Significance
Repair of chromosome breaks by polymerase theta-mediated end joining (TMEJ) requires short sequence identities in flanking DNA (microhomologies)—a sequence-context constraint that is unique among DNA repair pathways. Though microhomologies have a central role in TMEJ, it has been uncertain whether an organized mechanism to identify them even exists. Using a combination of chromosomal and extrachromosomal substrates, we describe how polymerase theta efficiently locates microhomologies when present, and creates them de novo when absent. We show how this generates a pattern of microhomology-mediated end joining products that is sufficiently distinct from other end joining pathways and that it can be used as a biomarker for TMEJ activity in cancer genomes.
Keywords: Pol theta, chromosome break repair, microhomology-mediated end joining, mutational signatures, BRCA
Abstract
DNA polymerase theta mediates an end joining pathway (TMEJ) that repairs chromosome breaks. It requires resection of broken ends to generate long, 3′ single-stranded DNA tails, annealing of complementary sequence segments (microhomologies) in these tails, followed by microhomology-primed synthesis sufficient to resolve broken ends. The means by which microhomologies are identified is thus a critical step in this pathway, but is not understood. Here we show microhomologies are identified by a scanning mechanism initiated from the 3′ terminus and favoring bidirectional progression into flanking DNA, typically to a maximum of 15 nucleotides into each flank. Polymerase theta is frequently insufficiently processive to complete repair of breaks in microhomology-poor, AT-rich regions. Aborted synthesis leads to one or more additional rounds of microhomology search, annealing, and synthesis; this promotes complete repair in part because earlier rounds of synthesis generate microhomologies de novo that are sufficiently long that synthesis is more processive. Aborted rounds of synthesis are evident in characteristic genomic scars as insertions of 3 to 30 bp of sequence that is identical to flanking DNA (“templated” insertions). Templated insertions are present at higher levels in breast cancer genomes from patients with germline BRCA1/2 mutations, consistent with an addiction to TMEJ in these cancers. Our work thus describes the mechanism for microhomology identification and shows how it both mitigates limitations implicit in the microhomology requirement and generates distinctive genomic scars associated with pathogenic genome instability.
DNA double-strand breaks (DSBs) in the chromosome are generated spontaneously, after exposure of cells to exogenous agents (e.g., ionizing radiation), and are induced during meiosis or development of the adaptive immune response (1). DSBs are also generated by nucleases, especially Cas9, as intermediates in genome engineering. They are usually repaired by the nonhomologous end joining (NHEJ) pathway, which joins the two ends of a break with minimal processing (2), or the homologous recombination (HR) pathway, which uses another DNA molecule as a template for repair (3). Impairment of either of these repair pathways—especially impairment in HR due to deficiency in BRCA1 or BRCA2—leads to genome instability, which can cause cell death or cancer (4).
Another DSB repair pathway is defined by its requirement for DNA polymerase theta (Pol θ, gene name POLQ) (5–7), and has been termed theta-mediated end joining (TMEJ). TMEJ overlaps with previously defined alternative nonhomologous end joining (Alt-NHEJ) and microhomology-mediated end joining (MMEJ) pathways (8–10), though the extent to which these definitions overlap is not clear. In mammals, TMEJ is both more frequent and essential for viability in cells deficient in NHEJ (11–13) or HR (14–16). A specific requirement in BRCA-deficient contexts for Pol θ has identified this protein as a therapeutic target in BRCA-deficient breast cancers (17). However, TMEJ mechanism is not well understood, and it is important to determine its role and relevance in NHEJ and HR proficient cells.
TMEJ and HR pathways engage a common intermediate, the 3′ ssDNA tails generated after resection of chromosome breaks (7, 11). Pol θ aligns these tails and anneals small 2- to 6-bp patches of complementary sequence (microhomologies), which is followed by removal of at least one nonhomologous tail, then microhomology-primed synthesis sufficient to resolve remaining gaps (Fig. 1A) (7, 18). Differing location and frequency of microhomologies at different break sites is thus expected to have an impact on pathway outcome. At a minimum, the extent of deletion associated with repair by TMEJ will be determined by the locations of microhomologies relative to the break site, and especially the means by which these microhomologies are identified. Breaks in microhomology-poor regions of the genome could also lead to impaired TMEJ activity, and consequently cell death or cancer-causing genome rearrangements.
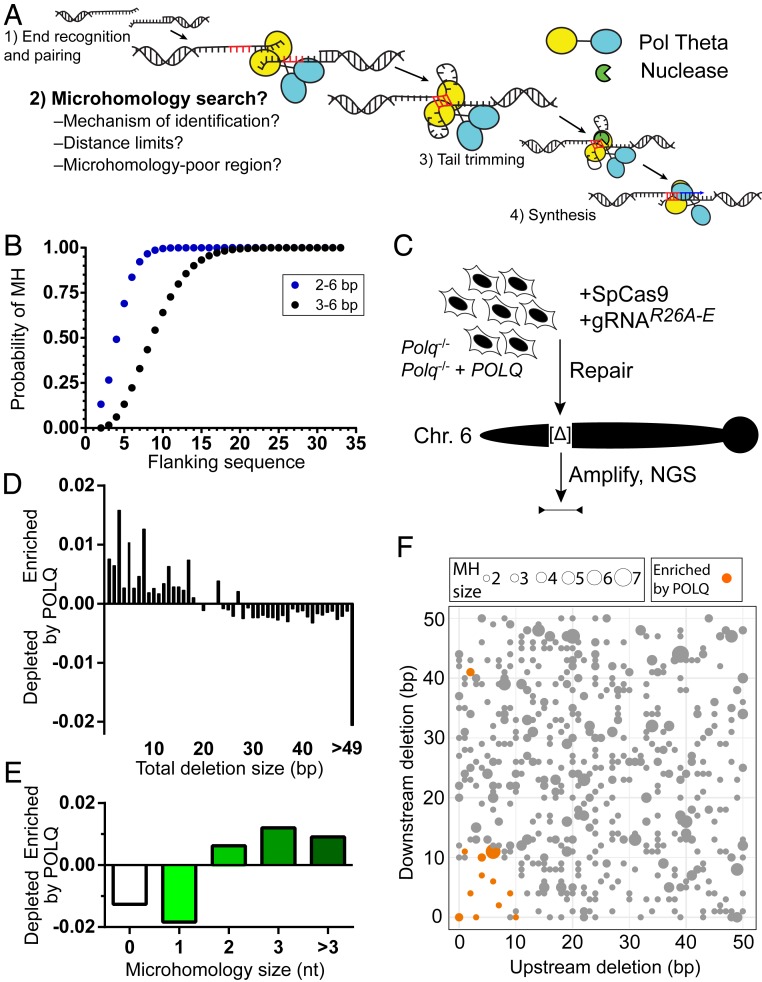
Fig. 1.
Characterization of Pol θ-dependent deletions after a chromosome break. (A) Steps required for TMEJ, emphasizing a critical role for microhomology identification. (B) The probabilities of finding 2- to 6-bp (blue) or 3- to 6-bp (black) microhomologies (MHs) were determined for sets of 100,000 randomly generated pairs of sequences of increasing size (flanking DNA sequence), from 2 to 33 nucleotides (nts). (C) Cas9 targeted to five different break sites in the Rosa26 locus (R26A-E) were separately introduced into transformed MEFs from Polq−/−-deficient mice engineered to express human POLQ or not. Chromosome break repair products were recovered 24 h later, amplified, and characterized by NGS. (D and E) The difference in the fraction of repair products with noted size of deletion (D) or microhomology (E) in POLQ-expressing cells vs. Polq−/− was averaged across all five break sites tested. (F) Filled circles denote the location of all microhomologies 2 bp or more relative to the break site for all five break sites, with microhomology size noted according to the size of the filled circle. Deletions enriched in cells expressing WT POLQ vs. Polq−/− cells in triplicate experiments are shown in orange and were identified using a two-tailed t test and the Benjamini–Hochberg procedure to adjust P values for multiple comparisons, with a false discovery rate of 0.05.
Here we explore the basis for the microhomology identification step in TMEJ by systematically defining Pol θ-dependent repair products at a series of Cas9-induced chromosome breaks, where the break sites were chosen to possess varied density of break site-proximal microhomologies. We then explored the mechanistic requirements for TMEJ that are suggested by these results using an extrachromosomal substrate assay, as this latter strategy allows for unambiguous assessment of TMEJ activity on systematically varied substrates. Our results support a bidirectional scanning mechanism that mitigates deletion associated with TMEJ, as this scanning mechanism efficiently identifies microhomologies only when they are within 15 bp of either side of the break site. Break site locations within the genome that are depleted of microhomologies within 15 bp can nevertheless still be effectively repaired by TMEJ. In such contexts, TMEJ requires one or more cycles of aborted synthesis that generate long microhomologies de novo, which now better support processive synthesis. These latter products possess locally templated insertions (TINS) that are highly characteristic of this pathway, and are consequently an effective biomarker for Pol θ/TMEJ activity in breast cancer genomes.
Results
Characterization of Pol θ-Mediated Deletions after a Chromosomal Break.
Prior studies indicate efficient Pol θ-mediated synthesis activity requires microhomologies of 3 bp or more (11, 18, 19). To assess how this requirement could impact repair, we employed in silico modeling to determine the likelihood of finding such microhomologies as a function of increasing distance from the break site. The frequency of a 3-bp or more microhomology in a set of random pairs of break site flanking sequences is 64% when considering 10 bp of flanking sequence, and 93% when considering 15 bp. Inclusion of 2-bp microhomologies is sufficient to increase the frequency within 10 bp to 99% (Fig. 1B), though as discussed below, 2-bp microhomologies are less able to promote processive repair synthesis by Pol θ.
To address how these limitations impact TMEJ, we focused on five break sites (termed R26A through R26E) within a 7-kb region that varied according to the density and length of microhomologies near the break site (SI Appendix, Fig. S1A). In particular, two closely located sites (within 500 bp) are unusually rich (R26A) vs. unusually poor (R26E) in terms of the availability of microhomologies within 15 bp.
We generated chromosome breaks at each of the five break sites by direct introduction of appropriately targeted Streptococcus pyogenes Cas9 ribonucleoprotein complexes into T antigen transformed mouse embryo fibroblasts (MEFs). We harvested cells 24 h later in an attempt to mitigate the contribution of repair after excessive nonspecific degradation of DNA ends, while ensuring that repair products with insertions and deletions accumulate at the majority of chromosomes. We amplified products without a phenotype-based screen, to ensure product spectra is limited only by whether the products can be amplified (i.e., retains primer sequences), then characterized products by next generation sequencing (NGS) (20) (Fig. 1C).
We first analyzed simple deletion products; i.e., deletions of flanking DNA without inserted sequences. We systematically identified those products enriched in human POLQ-expressing MEFs (Polq−/− MEFs complemented by expression of POLQ cDNA), relative to the Polq−/−-deficient isogenic parental cell line (7). Highly enriched products consisted exclusively of deletions <30 bp (Fig. 1D) that were associated with 2- to 6-bp microhomologies (microhomology-associated deletions [MHDs]) (Fig. 1E). The microhomologies chosen are also largely restricted to those within 10 to 15 bp of either side of the break site (Fig. 1F and SI Appendix, Table S1). Moreover, nearly all microhomologies present less than 10 bp from termini (7/8) are significantly enriched (Fig. 1F). In accord with a restriction to 15 bp flanking the break site, we observed no significantly enriched MHDs in repair products recovered from breaks at R26E, the site with few break site-proximal microhomologies (SI Appendix, Fig. S1A and Table S1). More broadly, MHD associated with large deletions (>15 bp from either end) are actually suppressed by Pol θ/TMEJ for all five break sites tested, including R26E (SI Appendix, Fig. S1B). TMEJ thus promotes more accurate repair.
Mechanism of Microhomology Search.
We addressed the mechanistic basis for microhomology choice described above by employing a cellular TMEJ assay that allows for systematic variation of the substrate. This assay employs extrachromosomal “pre-resected” substrates, consisting of double-stranded DNA fragments with ends that have 70-nt single-stranded DNA 3′ tails. The preresected tails block engagement of KU-dependent NHEJ, thus joining relies exclusively on Pol θ for efficient repair (joining efficiency reduced over 10-fold in Polq-deficient cells; e.g., Fig. 2A) (7, 11).
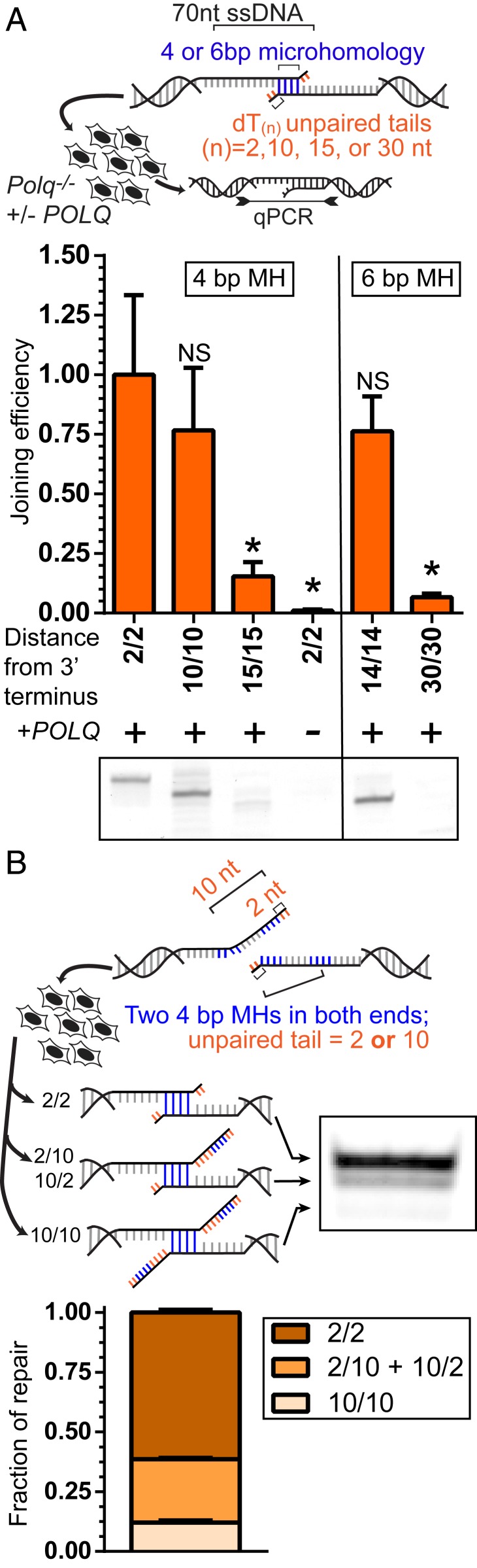
Fig. 2.
Mechanistic basis for microhomology usage by Pol θ. (A) The 691-bp dsDNA substrates with 70 nt 3′ ssDNA tails possessed microhomologies of 4 and 6 bp (4-bp MH, 6-bp MH) that were located in ssDNA tails 2, 10, 14, 15, and 30 nt from both head and tail 3′ termini (e.g., 2/2). Sequence 3′ of the microhomology was replaced with polyT(n) tracts. Substrates were introduced into the MEFs with and without POLQ expression described above. Head-to-tail end joining efficiencies in recovered DNA were determined by qPCR and normalized to the joining efficiency observed using the 2/2 substrate. Statistical significance was assessed by one-way ANOVA with Bonferroni correction to account for multiple comparisons; NS, not significant; *P < 0.05. Experiments on substrates with 4-bp MH and 6-bp MH substrates were performed independently. Electrophoresis of a representative end point PCR is shown to confirm preferential usage of terminal 4- and 6-bp MHs for repair (Bottom). (B) Identical 4-bp MHs were located 2 nt and 10 nt from both head and tail 3′ termini and introduced into the cells described above. Products were amplified and characterized by electrophoresis (representative experiment shown); the mean relative amounts of noted species were determined for three independent experiments. Error bars denote the SEM.
We first assessed if a distance restriction on the ability of Pol θ to identify microhomologies helped explain the chromosomal repair results described above. We compared activity on substrates where a defined microhomology was present at increasing distance from the two 3′ termini and replaced sequence downstream (3′) of the embedded microhomology with poly(dT) tracts, to ensure the absence of even trivial alternate microhomologies closer to the 3′ terminus. These substrates were introduced into the same isogenic MEFs with or without POLQ expression as described above, and repair was assessed by quantifying the efficiency of repair, as well as by characterizing repair product structure (Fig. 2A).
TMEJ was similarly efficient when a 4-nt microhomology was embedded 2 or 10 nt from both 3′ termini, but was only 15% of maximal levels when the microhomology was embedded 15 nt (identified as 2/2, 10/10, and 15/15; distances refer to the size of nonhomologous tail after microhomology alignment) (Fig. 2A). TMEJ was also reduced if only one copy of the microhomology was located 15 nt distal to the break (2/15) (SI Appendix, Fig. S2A). In accordordance with this result, we rarely observe significant enrichment by POLQ expression of chromosomal MHDs that are similarly asymmetric (Fig. 1F; considering nine potential MHDs with <3 bp deleted from one flank, and 9 to 15 bp from the other, only two are enriched/orange). These observations exclude a mechanism where one end is fixed, and search proceeds inward from the other (i.e., the microhomology search typically progresses inward from both termini, rather than only one).
A larger 6-bp microhomology allowed for partial rescue of repair efficiency when located more than 10 bp away (14/14; 75%, relative to 2/2), but repair was abolished for even this larger microhomology when it was located 30 bp distal from both 3′ termini (Fig. 2A). Therefore, the limitation of Pol θ-dependent chromosomal MHD to within 15 bp of break sites described above (Fig. 1F) is at least in part due to a reduced ability of Pol θ to use microhomologies further away (Fig. 2A).
Microhomologies that are 2 nt vs. 10 nt from both 3′ termini are used with similar efficiencies when they are the most 3′ proximal microhomologies present. We considered next whether they are also functionally equivalent when in competition. We designed a single TMEJ substrate where both ssDNA tails had two copies of the same microhomology, with one copy located 2 nt from the 3′ terminus, and the other copy 10 nt from the 3′ terminus (2/2 + 10/10) (Fig. 2B). Strikingly, the most proximal pair of microhomologies (2/2) was employed five times as frequently as any of the more distally located microhomologies (10/10, as well as 10/2 and 2/10) (Fig. 2B). Analysis of chromosomal data similarly reflected a strong preference for the more 3′ proximal of two equivalent microhomologies (SI Appendix, Fig. S2B). These results are indicative of a mechanism that “scans” for available microhomologies, initiating from the 3′ terminus.
Pol θ Generates Locally Templated Insertions.
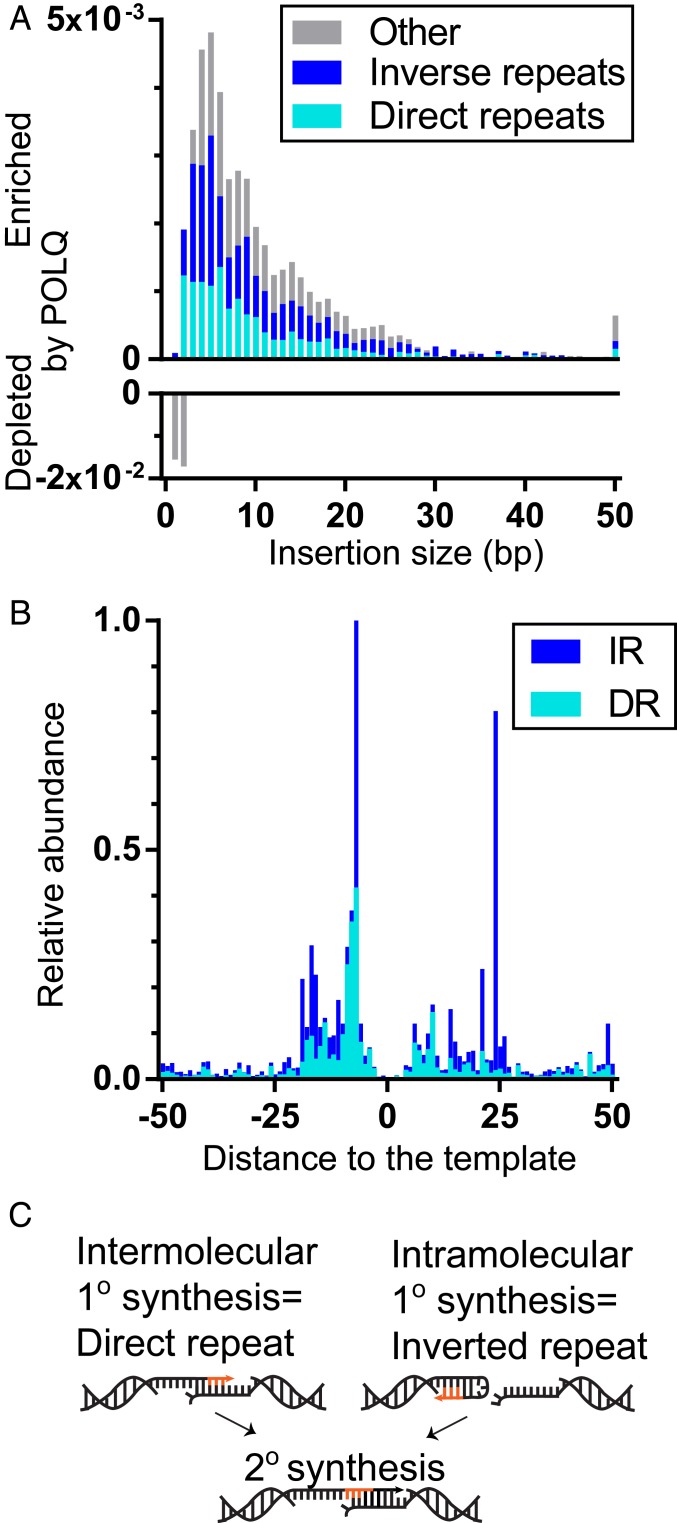
The experiments above focused on simple deletions (i.e., loss of flanking DNA without inserted sequences). We considered next the subset of chromosomal repair products that contain inserted sequence, whether such products contained deletion of flanking sequence or not. Insertion lengths varied between 1 and 157 bp. Pol θ increases the fraction of repair products with insertions over 2 bp, most clearly those between 3 and 6 bp (Fig. 3A). For most of these products, inserted sequences can be defined as repeated relative to nearby flanking DNA sequence (typically within 30 bp) (Fig. 3B). These are best explained if the insertions are products of template-dependent synthesis (TINS) (21–23). Most of the remaining insertions enriched in POLQ-expressing cells (gray in Fig. 3A) likely also employ a local template, but the length of the 1° synthesis tract was not sufficiently long to pass the 5-nucleotide minimum value we employed to exclude Pol θ-independent insertions. A small fraction (<0.1% of total repair) of insertions employed template distal to the break site, including other chromosomes. We identified two classes of TINS: synthesis of 5 bp or more of sequence that is directly repeated, relative to flanking DNA (direct repeats [DRs]), or synthesis of 5 bp or more of sequence that is the reverse complement of flanking DNA (inverse repeats [IRs]) (Fig. 3C). The former is consistent with a primary round of synthesis (1° synthesis) initiated from one broken, resected end, using the second resected end as a template (intermolecular synthesis), while the latter is consistent with 1° synthesis that was initiated from a hairpin (intramolecular synthesis).
Fig. 3.
Pol θ generates locally templated insertions. (A) The difference in the fraction of repair products with noted size of insertion in POLQ-expressing cells vs. Polq−/− was averaged across all five break sites. Insertions were defined as having 5 bp or more direct or inverse repeated sequence (DR, cyan; IR, blue) relative to flanking DNA (see also C). (B) The relative abundance of insertions with 5 bp or more repeated sequence is plotted according to the location of the repeat within flanking DNA for both DR (cyan) and IR (blue) insertions. (C) Model for generating direct repeat vs. inverted repeat templated insertions.
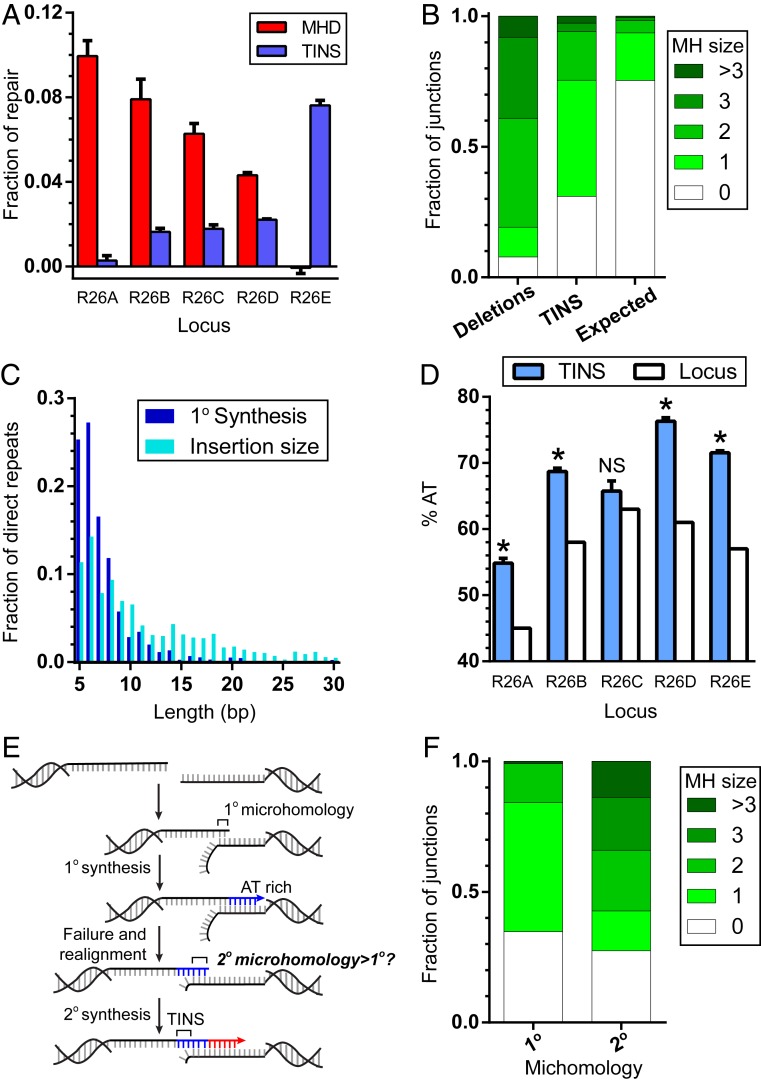
The frequency of TINS varied widely across different break sites. Notably, TINS frequency was almost undetectable at R26A, the break site that is unusually rich in break proximal microhomologies (Fig. 4A). In contrast, TINS accounted for the only significant class of Pol θ-dependent events at R26E (Fig. 4A), where flanking sequence is poor in microhomologies.
Fig. 4.
Characterization of TINS. (A) Fraction of repair events enriched by POLQ expression that generated deletions with 2 bp or more of microhomology within 15 bp of the break site (MHD, red), or a templated insertion larger than 2 bp (TINS, blue), for each of the five break sites tested. Bar represents the mean and error bars SEM from three biological replicates. (B) Average fraction of junctions associated with the indicated microhomology sizes in deletions significantly enriched by POLQ expression (deletions), in insertions larger than 2 bp, and directly repeated relative to flanking DNA (TINS), and as would be expected by chance, if microhomologies played no role (expected). (C) Fraction of products with insertions of 5 bp or more sequence directly repeated relative to flanking DNA, comparing total length of synthesized nucleotides (insertion size; cyan) to the length of the first round of synthesis (1° synthesis; dark blue). (D) Percent AT content in TINS defined as in C (blue), compared to AT content in the 100 bp surrounding the break site for each locus (locus; white). Bar represents the mean and error bars SEM for three biological replicates. Statistical significance was assessed by a one-sample t test on each locus individually; NS, not significant; *P < 0.05. (E) Model for the role of TINS in generating de novo (2°) microhomologies. (F) Fraction of repair events associated with the indicated microhomology sizes for products with 4 or 5 bp of TINS in R26E, for 1° microhomologies vs. 2° microhomologies (see also E).
Decreased availability of break site-proximal microhomologies thus correlates with both decreased MHDs and increased TINS. Prior work argues that both MHDs and TINS involve microhomology-mediated synthesis (5, 24). However, MHD are generated after a single round of alignment-directed synthesis that was sufficiently processive to complete repair. By comparison, TINS are generated when the initial round of alignment-directed synthesis fails before repair is complete and is followed by one or more additional rounds of alignment and synthesis (Fig. 3C). We sought to address next whether we can identify a mechanistic basis for failed synthesis.
We first confirmed that like MHDs, the 1° round of synthesis in TINS favors alignment at microhomologies; the frequency of use of microhomologies 2 bp or more in this context is higher than would be expected by chance (25% vs. 7%) (Fig. 4B). Preference for microhomologies is also sufficient to explain the nonrandom siting of synthesis initiation that is readily apparent in a map of the sources of templated insertions (Fig. 3B). However, microhomologies associated with TINS are generally shorter, relative to those associated with MHDs, where 93% of MHDs are associated with microhomologies 2 bp or more (Fig. 4B).
We further probed this correlation using specific microhomologies. We identified three different microhomologies that varied in size, but which were similarly located relative to the break site, then compared the relative frequencies of the two classes of events for each of the microhomologies (SI Appendix, Fig. S3A). A long, 6-bp microhomology (AGTCTT) in R26A led to MHDs 254-fold more often than TINS. An intermediate-sized 3-bp microhomology (TCC) in R26B led to MHDs 75-fold more frequently than TINS. Finally, a short 2-bp microhomology (AT) in R26E generated MHDs only 1.66-fold more frequently than TINS. A given microhomology consequently contributes to both classes of Pol θ-dependent events, but the extent that MHD is favored over TINS decreases as the size of the initial microhomology decreases.
We conclude the probability of an aborted round of synthesis (and resolution with TINS) is partly reliant on instability in the alignment of the 1° microhomology. Two characteristics of TINS argue the processivity of synthesis after initial alignment also has a critical role in determining whether this round of synthesis aborts. The 1° synthesis tracts in insertions are short—rarely more than 10 bp (Fig. 4C). Longer insertions, while present (insertion size, Fig. 4C), involve more than one round of templated insertions (e.g., SI Appendix, Fig. S3B). In addition, the AT content of TINS was enriched relative to flanking DNA for all five break sites tested (Fig. 4D). This presumably reflects less effective stabilization of alignments when synthesis proceeds through AT-rich flanking sequence (relative to GC-rich templates), more frequent disruption of the alignment, and consequently a requirement for a second alignment and round of synthesis.
Prior work has noted how unsuccessful rounds of synthesis in TINS can generate microhomologies de novo (2° microhomologies), and these 2° microhomologies can be employed in alignments that prime the next round of synthesis (Fig. 4E) (5, 25). We show here that these 2° microhomologies were typically longer (57% > 1 bp) than the initial, failed 1° microhomology-driven alignment (16% > 1 bp) (Fig. 4F). TINS is thus effectively an adaptive mechanism—in microhomology-poor regions it generated new, more stable microhomologies that increased the likelihood of successful repair.
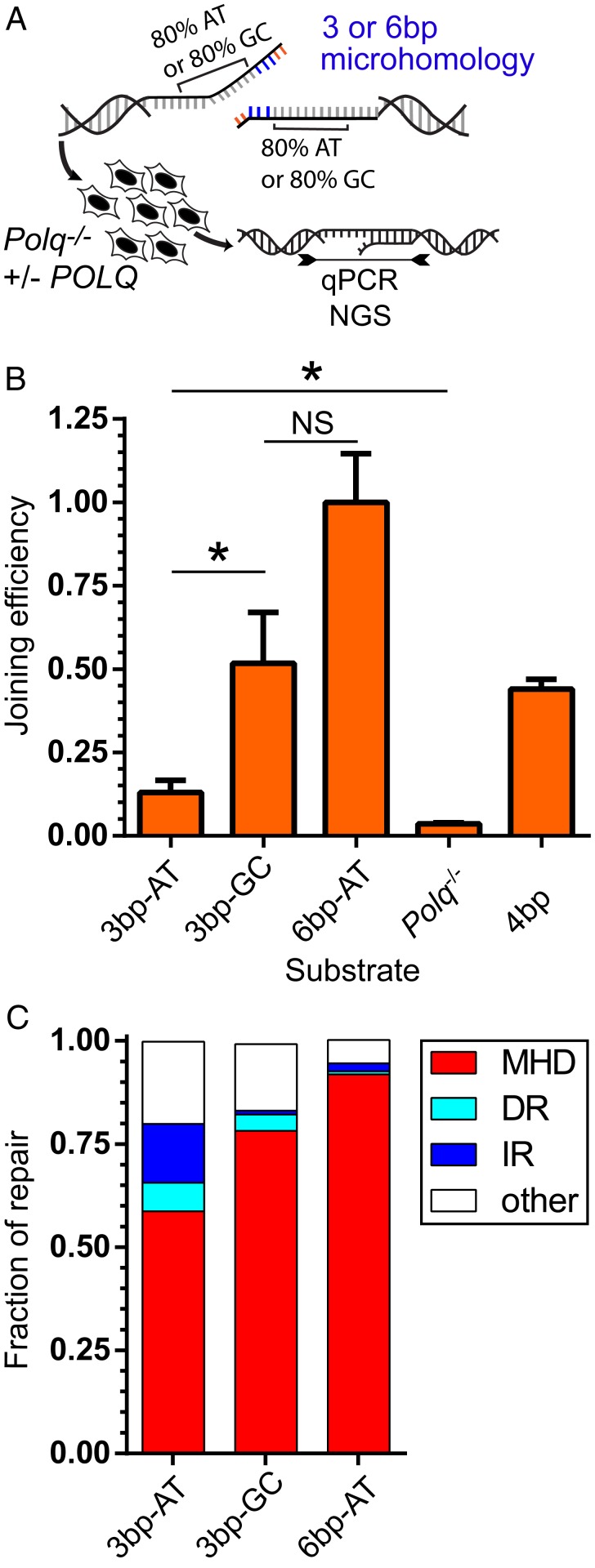
We employed extrachromosomal substrates to directly test if TINS are a function of both reduced size of 1° microhomology, as well as the AT content of the template flanking the microhomology. We assessed the importance of flanking DNA by generating two substrates with the same short, 3-bp microhomology (TAG), but highly divergent levels of flanking AT content; a substrate with 80% AT content downstream of the microhomology vs. a substrate with 80% GC content downstream of the microhomology (Fig. 5A). When flanking DNA was AT rich, joining was 25% as efficient as when flanking DNA was GC rich (Fig. 5B), and repair products generated using the substrate with AT-rich flanking DNA more often possessed TINS (Fig. 5C). As with chromosomal repair products, both direct repeat and inverted repeat TINS were frequent. These results are consistent with a critical role for ongoing synthesis in stabilizing a given alignment and enabling productive repair. We further used a variant of the substrate with AT-rich flanking DNA with a longer microhomology (6 bp) (Fig. 5A). The longer 1° microhomology was sufficient to promote higher joining efficiencies (Fig. 5B), as well as lower frequencies of TINS (Fig. 5C), relative to both of the substrates (AT rich and GC rich) with 3-bp microhomologies.
Fig. 5.
Frequency of TINS is dependent on both microhomology size and template AT content. (A) TMEJ substrates possessed the same 3-bp microhomology followed by 20 bp of template that was either 80% AT (3 bp-AT) or 80% GC (3 bp-GC), or which possessed the 80% AT content template, but a longer 6-bp microhomology (6 bp-AT). The efficiency of cellular TMEJ (B) was determined for each substrate as in Fig. 2A, and product structures (C) were characterized by NGS. (B) Joining efficiency was determined by qPCR and normalized to results using the 6 bp-AT substrate and compared to results using the original 2/2 4-bp (4 bp) substrate described in Fig. 2A. Bars represent the mean and error bars the SEM from three biological replicates. Statistical significance was assessed by one-way ANOVA with Bonferroni correction to account for multiple comparisons; NS, not significant; *P < 0.05. (C) TMEJ products were amplified and characterized by sequencing as MHD if possessing deletions at 2 bp or more of microhomology, DR, if products contain 5 bp or more of templated (directly repeated) synthesis or 5 bp or more of templated (inversely repeated, IR) synthesis. “Other” represents products inconsistent with MHD or TINS.
TINS Genomic Scars Are a Biomarker for Increased TMEJ Activity in BRCA Mutated Cancers.
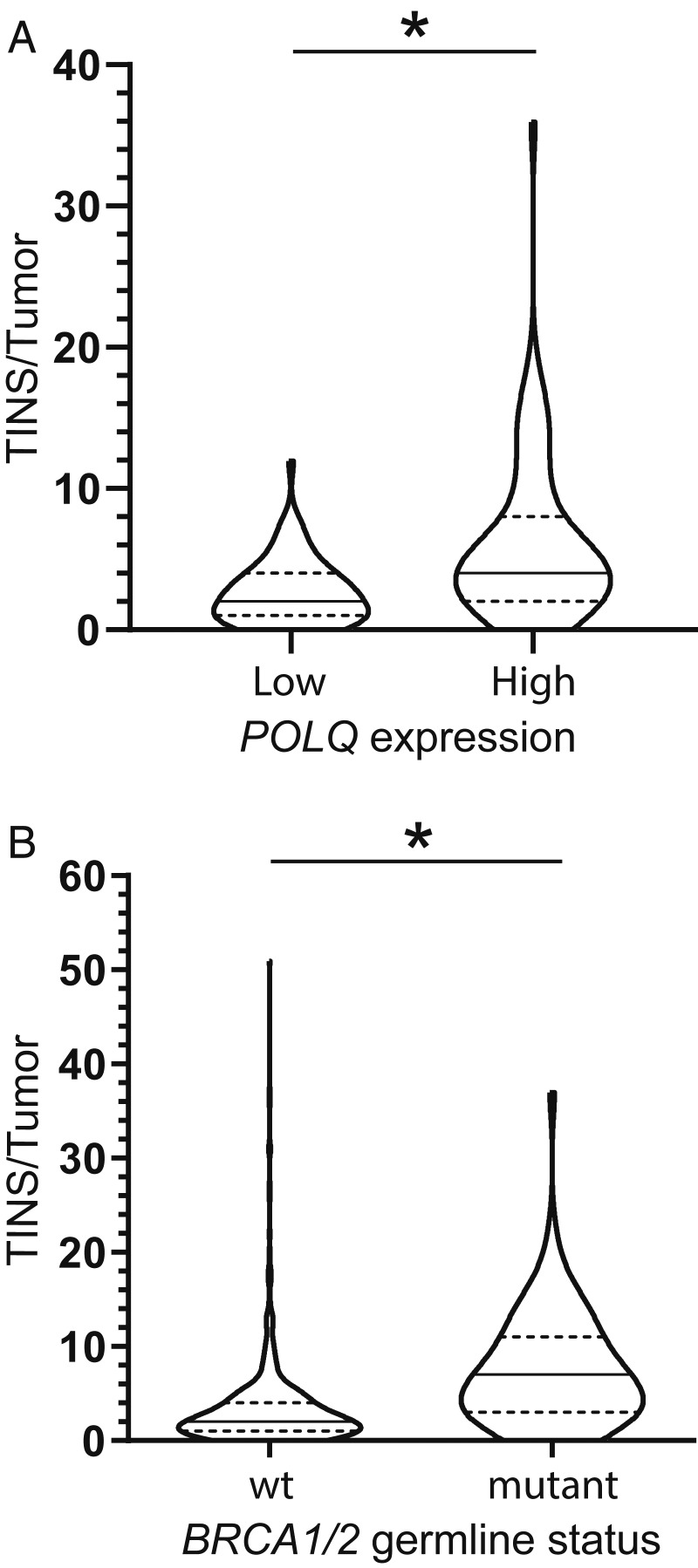
MHD are often—though not always (see, e.g., Fig. 4A)—the most frequent Pol θ-dependent repair products for a given chromosome break and serve as a biomarker for TMEJ activity (16). However, some MHDs are favored during NHEJ (SI Appendix, Fig. S4A), while yet others can be suppressed by Pol θ (SI Appendix, Fig. S1B). By comparison, TINS are a more characteristic product of Pol θ-dependent repair (SI Appendix, Fig. S4B), and consequently have been proposed as a more effective biomarker for Pol θ/TMEJ activity (26). To address this possibility, we assessed whether it was possible to correlate the frequency of TINS with POLQ expression levels in the genomes of breast cancers previously sequenced by International Cancer Genome Consortium (ICGC) (27) and made publicly available at https://dcc.icgc.org/releases/release_26/Projects/BRCA-EU. We determined that TINS were higher in the 65 cancers with high POLQ expression (top quartile), relative to the 66 cancers with low POLQ expression (bottom quartile) (P < 0.0001, Mann–Whitney U test) (Fig. 6A). Notably, we observe a similar correlation of TINS with POLQ expression (P = 0.0002, Mann–Whitney U test) after excluding cancers with germline mutations in BRCA1 or BRCA2, a possible confounding issue (SI Appendix, Fig. S4C) (discussed in the following paragraph). Moreover, inserted sequences identified as TINS in cancer genomes were AT rich, relative to flanking DNA (SI Appendix, Fig. S4D), in accord with our description of Pol θ-dependent TINS at Cas9-induced double-strand breaks (Fig. 4D).
Fig. 6.
TINS are increased in tumors with high POLQ expression and with BRCA mutations. (A and B) Number of insertions with templated synthesis 5 bp or more per tumor (TINS/tumor, defined as in Fig. 4A) for breast cancer genomes previously sequenced by ICGC were determined according to differing POLQ mRNA expression level (A) or BRCA1 or BRCA2 germline status (B). (A) POLQ expression in fragments per kilobase of transcript per million was normalized based on HPRT1, TBT, and GAPDH expression for 261 breast cancers. Tumors were categorized as expressing low (POLQ low; bottom quartile) or high levels of POLQ mRNA (POLQ high; top quartile). Statistical significance was assessed with a two-tailed Mann–Whitney U test; *P < 0.05. (B) Breast cancers were categorized as having germline BRCA1 or BRCA2 mutations (mutant, 75 cancers) or not (WT, 494 cancers).
Pol θ is required for viability in cancer cell lines deficient in BRCA1/2 (14, 15), and higher levels of POLQ expression are observed in BRCA1/2 mutated breast cancers (SI Appendix, Fig. S4E) (14, 28, 29). We show here that the genomes from the 75 breast cancer patients with germline BRCA mutations had a median of seven TINS/genome, a much higher frequency than that observed in the remaining patients with no germline BRCA mutations (two TINS/genome) (P < 0.0001, Mann–Whitney U test) (Fig. 6B). The ability to correlate BRCA1/2 deficiency with increased frequency of TINS, a biomarker more directly reflective of Pol θ/TMEJ activity than expression, provides further support that addiction to TMEJ is broadly associated with BRCA1/2-deficient cancers.
Discussion
Mechanistic Basis for Microhomology Identification.
TMEJ identifies microhomologies through a scanning mechanism that is initiated from the 3′ terminus and favors break site-proximal, 2-bp or larger microhomologies (Fig. 2B). Increased size of microhomology can modestly extend the distance searched beyond the 15-bp limit described above (Fig. 2A), and also impacts the extent proximal microhomologies are favored. Asymmetrically located pairs of microhomologies are used less efficiently than a symmetrically located pair of microhomologies (e.g., 10/10 vs. 2/15 substrates, Figs. 1F and 2A and SI Appendix, Fig. S2A), implying scanning is typically bidirectional.
Past work from our group and others has determined that in some contexts, Pol θ-dependent repair can include microhomologies more distal than 15 bp from 3′ termini. These contexts include cells deficient in regulation of end resection (cells defective in KU or 53BP1) or wild-type (WT) cells after a much longer recovery period than is used here (24 h) (11, 16, 30). Given similar results using preresected extrachromosomal substrates (where we can systematically vary microhomology availability relative to a definitive 3′ ssDNA terminus), we suggest this second class of Pol θ-dependent products reflects loss of 3′ terminal ssDNA before Pol θ could be engaged, rather than a fundamental change in the preference of Pol θ for break-proximal microhomologies. Loss of 3′ ssDNA after resection can be inferred by substitution of long tracts of flanking target DNA with donor-specific sequence in the majority of gene-targeting products (e.g., ref. 31), and can also be observed when using preresected extrachromosomal substrates (11). Possible mechanisms for loss of 3′ ssDNA termini include engagement of the Artemis endonuclease in the context of canonical NHEJ (32), or engagement of Pol δ-editing exonuclease activity in the context of homologous recombination (31, 33).
Moreover, there is little mechanistic rationale for use of microhomologies >15 bp from either side of the break site, since 1) for break sites rich in microhomologies in this region, the identification of these microhomologies is both efficient (Fig. 1F) and strongly favors the most break site-proximal microhomologies (Fig. 2B); and 2) for the 7% of break sites without a break site-proximal microhomology of 3 bp or more (Fig. 1B), iterative rounds of synthesis can generate microhomologies de novo that are now longer, and which support processive synthesis and successful repair (Fig. 4).
Mechanism of Template-Dependent Insertions.
Repair by simple Pol θ-dependent MHDs is unlikely to be efficient at the 7% of break sites that do not have microhomologies 3 bp or more within 15 nt of the break site, and instead generates near-compensatory levels of Pol θ-dependent products with locally templated insertions (TINS) (Fig. 4A). We show that both classes of Pol θ-mediated repair—MHD and TINS—are products of synthesis initiated from primers annealed to template at sites of short complementary sequence. TINS reflect instances of alignment and synthesis where synthesis was not sufficiently processive to allow for complete repair. Such failures are primarily due to alignments that rely on short, <3-bp microhomologies or no microhomology, consistent with in vitro data emphasizing the importance of alignments of larger microhomologies for processive synthesis (19, 34). However, failure is also more frequent if synthesis proceeds through AT-rich regions (Figs. 4D and 5), implying a contribution of ongoing synthesis to the stabilization of the aligned ends. Importantly, the initial aborted round of synthesis often generates a new, larger microhomology (Fig. 4F). This explains how TINS is compensatory in microhomology-poor, AT-rich regions—it is effectively an adaptive mechanism, as Pol θ can generate new microhomologies that are sufficiently stable to sustain processive synthesis and successful repair.
We show that TINS is a specific marker for TMEJ activity (SI Appendix, Fig. S4B). They are present at a much higher frequency in a panel of 75 breast cancers with germline mutations in BRCA1/2 (Fig. 6B), consistent with a requirement for TMEJ for viability of several BRCA1/2-defective cancer cell lines (14, 15). Indeed, Pol θ is required for viability in the context of a wide variety of DNA damage response defects, and thus is as an attractive target for therapy in as many as 30% of all breast tumors (16, 17). Assessment of TINS in tumor genomes thus holds promise as a biomarker for deciding when TMEJ should be targeted for cancer therapy.
Relationship of TMEJ to Other End Joining Repair.
We have defined the outcomes of TMEJ as 1) those deletion products associated with microhomologies 2 bp or more that are located within 15 bp of either side of the break site, as well as 2) products with 5 bp or more of synthesis that results in an insertion, and the synthesis employs template within 50 bp of either side of the break site.
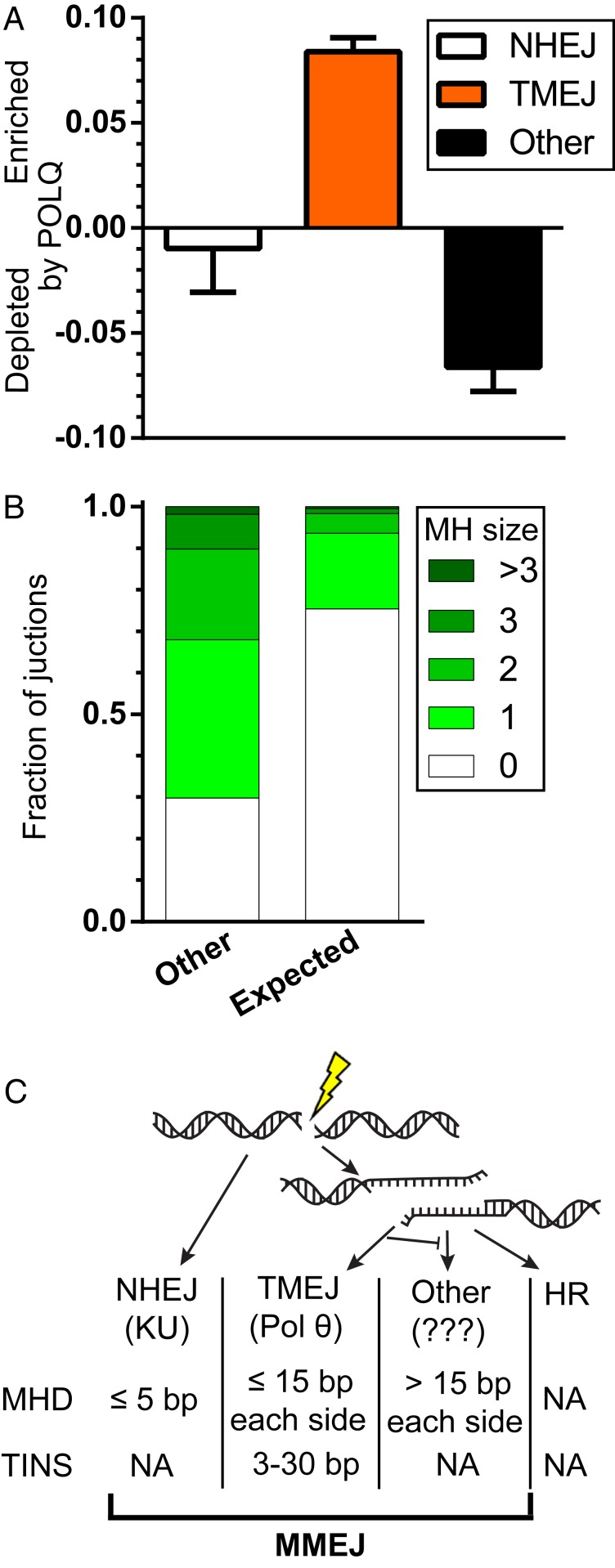
Loss of Pol θ does not typically lead to compensatory increases in the accurate, <5-bp deletion products that can be clearly linked to NHEJ (Fig. 7A) (30). Instead, repair is rechanneled to “other” products with larger deletions (>15 bp from either side of the break site; Fig. 7A) that also favor microhomologies, though to a lesser degree than TMEJ (Fig. 7B). The contribution of TMEJ to overall repair also varies little over the five break sites tested—an average of 8.4 ± 1.5% (SD)—despite wide variation in the availability of microhomologies. Taken together, our results imply 5 to 10% of Cas9-induced DSBs preferentially engage TMEJ and not NHEJ, likely because these DSB ends have been resected (Fig. 7C). As also noted above, less accurate end joining products that are enriched in Pol θ-deficient cells may at least in part reflect clipping of 3′ ssDNA tails generated by resection (e.g., by Artemis) (11, 32), followed by NHEJ or yet-undefined alternate end joining mechanisms.
Fig. 7.
TMEJ promotes genomic stability. (A) The extent of enrichment of repair product classes upon POLQ expression, averaged across all five break sites. Products were classified as NHEJ (white bar) if deleted sequence was less than 5 bp and possessed no or 1 bp of microhomology, as TMEJ (orange bar) if deleted sequence was less than 15 from either end and possessed 2 bp or more of microhomology, or had templated insertions larger than 2 bp, and as “other” (black bar) if deleted sequence from either flank exceed 15 bp. Bar represents the mean and error bars SEM from the five loci. (B) Average fraction of repair events associated with the indicated microhomology sizes in deletions larger than 15 bp from both sides of the break (other) or as would be expected by chance, if microhomologies played no role (expected). (C) Following a DSB, ends are either ligated through the NHEJ pathway (dependent on the KU heterodimer) or resected to form 3′ ssDNA tails. These are substrates for TMEJ (dependent on Pol θ), HR, or a third, undescribed form of end joining that also favors microhomologies. Different end joining pathways have different mutational outcomes (MHD of the indicated sizes or TINS).
A role for microhomologies in mammalian end joining has long been clear (35). Our work shows they have a variety of sources. Some MHDs are generated by both Pol θ/TMEJ and NHEJ (those with less than 5 bp of deletion; SI Appendix, Fig. S4A), while others are primarily attributable to Pol θ/TMEJ (between 5 and 15 distal to either flank; Fig. 1F). Yet a third class (those suppressed by TMEJ in otherwise WT cells, and more than 15 bp from either strand-break terminus; other, Fig. 7A) has distinct and probably complex genetic requirements. MHDs are thus generated by at least three genetically distinguishable mechanisms, highlighting a critical flaw in the frequent attribution of such products to a single MMEJ pathway (Fig. 7C).
Implicit to a requirement for microhomologies during end joining is 1) an associated loss of genomic information, and 2) potentially impaired repair in contexts where microhomologies are hard to find. We describe here how an elegant search mechanism largely overcomes these limitations in a manner that in most contexts—e.g., in cells proficient in canonical pathways, and which appropriately regulate end resection—least threatens genome stability. At the same time, cancer-causing deficiencies in BRCA genes leads to excessive engagement of TMEJ and “addiction” of cells to this pathway, which can help drive cancer progression.
Materials and Methods
Cell Lines.
Polq−/− MEFs were generated and immortalized with T antigen as described (7) from Polq-null mice generated by conventional knockout (36) that were obtained from The Jackson Laboratory and maintained on a C57BL/6J background. Pol θ was expressed by introducing WT POLQ human cDNA by lentiviral infection, with cells maintained in medium containing 4 μg/mL of puromycin. Cell culture conditions and validation of mycoplasma-free cultures were as described in SI Appendix.
Extrachromosomal Assay.
As in a previous report (11), extrachromosomal substrates consist of a 557-bp core DNA duplex ligated to head and tail caps with end structures that were varied as described in each figure and using oligonucleotide sequences as described in SI Appendix, Table S2); a more detailed description of substrate generation methods can be found in SI Appendix, Supplementary Methods. A total of 75 ng of these substrates was electroporated into 200,000 cells with 1 μg of pMAX-GFP (Lonza) using the Neon system (Invitrogen) in a 10-μL tip with a 1,350 V, 30-ms pulse (three pooled electroporations formed a biological replicate) and incubated for 1 h at 37 °C. Cells were washed with Hank’s balanced saline solution and incubated with 25 U of benzonase (Sigma) for 10 min at 37 °C. DNA was purified using the QIAamp DNA mini kit (QIAGEN), and products were analyzed using SYBR green qPCR or a 30-cycle end point PCR (primers described in SI Appendix, Table S3) and run on a 6% polyacrylamide gel. Each experiment consisted of three replicates of the above protocol. Quantification of gel bands was done with ImageQuant 8.1
Chromosomal Assay.
Chromosomal DSB repair assays were performed using Cas9-gRNA RNP complexes assembled from Cas9 purified after overexpression in bacteria (37) (Addgene 69090), as well as annealed tracrRNA and target-specifying crRNA (see SI Appendix, Table S4 for gRNA target sequence) with stabilizing modifications (Alt-R, IDT). A total of 7 pmol of Cas9 was incubated at room temperature with 8.4 pmol of annealed crRNA+tracrRNA for 30 min, and electroporated into 200,000 cells with 32 ng of pMAX-GFP as described above (three pooled electroporations formed a biological replicate). Cells were incubated for 24 h and DNA was harvested with the QIAamp DNA mini kit (QIAGEN). Each experiment consisted of three biological replicates. DNA equivalent to 60,000 genomes was amplified for 24 cycles using primers purified by polyacrylamide gel electrophoresis (IDT) that included a 6-bp barcode, a spacer sequence of varying length (1 to 8 bp) to increase library diversity, and 21 (forward primer) or 22 (reverse primer) bp of Illumina adapter sequence (sequences in SI Appendix, Table S5). Amplicons were then purified using a 2% agarose (Lonza) gel and the QIAquick Gel Extraction Kit (QIAGEN), recovered DNA amplified with secondary NGS PCR primers (SI Appendix, Table S5) for five cycles, and purified with AMPure XP beads (Beckman Coulter). Libraries were sequenced using a 300-cycle MiniSeq Mid Output Kit (R26A) or an iSEq. 100 i1 Kit (R26B-E), including 20% of PhiX Control v3 DNA (Illumina). The number of reads analyzed per sample is reported in SI Appendix, Table S6, and analysis methods are described in SI Appendix. Deindexed, filtered, and characterized junctions are provided in Datasets S1–S5. Data were analyzed using CLC Genomics Workbench 8 and Microsoft Excel.
Statistical Analysis.
To identify MHDs significantly depleted in cells lacking Polq, we first identified the set of deletion products where each product was represented at a mean frequency greater than 1 × 10−5 in cells expressing POLQ. We then compared the frequencies for each product from three biological replicates for both Polq−/− cells vs. Polq−/− + POLQ cells using a two-tailed t test without sample pairing, and employed the Benjamini–Hochberg method with a false discovery rate of 0.05 to limit false positives that arise from making multiple comparisons. Calculations were performed in Microsoft Excel. Other experiments employed statistical tests as indicated in the figure legends using GraphPad Prism 8.
Data Availability.
All raw fastq files are available at NCBI SRA (accession number PRJNA605803). The tables of results after additional analysis of the deindexed junctions are available upon request.
Supplementary Material
Acknowledgments
Our studies were supported by National Cancer Institute grant CA222092 (D.A.R. and G.P.G.), Department of Defense grant W81XWH-18-1-004 (D.A.R. and G.P.G.) and T32CA009156 (J.-E.C.)
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All raw fastq files are available at National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (accession number PRJNA605803).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921791117/-/DCSupplemental.
References
- 1.Chapman J. R., Taylor M. R. G., Boulton S. J., Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Waters C. A., Strande N. T., Wyatt D. W., Pryor J. M., Ramsden D. A., Nonhomologous end joining: A good solution for bad ends. DNA Repair (Amst.) 17, 39–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jasin M., Rothstein R., Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 5, a012740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venkitaraman A. R., Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108, 171–182 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Chan S. H., Yu A. M., McVey M., Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6, e1001005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roerink S. F., van Schendel R., Tijsterman M., Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 24, 954–962 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yousefzadeh M. J., et al. , Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 10, e1004654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulton S. J., Jackson S. P., Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 15, 5093–5103 (1996). [PMC free article] [PubMed] [Google Scholar]
- 9.Kabotyanski E. B., Gomelsky L., Han J. O., Stamato T. D., Roth D. B., Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 26, 5333–5342 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma J.-L., Kim E. M., Haber J. E., Lee S. E., Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23, 8820–8828 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyatt D. W., et al. , Essential roles for polymerase θ-Mediated end joining in the repair of chromosome breaks. Mol. Cell 63, 662–673 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saito S., Maeda R., Adachi N., Dual loss of human POLQ and LIG4 abolishes random integration. Nat. Commun. 8, 16112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zelensky A. N., Schimmel J., Kool H., Kanaar R., Tijsterman M., Inactivation of Pol θ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat. Commun. 8, 66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceccaldi R., et al. , Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 518, 258–262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mateos-Gomez P. A., et al. , Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 518, 254–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng W., et al. , Genetic determinants of cellular addiction to DNA polymerase theta. Nat. Commun. 10, 4286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins G. S., Boulton S. J., Beyond PARP-POLθ as an Anticancer Target. Science 359, 1217–1218 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Kent T., Chandramouly G., McDevitt S. M., Ozdemir A. Y., Pomerantz R. T., Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol. 22, 230–237 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He P., Yang W., Template and primer requirements for DNA Pol θ-mediated end joining. Proc. Natl. Acad. Sci. U.S.A. 115, 7747–7752 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvajal-Garcia J., Cho J.-E., Ramsden D. A., High throughput sequencing data from mechanistic basis for microhomology identification and genome scarring by polymerase theta. R26A, R26B, R26C-E+extrachromosomal. NCBI. https://www.ncbi.nlm.nih.gov/sra/PRJNA605803. Deposited 11 February 2020. [DOI] [PMC free article] [PubMed]
- 21.Yoshida H., et al. , Analysis of the joining sequences of the t(15;17) translocation in human acute promyelocytic leukemia: Sequence non-specific recombination between the PML and RARA genes within identical short stretches. Genes Chromosomes Cancer 12, 37–44 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Jäger U., et al. , Follicular lymphomas’ BCL-2/IgH junctions contain templated nucleotide insertions: Novel insights into the mechanism of t(14;18) translocation. Blood 95, 3520–3529 (2000). [PubMed] [Google Scholar]
- 23.Khodaverdian V. Y., et al. , Secondary structure forming sequences drive SD-MMEJ repair of DNA double-strand breaks. Nucleic Acids Res. 45, 12848–12861 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Schendel R., van Heteren J., Welten R., Tijsterman M., Genomic scars generated by polymerase theta reveal the versatile mechanism of alternative end-joining. PLoS Genet. 12, e1006368 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu A. M., McVey M., Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 38, 5706–5717 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schimmel J., van Schendel R., den Dunnen J. T., Tijsterman M., Templated insertions: A smoking gun for polymerase theta-mediated end joining. Trends Genet. 35, 632–644 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Nik-Zainal S., et al. , Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins G. S., et al. , Overexpression of POLQ confers a poor prognosis in early breast cancer patients. Oncotarget 1, 175–184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemée F., et al. , DNA polymerase θ up-regulation is associated with poor survival in breast cancer, perturbs DNA replication, and promotes genetic instability. Proc. Natl. Acad. Sci. U.S.A. 107, 13390–13395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schimmel J., Kool H., van Schendel R., Tijsterman M., Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 36, 3634–3649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kan Y., Ruis B., Takasugi T., Hendrickson E. A., Mechanisms of precise genome editing using oligonucleotide donors. Genome Res. 27, 1099–1111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biehs R., et al. , DNA double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Mol. Cell 65, 671–684.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anand R., Beach A., Li K., Haber J., Rad51-mediated double-strand break repair and mismatch correction of divergent substrates. Nature 544, 377–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Black S. J., et al. , Molecular basis of microhomology-mediated end-joining by purified full-length Polθ. Nat. Commun. 10, 4423 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth D. B., Wilson J. H., Nonhomologous recombination in mammalian cells: Role for short sequence homologies in the joining reaction. Mol. Cell. Biol. 6, 4295–4304 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shima N., Munroe R. J., Schimenti J. C., The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol. Cell. Biol. 24, 10381–10389 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin S., Staahl B. T., Alla R. K., Doudna J. A., Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw fastq files are available at NCBI SRA (accession number PRJNA605803). The tables of results after additional analysis of the deindexed junctions are available upon request.