PEG)2 cross-linked adducts in the presence (Right) of ARE/ETS DNA. (C) ARΔNTD binding to fluorescein-labeled ARE/Scr DNA with increasing concentrations of ERG. (D, Top) sequence of fluorescein-labeled ERG peptide. Putative AIMs in the ERG ETS domain are underlined. (D, Bottom) ARΔNTD binding to N-terminal fluorescein-labeled ERG peptides in the presence or absence of unlabeled ARE dsDNA. Peptides correspond to ERG WT or mutant AIMs or neighboring alpha-helix 2 (α2). NB indicates no binding detected at 2 µM. (E) AR binding to fluorescein-labeled ARE/ETS DNA in the presence and absence of ERG peptides. (F) ARΔNTD binding to fluorescein-labeled ARE/ETS DNA in the presence of full-length and increasing concentrations of ERGAIM peptide. (G) ARΔNTD binding to a fluorescein-labeled peptide corresponding to AF1 of the AR NTD in the presence or absence of unlabeled ARE dsDNA and full-length ERG. Data in C–G were acquired by fluorescence polarization and are shown as mean ± SD (n = 3 technical replicates; one way ANOVA, with orange and blue asterisks comparing to either AR and ERG, respectively). ****P ≤ 0.0001; ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05; ns, not significant, P > 0.05.
We next queried how AR and ERG interacted at the protein level using the cysteine cross-linker 1,8-bismaleimido-diethyleneglycol [BM(PEG)2]. Cross-linking either AR or ERG alone resulted in a laddering of AR or ERG adducts, respectively. Strikingly, when combined together, we observed preferential accumulation of intermediate-sized cross-linked species (Fig. 2B and SI Appendix, Fig. S2A). Western blotting confirmed both AR and ERG were present in these cross-linked species (SI Appendix, Fig. S2B). These trends were maintained with other cysteine cross-linkers and the primary amine cross-linker bis(sulfosuccinimidyl)suberate (BS3) (SI Appendix, Fig. S2 B and C). ERG similarly augmented cross-linking of FL AR (SI Appendix, Fig. S2D). To gauge the stoichiometry of this interaction, we titrated increasing amounts of full-length ERG relative to AR and measured AR binding to dsDNA. Although substoichiometric levels of ERG could modestly enhance AR binding to DNA, maximal stimulation was observed with equimolar ERG. No additive effect was seen with excess ERG (Fig. 2C). Together with providing further evidence of an AR/ERG complex, the formation of the ternary complex suggests the presence of unique interfaces accessible for cross-linking.
To identify the AR-interacting region of ERG, we first queried the protein sequence of ERG for hydrophobic stretches resembling the canonical LXXLL motif found in AR coregulators. We identified two overlapping sequences within the first alpha-helix of the ETS domain—WQFLL and LLELL—and tested whether a 17-residue peptide corresponding to this helix could directly bind AR. The peptide bound AR independent of DNA at low nanomolar Kd (Fig. 2D), an affinity similar to that observed for several AR coactivating peptides (34). Mutating several residues in the putative AIMs to alanine abolished binding, and a similar sized peptide corresponding to residues in the neighboring alpha-helix (⍺2) failed to bind AR.
We next tested whether the AIM-containing peptide was sufficient to stimulate AR. The AIM-containing peptide modestly (∼1.5-fold) but consistently inhibited AR binding to DNA. In contrast, the mutated AIM and ⍺2 peptides had no effect (Fig. 2E). This suggests that, while the ERG AIM peptide can bind AR, it is not sufficient to allosterically activate AR binding to DNA. Furthermore, excess AIM-containing peptide reversed the stimulatory effect of full-length ERG on AR DNA binding in a dose-dependent fashion using dsDNA templates with the ETS site intact (Fig. 2F) or scrambled (SI Appendix, Fig. S2E).
Because equimolar amounts of ERG AIM peptide failed to reverse the stimulatory effects of full-length ERG, we postulated that additional surfaces of ERG could possibly interact with AR. We individually mutated all five cysteines in ERG to serines to evaluate whether any of these substitutions abrogated the formation of AR-ERG sulfhydryl cross-links (SI Appendix, Fig. S2F). Consistent with our model, C312S resulted in the greatest attenuation of AR-ERG cross-links. This residue is the only cysteine in the ETS domain and is located in a loop adjacent to helix 1, proximal to the AIM. However, C28S and C77S also formed fewer AR-ERG cross-links, suggesting the ERG N terminus may also interact with AR.
We next measured how binding of the ERG AIM compared with that of the autoregulatory interaction between AF1 in the AR NTD and AF2 in the LBD. A fluorescein-labeled peptide containing the LBD-interacting motif in AF1 (FQNLF) bound ΔNTD AR with more than an order of magnitude reduced affinity compared to the ERG AIM peptide in the presence and absence of DNA (Fig. 2G), consistent with previous studies reporting micromolar affinities between AF1 and the AR LBD (10, 35). We measured a further decrease in AF1 binding to ΔNTD AR when full-length ERG was present (Fig. 2G), consistent with our observation that ERG activates full-length AR’s DNA binding activity.
Identification of Residues Important for ERG-Mediated Stimulation of AR Activity.
To further define the AR-interacting surface of ERG, we used structural modeling to identify helix 1 residues within the two distinct AIMs that, when mutated, likely compromise binding to AR (Fig. 3A). We introduced single and double alanine substitutions within the two putative AIMs of full-length ERG to assess their impact on AR binding to DNA. In four of the five ERG mutants tested, we observed defects in AR binding quantitatively on two ARE dsDNAs (Fig. 3B and SI Appendix, Fig. S3C), and qualitatively through gel shifts, with AIM mutants resembling AR alone, consistent with impaired association between AR and ERG (Fig. 3C and SI Appendix, Fig. S3D).
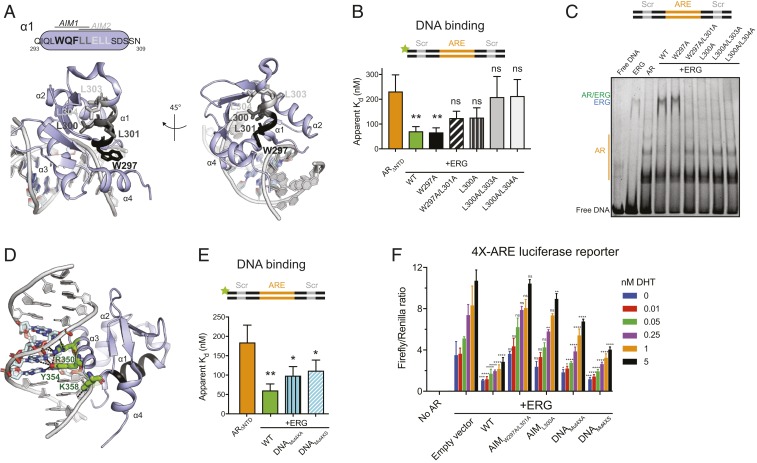
Fig. 3.
Identification of residues important for ERG-mediated stimulation of AR activity. (A) Mapping of AIMs onto helix 1 in the crystal structure of ERG bound to ETS dsDNA (Protein Data Bank [PDB]: 4IRI). Candidate AR-interacting residues are shown as sticks. Black residues correspond to putative AIM1, and light gray AIM2; dark gray is common to both motifs. This and other structure figures were generated with PyMol. (B) AR binding to fluorescein-labeled ARE/Scr DNA in the presence of WT and AIM mutant ERG. (C) EMSA of ARE/Scr dsDNA in the presence of ARΔNTD and WT and ERG variants. Gel (4 to 20% TBE PAGE) is stained with Sybr Gold. (D) ERG DNA binding residues targeted for mutagenesis (green sticks) mapped onto the structure in A. (E) AR binding to fluorescein-labeled ARE/Scr DNA in the presence of WT and DNA binding mutant ERG variants. Data in B and E were acquired by fluorescence polarization and are shown as mean ± SD (n = 3 technical replicates; one-way ANOVA). (F) Activation of a 4×-ARE firefly luciferase reporter by AR-VP16 and regulation of AR by WT and ERG mutants in HEK293 cells. Data were acquired 24 h after transfection and normalized to a control Renilla luciferase reporter. Data are shown as mean ± SD (n = 3 biological replicates; two-way ANOVA). ****P ≤ 0.0001; ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05; ns, not significant, P > 0.05.
We detected no difference in ERG expression or in binding to ETS dsDNA with three AIM 1 mutants (W297A, W297A/L301A, and L300A) (SI Appendix, Fig. S3 A, B, and E); however, mutants in AIM 2 residues (L303A or L304A) expressed poorly and failed to produce functional protein. Indeed, the crystal structure of ERG shows that L303 and L304 are an integral part of a conserved network of hydrophobic residues that form the core domain common to ETS family members (Fig. 3A) (36). Thus, the inability of these mutant proteins to stimulate AR is incidental due to protein destabilization resulting from disruption of critical intramolecular contacts, rather than a direct effect on AR binding. We therefore conclude the WXXLF motif in AIM 1 is largely responsible for ERG interaction with AR.
Having identified ERG mutants deficient in AR interaction but competent for DNA binding, we next sought to isolate mutants that failed to bind DNA but could stimulate AR. Because a previously reported mutant in the DNA binding domain of ERG (R350K) still retained residual DNA binding activity (37), we targeted three additional DNA binding residues to further inhibit base-specific and DNA backbone contacts (Fig. 3D). These two quadruple mutants, DNAMut4XA (K338E/R350K/Y354A/K358E) and DNAMut4XS (K338E/R350K/Y354S/K358E) both expressed comparably to wild-type (WT) ERG (SI Appendix, Fig. S3A) and, as predicted, failed to bind DNA (SI Appendix, Fig. S3E). When combined with AR, both ERG mutants stimulated AR, albeit not to the same extent as WT (Fig. 3E). In light of our earlier observation that ERG activates AR binding to dsDNA templates independent of an ETS site, the partial activation seen using an ERG mutant incapable of DNA binding could be explained by allosteric effects that compromise the AR-interacting surface.
To determine whether ERG separation of function mutants can selectively modulate AR transcriptional activity in vivo, we overexpressed ERG WT, AIM, or DNA binding mutants in the presence of an AR-VP16 fusion protein in HEK293 cells. Fusion to the VP16 transactivator induces constitutive nuclear localization and a robust, ligand-dependent transcriptional readout that is independent of coactivators (3, 38). AR activity is quantified with a luciferase reporter driven by four tandem AREs in a DHT-dependent fashion. Using this system, coexpression of WT ERG and ERG DNA binding mutants repressed AR transactivation, whereas ERG allele mutants defective in AR interaction (AIM mutants) had no effect (Fig. 3F). These results provide evidence that ERG modulates AR transcriptional activity in cells (in vivo) through direct interaction with AR via the AIM interface in helix 1, as delineated in our in vitro reconstitution studies.
Implications for AR Interaction with Other ETS Factors Altered in Prostate Cancer.
Although the TMPRSS2-ERG fusion is the most frequent ETS aberration in prostate cancer, genomic alterations impacting other ETS family members have also been identified, comprising up to 70% of PCa cases. These include oncogenic alterations in ETV1, ETV4, FLI1, ETV5, ELK4, ERF, and ELF1 (16, 18, 21, 39–43) that result in overexpression of factors silent in normal tissue, or decreased levels of tumor suppressors. To query whether the interaction between AR and the ERG AIM in helix 1 extends to other ETS family members, we performed a comparative analysis of known crystal structures and protein sequences of their ETS domains. In four structures of ETS proteins altered in PCa (36, 44–46), the AIM residues align well, are solvent exposed, and face helix 4 (Fig. 4A). The core ETS domain fold comprised of helices 1 through 3 exhibits little conformational variability; however, helix 4 is conformationally dynamic, with little structural and sequence similarity among ETS factors. Indeed, plasticity at the ERG interface between helices 1 and 4 has been observed by NMR and in several crystal structures (36).
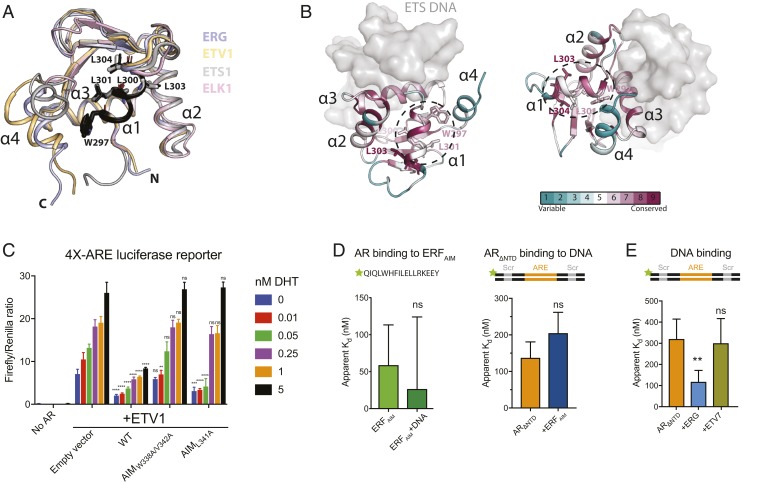
Fig. 4.
AR interaction with other ETS factors altered in prostate cancer. (A) Alignment of crystal structures of ETS factors altered in PCa from four distinct subfamilies shows conservation of the AIM (black) and conformational plasticity at the interface between α1 and α4. PDB codes: ERG, 4IRI; ETV1, 4BNC; ETS1, 2NNY; ELK1, 1DUX. (B) ERG AIM residues are conserved. Surface conservation of all ETS family members mapped onto the crystal structure of ERG and calculated with ConSurf (57). Predicted AR-interacting surface is highlighted in black dashed ellipse. (C) Activation of a 4×-ARE firefly luciferase reporter by AR-VP16 and regulation of AR by WT and ETV1 AR-interacting mutants in HEK293 cells. Data were acquired 24 h after transfection and normalized to a control Renilla luciferase reporter. Data are shown as mean ± SD (n = 3 biological replicates). (D, Left) AR binding to a fluorescein-labeled ERFAIM peptide. (D, Right) AR binding to fluorescein-labeled ARE/ETS DNA in the presence and absence of the ERFAIM peptide. (E) ARΔNTD binding to fluorescein-labeled ARE/Scr DNA in the absence or presence of the ETS factors ERG and ETV7. Data in D and E were acquired by fluorescence polarization and are shown as mean ± SD (n = 3 technical replicates; t test for D, one-way ANOVA for E). ****P ≤ 0.0001; ***P ≤ 0.001; **P ≤ 0.01; ns, not significant, P > 0.05.
We next mapped the sequence alignments of all 28 ETS factors onto the crystal structure of ERG to evaluate the sequence conservation of the putative AR-interacting region. Not surprisingly, residues comprising the network of intramolecular contacts forming the shared winged helix-turn-helix fold are the most highly conserved (Fig. 4B and SI Appendix, Fig. S4). However, the solvent-exposed AIM residues of helix 1 also display relatively high conservation, despite not being involved in protein stabilizing contacts (Fig. 4B). Helix 4, which is proximal to the AIM surface, shows no conservation, consistent with its conformational flexibility and potential to be displaced upon AR interaction.
To experimentally determine whether the AIM of ETS factors aberrantly amplified in PCa can similarly impact AR transactivation, we focused on ETV1, the second most frequent ETS translocation in PCa that has also been reported to reprogram the AR cistrome (18, 41). We measured DHT-dependent AR activity in the presence or absence of WT ETV1 and alanine mutants within its putative AIM, which bears the native sequence WQFLV. As with ERG (Fig. 3F), WT ETV1 exhibited a repressive effect on the AR-VP16 fusion, whereas the two AIM mutants failed to repress AR at high concentrations of DHT, when AR activity is most robust (Fig. 4C).
ERF is a recently identified tumor suppressor in PCa whose loss of function phenocopies ERG translocation in ERG negative PCa (21). Like ERG, ERF contains a putative AIM, WHFIL, which follows the pattern of WXXϕϕ in the ERG and ETV1 AIMs, where ϕ signifies a hydrophobic residue. Using the in vitro DNA binding assay, we found that the ERF AIM binds ΔNTD AR with nanomolar affinity, albeit slightly less than the ERG AIM, and similarly repressed the ability of AR to bind DNA (Fig. 4D). Finally, we recombinantly expressed and purified full-length ETV7 (SI Appendix, Fig. S5 A and B) (43), an ETS family member implicated in hematological malignancies that lacks the AIM sequence (WDYVY) and observed no effect on AR DNA binding (Fig. 4E). Collectively, these results implicate the WXXϕϕ motif in ETS proteins as an important mediator of AR activity.
Discussion
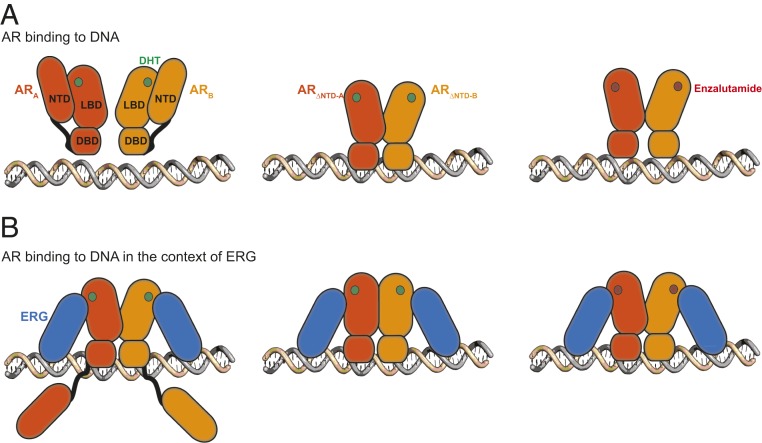
Progress in understanding the mechanism by which AR influences prostate cancer progression has been limited by the lack of biochemical systems to study the interaction of multidomain AR with DNA or with coactivators using purified proteins. Through optimization of conditions to express and purify active, multidomain AR from bacteria, we present here direct biochemical evidence that NTD of AR is autoinhibitory; that binding of the antiandrogen enzalutamide allosterically impedes DNA binding; and that ERG can stimulate AR through a direct interaction, independent of its own DNA binding activity, even in the face of both modes of inhibition (Fig. 5). Our model suggests that the effect of ERG on AR transactivation can be dictated through specific protein–protein interactions, rather than being a purely DNA-mediated composite effect of two adjacent transcription factors (47). We postulate that AR’s default autoinhibited state serves as a mechanism for selective tuning of AR transactivation upon interaction with relevant coactivators and corepressors within the proper genomic context. Disruption of this equilibrium through ERG amplification in prostate cancer could generate an imbalance of ERG/AR complexes relative to other coregulators, with resulting alteration of the AR cistrome and transcriptome.
Fig. 5.
A model of AR binding to DNA in the context of ERG. (A) AR binding to ARE DNA is autoinhibited by its NTD (Left and Middle). DHT is shown as green circles. Enzalutamide (red circles) impairs AR binding to DNA (Right). (B) ERG relieves AR NTD-mediated autoinhibition and stimulates AR binding to DNA (Left and Middle), even in the presence of enzalutamide (Right).
We identified an AIM within helix 1 of the ERG ETS domain that is partly responsible for the cooperative stimulation of AR by ERG. We propose that ERG binds to the AR AF2 through its AIM and displaces the intramolecular N/C interaction between the AR AF1 and AF2, thereby facilitating cooperative DNA binding and transactivation of AR/ERG codependent target genes. Competitive binding for AF2 and displacement of AF1 has been proposed for other AR transcriptional coactivators in cell-based assays (27, 48).
The identified ERG AIM diverges from the canonical LXXLL nuclear hormone receptor coregulator motif, with the sequence WXXLL. The AR AF2 is unique compared to other nuclear hormone receptors in its ability to preferentially bind divergent sequences with higher affinity, including FXXLF and WXXLF sites (10, 35, 49, 50). It stands to reason that AR activity can be modulated by other protein binding partners with similar hydrophobic motifs. Furthermore, it remains to be seen if ERG can interact with other related type I nuclear hormone receptors.
Conversely, solvent-exposed hydrophobic patches akin to the ERG AIM have been reported to mediate protein–protein interactions with other transcription factors within closely related winged helix transcription factors (51, 52). Interestingly, examination of primary sequences revealed that the AIM sequence was conserved in other ETS factors altered in prostate cancer (ETV1, ETV4, and ERF) but less so in ETS factors altered in hematological malignancies such as ETV6, ETV7, SPI1, and SPIB (SI Appendix, Fig. S4). We cannot exclude the possibility that additional surfaces unique to ERG contribute to AR interaction, as suggested by our cross-linking data (SI Appendix, Fig. S2F). An AIM-independent interaction with AR could potentially bear relevance for AR-V7, a clinically relevant AR splice variant lacking the LBD (53) that has recently been found to be enriched at AREs with flanking ETS motifs by ChIP-seq (54).
Our reconstituted system revealed, unexpectedly, that cooperative binding between ERG and AR is not exclusively dictated by the DNA sequence. Specifically, we found that ERG stimulates AR binding to high affinity palindromic ARE DNA consensus sites, as well as lower affinity ARE half sites, independent of an ETS consensus motif. The optimal 6- to 7-base pair spacing between AR and ETS binding sites that we observed on AR-ERG codependent genes in cells and in our synthetic dsDNA templates is intriguing and invokes at least two possible models: 1) that DNA can assist in orienting the two proteins for subsequent protein–protein interaction, and 2) that ERG can stimulate AR’s binding to DNA independent of ERG’s own DNA binding, consistent with recent evidence suggesting that AR and ERG form long-range interactions through chromosomal looping (55). Our biochemical observations also provide insight to previous findings in cancer cells that ERG amplification expands the AR cistrome to genes that otherwise have low AR occupancy (18–20), and that ETS factors can modulate AR binding to DNA when AR is otherwise impaired (Figs. 1E, 3F, and 4C and SI Appendix, Fig. S1G). The consequences of this cooperative interaction in enhancing AR activity and altering its sequence specificity within the context of the genetic and epigenetic landscape of CRPC will be a subject of future research.
Materials and Methods
Recombinant Protein Expression and Purification.
Genes corresponding to full-length human ETV7 and mouse AR were purchased as synthesized genes and codon optimized for expression in Escherichia coli (GeneArt Gene Synthesis, Thermo Fisher) and further cloned into pRSF-Duet1 or pET-Duet1 (Novagen) bearing an N-terminal Smt3 tag. Full-length human ERG isoform 2 was PCR amplified from a construct described previously (56) and cloned into pRSF-Duet1 with an N-terminal Smt3 fusion tag. The ERG point mutants C28S, C77S, C92S, C176S, C312S, W297A, W297A/L301A, L300A, L300A/L303A, L300/L304A, K338E/R350K/Y354A/K358E, and K338E/R350K/Y354S/K358E were generated by PCR-based site-directed mutagenesis with the following forward primers: C28S: tcgttgtttgagagtgcctacggaacgcca; C77S: atcaaaatggaaagtaaccctagccaggtg; C92S: tctcctgatgaaagcagtgtggccaaaggc; C176S: gggaaggaactgagcaagatgaccaaggac; C312S: tccaactccagcagcatcacctgggaaggc; W297A: cagatccagcttgcgcagttcctcctggagctc; W297A/L301A: cttgcgcagttcctcgcggagctcctgtcggacagc; L300A: ctttggcagttcgccctggagctcctgtcggac; L300A/L303A: cagttcgccctggaggccctgtcggacagctccaac; L300A/L304A: ttcgccctggagctcgcgtcggacagctccaactcc; K338E: cggcgctggggagagcgggagagcaaacccaac; R350K: gtagtaacggagggccttgctgagcttatc; Y354A: agcgaggccctccgtgcctactatgacgagaac; Y354S: agcgaggccctccgttcctactatgacgagaac; and K358E: ctccgttactactatgacgagaacatcatgacc. Expression plasmids were transformed into BL21DE3 codon plus cells (Novagen) and protein expression induced by addition of 0.1 mM isopropyl-β-D-thiogalactoside and overnight shaking at 16 °C. Cells were lysed by sonication or French press and supernatants purified by Ni-NTA (Qiagen), followed by affinity purification on heparin Hi-Trap (GE Healthcare), overnight cleavage of the Smt3 tag by Ulp1, and final purification by size exclusion chromatography on either Superdex 200 or Superdex 75 (GE Healthcare) in a final buffer of 350 mM NaCl, 40 mM Hepes pH 7.5, 1 mM Tris(2-carboxyethyl) phosphine (TCEP) for ETS proteins, and 350 mM NaCl, 40 mM Hepes pH 7.5, 1 mM TCEP, 5% glycerol, and either 20 μM DHT or enzalutamide for AR constructs.
DNA Binding Assays.
Unlabeled and 5′ fluorescein-labeled duplex DNAs were purchased from IDT and had the following sequences, with ARE sites in bold and ETS sites in italics: ARE: 5′ CCAGAACATCATGTTCTC
3′; ARE/Scr: 5′ TACCTAGCGTGGCCAGAACATCATGTTCTCCGGTGCGATCCAG 3′; ARE/ETS 6 bp: 5′ TACCGGAAGTGGCCAGAACATCATGTTCTCCGGTGAAGGCCAG 3′; ARE/ETS 10 bp: 5′ TACCGGAAGTGGCTACCCAGAACATCATGTTCTCCCATCGGTGAAGGCCAG 3′; ARE/ETS 18 bp: 5′ TACCGGAAGTGGCTACCCTAGTGGCCAGAACATCATGTTCTCCGGTGATCCCATCGGTGAAGGCCAG 3′; and ARE-Half-Site/Scr: 5′ AGACCTAGCGTGGCCAGAACATCATTAAGCCCGGTGCGATCCAG 3′. Binding buffer consisted of 150 mM NaCl, 40 mM Tris pH 8.0, 1 mM TCEP, 20 μM DHT or enzalutamide, 10% glycerol, and 0.01% Nonidet P-40. Unless otherwise indicated, equimolar amounts of AR and ERG were preincubated on ice for 30 min before mixing with the indicated dsDNA. For DNA gel shift assays, 200 nM of unlabeled DNA was incubated with either 250 nM or 1 μM (Fig. 2A) or 250 nM (Fig. 3C) of total protein on ice for 1 h. Gel shifted products were resolved on 10% or 4 to 20% Tris/borate/EDTA (TBE) PAGE and DNA stained with Sybr Gold (Thermo Fisher). For fluorescence polarization experiments measuring DNA binding, 100 nM fluorescein-labeled dsDNA was incubated for 30 min on ice with increasing concentrations of the indicated protein (0 to 4 μM final concentration). For competition assays, AR and ERG were preincubated on ice for 30 min before addition of the indicated amounts of unlabeled peptides for an additional 30 min before initiating DNA binding. For assays measuring DNA binding in the presence of unlabeled peptide, peptides were added in 4-molar excess to AR. Data from triplicate experiments were analyzed, and when applicable a model for receptor depletion was used to calculate apparent Kd values with Prism, GraphPad Software.
Peptide Binding by Fluorescence Polarization.
Unlabeled and N-terminally fluorescein-labeled peptides were purchased from Genscript and had the following sequences: ERGα1-AIM: QIQLWQFLLELLSDSSN; ERGα2: MTDPDEVARRWGERKSK; ERGα1-AIM-Mut: QIQLAQFAAEAASDSSN; ERF: QIQLWHFILELLRKEEY; and AF1Pep: KTYRGAFQNLFQSVREA. Using the binding buffer described above for DNA binding assays, 200 nM N-terminally labeled fluorescein peptides were incubated for 1 h on ice with increasing concentrations of ΔNTD AR (0 to 4 μM) in the presence or absence of equimolar ARE/Scr dsDNA and equimolar full-length ERG when indicated. Data from triplicate experiments were analyzed, and when applicable a model for receptor depletion was used to calculate apparent Kd values with Prism, GraphPad Software.
Protein Cross-Linking.
AR, ERG, or both were incubated in the presence or absence of unlabeled ARE/ETS dsDNA (6-bp insertion) at a final concentration of 10 μM for 1 h on ice and then dialyzed to 150 mM NaCl, 40 mM Hepes pH 7.5, 1 mM TCEP, 20 μM DHT, and 0.01% Nonidet P-40. Protein cross-linking was performed with the indicated concentration of cross-linker for 1 h on ice before being quenched with either 25 mM dithiothreitol for malemide cross-linkers BMOE, BM(PEG)2, and BM(PEG)3, or 50 mM Tris⋅HCl pH 8.0 for the amine cross-linker BS3. Cross-link identity was validated through Western blotting of AR (Abcam ab52615) and ERG (Santa Cruz sc-354). Total cross-links were resolved by SDS/PAGE and stained by Sypro Ruby (Bio-Rad). Quantitation of cross-linking was performed using densitometry (ImageJ).
AR Reporter Assays.
WT ERG, AIM, and DNA binding mutants were cloned into pCDNA3. WT ETV1 was purchased from Addgene (74981) and AIM point mutations corresponding to W338A/V342A and L341A were generated by PCR-mediated site-directed mutagenesis using the following forward primers: W338A/V342A: ggatcacttcagctcgcgcagtttttggcagctcttctggatgaccc and L341A: cagctctggcagtttgcggtagctcttctggatgacc. WT and mutant ETV1 were then cloned into pCDNA3.1. AR-VP16 and the 4×-ARE firefly luciferase reporter have been previously described (3). pRL-TK (Promega) was used as a Renilla luciferase normalization control. Plasmids were transfected into HEK293 cells in triplicate using Lipofectamine 2000 (Thermo Fisher) in the presence of various amounts of DHT, and luciferase activity was read 24 to 36 h after transfection using Dual Glo reagent (Promega). For quantification of the 4×-ARE reporter, firefly luciferase activity was normalized to Renilla and analyzed using two-way ANOVA, with ****P < 0.0001; n.s. (not significant), P > 0.05. Data presented as mean ± SD from n = 3 experiments.
Data Availability.
All data, associated protocols, methods, and sources of materials can be accessed in the text or SI Appendix.
Supplementary Material
Acknowledgments
This research was supported in part by the Department of Defense under award number W81XWH-18-1-0182 (E.V.W.); and National Cancer Institute grants CA193837, CA155169, CA224079, CA092629, CA160001, and CA008748 (C.L.S.). C.L.S. is an investigator of the Howard Hughes Medical Institute. We thank Shaun K. Olsen, Phillip J. Iaquinta, Brett S. Carver, and members of the C.L.S. and the S.K. laboratories for stimulating discussions. The content is solely the responsibility of the authors and does not represent the official views of the NIH.
Footnotes
Competing interest statement: C.L.S. serves on the Board of Directors of Novartis, is a cofounder of Overcoming Resistance in Cancer (ORIC) Pharmaceuticals, and coinventor of enzalutamide and apalutamide. He is a science advisor to Agios, Beigene, Blueprint, Column Group, Foghorn, Housey Pharma, Nextech, KSQ Therapeutics, Petra, and PMV Pharma.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922159117/-/DCSupplemental.
References
- 1.Watson P. A., Arora V. K., Sawyers C. L., Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 15, 701–711 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Bono J. S. et al.; COU-AA-301 Investigators , Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 364, 1995–2005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran C., et al. , Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324, 787–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson D., et al. , Integrative clinical genomics of advanced prostate cancer. Cell 162, 454 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Tan M. H., Li J., Xu H. E., Melcher K., Yong E. L., Androgen receptor: Structure, role in prostate cancer and drug discovery. Acta Pharmacol. Sin. 36, 3–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eftekharzadeh B., et al. , Hsp70 and Hsp40 inhibit an inter-domain interaction necessary for transcriptional activity in the androgen receptor. Nat. Commun. 10, 3562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaufele F., et al. , The structural basis of androgen receptor a ctivation: Intramolecular and intermolecular amino-carboxy interactions. Proc. Natl. Acad. Sci. U.S.A. 102, 9802–9807 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Royen M. E., van Cappellen W. A., de Vos C., Houtsmuller A. B., Trapman J., Stepwise androgen receptor dimerization. J. Cell Sci. 125, 1970–1979 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Brooke G. N., Parker M. G., Bevan C. L., Mechanisms of androgen receptor activation in advanced prostate cancer: Differential co-activator recruitment and gene expression. Oncogene 27, 2941–2950 (2008). [DOI] [PubMed] [Google Scholar]
- 10.He B., et al. , Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol. Cell 16, 425–438 (2004). [DOI] [PubMed] [Google Scholar]
- 11.He B., Kemppainen J. A., Voegel J. J., Gronemeyer H., Wilson E. M., Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH(2)-terminal domain. J. Biol. Chem. 274, 37219–37225 (1999). [DOI] [PubMed] [Google Scholar]
- 12.He B., Wilson E. M., Electrostatic modulation in steroid receptor recruitment of LXXLL and FXXLF motifs. Mol. Cell. Biol. 23, 2135–2150 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isaacs J. T., Resolving the coffey paradox: What does the androgen receptor do in normal vs. malignant prostate epithelial cells? Am. J. Clin. Exp. Urol. 6, 55–61 (2018). [PMC free article] [PubMed] [Google Scholar]
- 14.Pomerantz M. M., et al. , The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat. Genet. 47, 1346–1351 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor B. S., et al. , Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomlins S. A., et al. , Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research Network , The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y., et al. , ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat. Med. 19, 1023–1029 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao N., et al. , Aberrant expression of ERG promotes resistance to combined PI3K and AR pathway inhibition through maintenance of AR target genes. Mol. Cancer Ther. 18, 1577–1586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai C., et al. , ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. J. Clin. Invest. 123, 1109–1122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bose R. et al.; International SU2C/PCF Prostate Cancer Dream Team , ERF mutations reveal a balance of ETS factors controlling prostate oncogenesis. Nature 546, 671–675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J., et al. , An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 17, 443–454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q., et al. , A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol. Cell 27, 380–392 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson S., Qi J., Filipp F. V., Refinement of the androgen response element based on ChIP-Seq in androgen-insensitive and androgen-responsive prostate cancer cell lines. Sci. Rep. 6, 32611 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stelloo S., et al. , Endogenous androgen receptor proteomic profiling reveals genomic subcomplex involved in prostate tumorigenesis. Oncogene 37, 313–322 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Mounir Z., et al. , ERG signaling in prostate cancer is driven through PRMT5-dependent methylation of the Androgen Receptor. eLife 5, e13964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He B., Bowen N. T., Minges J. T., Wilson E. M., Androgen-induced NH2- and COOH-terminal interaction inhibits p160 coactivator recruitment by activation function 2. J. Biol. Chem. 276, 42293–42301 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Huang W., et al. , Multidomain architecture of estrogen receptor reveals interfacial cross-talk between its DNA-binding and ligand-binding domains. Nat. Commun. 9, 3520 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandra V., et al. , Multidomain integration in the structure of the HNF-4α nuclear receptor complex. Nature 495, 394–398 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandra V., et al. , The quaternary architecture of RARβ-RXRα heterodimer facilitates domain-domain signal transmission. Nat. Commun. 8, 868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asangani I. A., et al. , Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510, 278–282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheskis B. J., Karathanasis S., Lyttle C. R., Estrogen receptor ligands modulate its interaction with DNA. J. Biol. Chem. 272, 11384–11391 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Massie C. E., et al. , New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Rep. 8, 871–878 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu C. L., et al. , Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis. Mol. Oncol. 8, 1575–1587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Estébanez-Perpiñá E., et al. , The molecular mechanisms of coactivator utilization in ligand-dependent transactivation by the androgen receptor. J. Biol. Chem. 280, 8060–8068 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Regan M. C., et al. , Structural and dynamic studies of the transcription factor ERG reveal DNA binding is allosterically autoinhibited. Proc. Natl. Acad. Sci. U.S.A. 110, 13374–13379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandoval G. J., et al. , Binding of TMPRSS2-ERG to BAF chromatin remodeling complexes mediates prostate oncogenesis. Mol. Cell 71, 554–566.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masiello D., Cheng S., Bubley G. J., Lu M. L., Balk S. P., Bicalutamide functions as an androgen receptor antagonist by assembly of a transcriptionally inactive receptor. J. Biol. Chem. 277, 26321–26326 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Helgeson B. E., et al. , Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 68, 73–80 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Makkonen H., et al. , Identification of ETS-like transcription factor 4 as a novel androgen receptor target in prostate cancer cells. Oncogene 27, 4865–4876 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Baena E., et al. , ETV1 directs androgen metabolism and confers aggressive prostate cancer in targeted mice and patients. Genes Dev. 27, 683–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Budka J. A., Ferris M. W., Capone M. J., Hollenhorst P. C., Common ELF1 deletion in prostate cancer bolsters oncogenic ETS function, inhibits senescence and promotes docetaxel resistance. Genes Cancer 9, 198–214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sizemore G. M., Pitarresi J. R., Balakrishnan S., Ostrowski M. C., The ETS family of oncogenic transcription factors in solid tumours. Nat. Rev. Cancer 17, 337–351 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Cooper C. D., Newman J. A., Aitkenhead H., Allerston C. K., Gileadi O., Structures of the Ets protein DNA-binding domains of transcription factors Etv1, Etv4, Etv5, and Fev: Determinants of DNA binding and redox regulation by disulfide bond formation. J. Biol. Chem. 290, 13692–13709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamber E. P., et al. , Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization. EMBO J. 27, 2006–2017 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mo Y., Vaessen B., Johnston K., Marmorstein R., Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA. Nat. Struct. Biol. 7, 292–297 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Morgunova E., Taipale J., Structural perspective of cooperative transcription factor binding. Curr. Opin. Struct. Biol. 47, 1–8 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Bai S., He B., Wilson E. M., Melanoma antigen gene protein MAGE-11 regulates androgen receptor function by modulating the interdomain interaction. Mol. Cell. Biol. 25, 1238–1257 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He B., Kemppainen J. A., Wilson E. M., FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 275, 22986–22994 (2000). [DOI] [PubMed] [Google Scholar]
- 50.van de Wijngaart D. J., et al. , Novel FXXFF and FXXMF motifs in androgen receptor cofactors mediate high affinity and specific interactions with the ligand-binding domain. J. Biol. Chem. 281, 19407–19416 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Meinhart A., Blobel J., Cramer P., An extended winged helix domain in general transcription factor E/IIE alpha. J. Biol. Chem. 278, 48267–48274 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Harami G. M., Gyimesi M., Kovács M., From keys to bulldozers: Expanding roles for winged helix domains in nucleic-acid-binding proteins. Trends Biochem. Sci. 38, 364–371 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Watson P. A., et al. , Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc. Natl. Acad. Sci. U.S.A. 107, 16759–16765 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai L., et al. , ZFX mediates non-canonical oncogenic functions of the androgen receptor splice variant 7 in castrate-resistant prostate cancer. Mol. Cell 72, 341–354.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z., et al. , An AR-ERG transcriptional signature defined by long-range chromatin interactomes in prostate cancer cells. Genome Res. 29, 223–235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.King J. C., et al. , Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat. Genet. 41, 524–526 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ashkenazy H., et al. , ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, associated protocols, methods, and sources of materials can be accessed in the text or SI Appendix.