Significance
Using forward genetic analysis, we identified a previously unknown regulator of mouse lymphopoiesis, RABL3. The small GTPase RABL3 is also necessary for embryonic development. We found that GPR89, a putative GPCR or ion channel, interacts with, and is stabilized by, RABL3, and mice with hypomorphic mutations in either protein displayed B cell and T cell deficiencies. We crystallized RABL3 and showed it forms a homodimer with subunits similar to other small G proteins. However, the conformation of mutated RABL3 is abnormal and may disrupt effector binding. Critical proteins that control hematopoiesis remain to be discovered, and RABL3 is one such example.
Keywords: small GTPase, lymphopoiesis, GPR89, X-ray crystallography
Abstract
The small GTPase RABL3 is an oncogene of unknown physiological function. Homozygous knockout alleles of mouse Rabl3 were embryonic lethal, but a viable hypomorphic allele (xiamen [xm]) causing in-frame deletion of four amino acids from the interswitch region resulted in profound defects in lymphopoiesis. Impaired lymphoid progenitor development led to deficiencies of B cells, T cells, and natural killer (NK) cells in Rabl3xm/xm mice. T cells and NK cells exhibited impaired cytolytic activity, and mice infected with mouse cytomegalovirus (MCMV) displayed elevated titers in the spleen. Myeloid cells were normal in number and function. Biophysical and crystallographic studies demonstrated that RABL3 formed a homodimer in solution via interactions between the effector binding surfaces on each subunit; monomers adopted a typical small G protein fold. RABL3xm displayed a large compensatory alteration in switch I, which adopted a β-strand configuration normally provided by the deleted interswitch residues, thereby permitting homodimer formation. Dysregulated effector binding due to conformational changes in the switch I–interswitch–switch II module likely underlies the xm phenotype. One such effector may be GPR89, putatively an ion channel or G protein-coupled receptor (GPCR). RABL3, but not RABL3xm, strongly associated with and stabilized GPR89, and an N-ethyl-N-nitrosourea (ENU)-induced mutation (explorer) in Gpr89 phenocopied Rabl3xm.
To identify proteins with previously unrecognized and nonredundant functions in immunity, we carried out a forward genetic screen in third-generation (G3) mice carrying N-ethyl-N-nitrosourea (ENU)-induced germline mutations. Several mice from a single pedigree showed reduced frequencies of CD3+ T cells (SI Appendix, Fig. S1A), an increased CD4+-to-CD8+ T cell ratio (SI Appendix, Fig. S1B), higher CD44 expression in CD8+ T cells (SI Appendix, Fig. S1C), and an increased B cell-to-T cell ratio (SI Appendix, Fig. S1D) compared to wild-type (WT) mice. The phenotype was named “xiamen” (xm). Automated meiotic mapping (1) indicated that all aspects of the xm phenotype correlated with homozygosity for a mutation in Rabl3 (Fig. 1A), encoding Rab-like protein 3 (RABL3). The xm mutation resulted in an A-to-G transition eight nucleotides from the 3′ end of Rabl3 exon 2, which generated a cryptic donor splice site utilized to the exclusion of the normal intron 2 splice donor site (Fig. 1B). Twelve nucleotides were deleted from exon 2, resulting in an in-frame deletion of four amino acids (aa) (aa 43 to 46) from the 236-aa RABL3 protein (SI Appendix, Fig. S2 A–C). The xm mutation resulted in lower RABL3 expression compared to WT RABL3 (SI Appendix, Fig. S2D).
Fig. 1.
The xiamen phenotype. (A) Manhattan plot. −Log10 P values plotted vs. the chromosomal positions of mutations identified in the G1 founder of the affected pedigree. Y, Y chromosome. (B) Schematic of xiamen mutation in Rabl3. (C) Representative photograph of 8-wk-old female xm/− mouse and WT littermate. (D) Body weights of 12-wk-old mice (n = 7–23 mice per genotype). (E–H) Complete blood count testing. White blood cells (WBC) (E), lymphocytes (F), monocytes (G), platelets (H), and neutrophils (I) in 8-wk-old xm/− mice and WT littermates. Data are representative of two independent experiments with seven mice per genotype. K, thousands. (J–W) Immune phenotypes of Rabl3+/+, Rabl3+/−, or Rabl3xm/- mice. (J) Frequency of CD3+ T cells in the peripheral blood. (K) Ratio of CD4+ to CD8+ peripheral blood T cells. (L and M) Mean florescence intensity (MFI) of CD44 (L) and CD62L (M) in CD3+ T cells from peripheral blood. (N) Ratio of B cells to T cells (B:T) in peripheral blood. B220 (O), IgD (P), and IgM (Q) MFI in peripheral blood B cells. (R) Serum βGal-specific IgG level in mice immunized with T cell-dependent antigen rSFV-βGal. Data presented as absorbance at 450 nm. (S) Quantitative analysis of the β-gal–specific cytotoxic T cell killing response in mice immunized with rSFV-βGal. (T) Serum NP-specific IgM level in mice immunized with T cell-independent antigen NP-Ficoll. Data presented as absorbance at 450 nm. (U) Viral load in spleens of mice 7 d after MCMV infection. (V) Quantitative analysis of NK cell killing response toward B2m-deficient target cells 3 d after target cell injection. (W) Frequency of NK cells in peripheral blood. (J–W) Data are representative of one experiment with 6 to 23 mice per genotype; data points represent individual mice. Error bars indicate SD. *P < 0.05; ***P < 0.001; NS, not significant. bp, base pairs.
To confirm causation, we used CRISPR/Cas9 gene targeting to knock out Rabl3. However, no Rabl3−/− mice were born from crosses of heterozygotes, suggesting that complete ablation of this gene is embryonic lethal (P = 2.54 × 10−8, χ2 test; n = 104 mice: 37 Rabl3+/+, 67 Rabl3+/−, 0 Rabl3−/−). We therefore crossed Rabl3xm/+ mice to CRISPR/Cas9-targeted Rabl3 heterozygotes (Rabl3+/−), which were phenotypically normal in both cases, to generate Rabl3 compound heterozygotes (Rabl3xm/−; hereafter xm/−) with simple heterozygosity for all residual ENU-induced mutations. Offspring with the xm/− genotype were born at slightly below the expected Mendelian frequency (P = 0.04689, χ2 test; n = 200 mice: 57 Rabl3+/+, 108 Rabl3+/− or Rabl3xm/+, 35 Rabl3xm/−) and had no obvious external deformities although they were ∼30% smaller than their WT littermates (Fig. 1 C and D). The thymi and spleens of xm/− mice were smaller and contained fewer cells than those of WT littermates (Fig. 2A). Complete blood count testing indicated that xm/− mice had reduced numbers of white blood cells (Fig. 1E) and lymphocytes (Fig. 1F); numbers of monocytes, platelets, and neutrophils were normal (Fig. 1 G–I).
Fig. 2.
Impaired thymocyte development caused by a Rabl3 mutation in mice. (A) Numbers of each thymocyte or T cell population in 8-wk-old xm/− and WT littermates. (B and C) Representative flow cytometry plots of thymocytes in xm/− mice and WT littermates. Numbers adjacent to outlined regions indicate percent cells in each ± SD (n = 5 xm/− mice, 8 WT littermates). (D) Flow cytometry analysis of CD44 expression (mean fluorescence intensity [MFI]) on thymic or splenic T cells in 8-wk-old xm/− mice and WT littermates. (E and F) Annexin V staining of CD8+ (E) or CD4+ T cells (F) in peripheral blood obtained from 8- to 12-wk-old xm/− mice or WT littermates (n = 11–18 mice per genotype). (G) IL-7Rα expression on CD4+ or CD8+ T cells in peripheral blood obtained from 8-wk-old xm/− mice and WT littermates (n = 4–10 mice per genotype). (H) Antigen-specific CD8+ T cells induced by immunization of xm/− mice and WT littermates with OVA/alum (n = 5–18 mice per genotype). (I and J) Impaired homeostatic expansion of xm/− T cells. CellTrace Far Red-stained pan T cells isolated from the spleen of xm/− or WT littermates were adoptively transferred into sublethally irradiated (8.5 Gy) WT hosts (CD45.1). Representative flow cytometric histograms (I) and frequency (J) of proliferated dye-positive T cells in the spleen of irradiated recipients, 7 d after transfer (n = 5 mice per group). Data are representative of one experiment (H), two independent experiments with five to eight mice per genotype (A, D, I, and J), or three independent experiments (E–G). (A, D–H, and J) Data points represent individual mice. Error bars indicate SD. **P < 0.01; ***P < 0.001; NS, not significant. DP, double positive; SP, single positive; tet+, tetramer positive.
The xm/− mice recapitulated the phenotypes observed in xm/xm mice (Fig. 1 J–N), validating the Rabl3 mutation as causative. Further analysis showed xm/− mice had many fewer CD4+ and CD8+ T cells, total T cells, and total B cells per microliter of blood (SI Appendix, Fig. S1 E–H). Additional phenotypes, including reduced expression of CD62L on xm/− T cells, were observed, suggestive of an effector memory phenotype, consistent with the elevated expression of CD44. We also detected reduced surface B220 (Fig. 1O) and immunoglobulin D (IgD) (Fig. 1P) expression, with a concomitant increase in IgM expression (Fig. 1Q) on B cells in the peripheral blood of xm/− compared to WT mice, suggesting aberrant B cell development. In response to immunization with recombinant Semliki Forest virus-encoded β-galactosidase (rSFV-βgal), xm/− mice displayed impaired antigen-specific antibody production and cytotoxic T lymphocyte (CTL) killing activity compared to WT littermates (Fig. 1 R and S). However, the T cell-independent antibody response to 4-hydroxy-3-nitrophenylacetyl-Ficoll (NP-Ficoll) was normal (Fig. 1T). Xm/− mice challenged with a sublethal dose of mouse cytomegalovirus (MCMV) had elevated viral titers in the spleen 5 d after infection (Fig. 1U), probably as a consequence of impaired natural killer (NK) cytolytic function (Fig. 1V); NK cell frequencies in the blood were normal (Fig. 1W). Thus, mutation of Rabl3 had a broad effect on mature lymphocyte frequencies and functions.
We examined whether RABL3 is necessary for lymphocyte development. Given the severe peripheral T cell deficiency, we tested whether thymocyte development occurred normally in xm/− mice. We found that all immature T cell populations were reduced in number in xm/− thymi (Fig. 2A) despite relatively normal frequencies (Fig. 2B). We also observed changes in the proportions of double negative (DN) subsets in xm/− thymi (Fig. 2C). These findings suggest that Rabl3 mutation impairs T cell differentiation in the thymus. Mature CD4+ and CD8+ T cells in the spleens of xm/− mice showed increased expression of the surface glycoprotein CD44, which encompasses recently activated, expanding, and memory phenotype cells (Fig. 2D); however, this was not evident on developing thymocytes (Fig. 2D). An increased percentage of xm/− peripheral CD8+ and CD4+ T cells was apoptotic as measured by annexin V staining under steady-state conditions compared to the corresponding WT cells (Fig. 2 E and F). In addition, reduced interleukin (IL)-7Rα expression was detected in peripheral T cells of xm/− mice compared to WT littermates (Fig. 2G). Nonetheless, the antigen-specific CD8+ T cell response of xm/− mice to immunization with aluminum hydroxide-precipitated ovalbumin (OVA/alum) was normal (Fig. 2H).
It was unclear whether the severe T cell deficiency in xm/− mice was solely a consequence of impaired development or whether there was a survival defect in mature T cells. We therefore measured in vivo T cell survival by injecting both unirradiated and sublethally irradiated recipient mice with a mixture containing equal numbers of dye-labeled WT (CD45.1) and xm/− (CD45.2) splenic pan T cells. Seven days after adoptive transfer, we found that the spleens of recipient mice contained far fewer xm/− donor T cells than WT donor T cells (Fig. 2 I and J), and xm/− cells that were recovered appeared to have proliferated less compared to WT donor cells based on dye dilution (Fig. 2I). In addition, the frequency of xm/− donor T cells was significantly lower than that of WT donor T cells in spleens from the unirradiated recipients (Fig. 2 I and J). These results indicate a T cell intrinsic survival defect under lymphocyte-replete conditions that worsened during homeostatic expansion.
Additional analysis showed the frequency of pre-pro-B, pro-B, and pre-B cells in the bone marrow appeared comparable in xm/− mice and WT littermates (Fig. 3A); however, the absolute numbers of cells in these populations were reduced (Fig. 3H) due to overall reduced cell counts in the bone marrow (Fig. 3K). The frequency of immature B cells in the bone marrow was similar in xm/− mice and their WT littermates (B220+IgM+IgD−) (Fig. 3B), but absolute numbers were decreased (Fig. 3H), and few xm/− B cells progressed to the mature recirculating stage (B220+IgM+IgD+) (Fig. 3 B and H). In the spleen, xm/− mice had reduced frequencies and numbers of transitional, follicular, and marginal zone B cells (Fig. 3 C–F and H). In contrast, we observed increased frequencies and numbers of B1a and B1b cells in the peritoneal cavity of xm/− mice (Fig. 3 G and H), consistent with the normal TI antibody response in these animals (Fig. 1T). These data suggest that Rabl3 mutation affected B cell development.
Fig. 3.
Impaired B cell development caused by a Rabl3 mutation in mice. (A–G) Representative flow cytometry plots showing B cell development in the bone marrow (BM) (A and B), spleen (C–F), and peritoneal cavity (G) from 8-wk-old xm/− mice and WT littermates. Each B cell subset was gated as follows: pre-pro-B: B220lowBP-1−CD24low, pro-B: B220lowBP-1−CD24+, pre-B: B220lowBP-1+CD24+, immature B: B220+IgM+IgD−, transitional B: B220+IgMhighIgDlow, mature recirculating B: B220+IgD+IgM+, T1: B220+CD93+IgMhighCD23−, T2: B220+CD93+IgMhighCD23+, marginal zone precursor (MZP): B220+CD93−IgM+CD21+CD2a3high, T3: B220+CD93+IgMlowCD23+, follicular B (FOB): B220+CD93−IgM+CD21+CD23high, marginal zone B (MZB): B220+CD93−IgM+CD21+CD23low, B2: B220+CD19+, and B1: B220lowCD19+. Numbers adjacent or inside outlined regions represent percent cells in each ±SD (n = 5 xm/− mice, 8 WT littermates). (H) Numbers of each B cell subset in 8-wk-old xm/− mice and WT littermates. (I–K) Representative flow cytometry plots (I) and quantitative analysis (J and K) of the HSC and progenitor populations in the bone marrow of xm/− and WT littermates. (L) Repopulation of HSC and progenitor populations in competitive bone marrow chimeras. A 1:1 mixture of xm/− BM (CD45.2) cells and congenic WT BM (C57BL/6J; CD45.1) competitor cells was injected into lethally irradiated Rag2−/− recipients. Donor chimerism levels in the peripheral blood were assessed using congenic CD45 markers at 8 wk posttransplant (n = 6 recipients per group). (A–L) Data are representative of two independent experiments with three to eight mice per genotype. (H and J–L) Data points represent individual mice. Error bars indicate SD. *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. LSK, Lin−Sca-1+cKit+; LT-HSC, long-term hematopoietic stem cell; PBMC, peripheral blood mononuclear cells.
In addition to B and T cell deficiency, xm/− mice displayed reduced numbers of NK and NK1.1+ T cells in the spleen compared to WT littermates (SI Appendix, Fig. S3A). Normal numbers of myeloid cells were present in the spleens of xm/− mice (SI Appendix, Fig. S3 B and C). Finally, innate immune signaling in peritoneal macrophages was normal, as indicated by cytokine secretion in response to various stimuli (SI Appendix, Fig. S4). All together, these results indicate that RABL3 is essential for lymphopoiesis in mice.
The lymphocyte-specific effect of Rabl3 mutation led us to investigate hematopoietic stem and progenitor cell populations in the bone marrow. First, we found that xm/− mice had reduced numbers of bone marrow cells compared to WT littermates (Fig. 3K). The frequency of the LK+ population, which includes common myeloid progenitors (CMPs), megakaryocyte–erythrocyte progenitors (MEPs), and granulocyte–macrophage progenitors (GMPs), was increased in xm/− mice whereas the proportion and number of cells in each subpopulation were not affected (Fig. 3 I–K). However, we found that the frequency of lymphoid-primed multipotent progenitors (LMPPs) (Fig. 3J) and number of common lymphoid progenitors (CLPs) (Fig. 3K), which are the precursors of lymphocytes, were decreased in xm/− bone marrow compared to those from WT littermates (Fig. 3 J and K). These data are consistent with a specific impairment in lymphocyte development apparent as early as the LMPP stage in Rabl3 mutant mice.
We examined lymphocyte development in mixed bone marrow chimeras. We injected lethally irradiated Rag2−/− recipients with a 1:1 mixture of xm/− (CD45.2) and WT (CD45.1) bone marrow cells. Xm/− cells were at a striking disadvantage in repopulating the lymphoid lineage (CD45.2) (Fig. 3L). However, xm/− and WT cells were equally effective in repopulating the myeloid lineages (Fig. 3L). We also generated reciprocal bone marrow chimeras to determine if the xm/− phenotype was intrinsic to the hematopoietic compartment. Lymphocyte deficiency was observed in WT recipients (CD45.1) of xm/− bone marrow (CD45.2) whereas lymphocyte reconstitution occurred normally in xm/− recipients (CD45.2) of WT bone marrow (CD45.1) (SI Appendix, Fig. S5). These results indicate that normal lymphocyte development depends on RABL3 function in the hematopoietic compartment.
RABL3 is classified as a Rab-like small GTPase (2) although its sequence bears minimal similarity to proteins of the Ras superfamily, including the Rab proteins with which it shares at most 30% identity (SI Appendix, Fig. S6). RABL3 is highly conserved in mammals, with 92% amino acid sequence identity in humans and mice. The guanosine triphosphate (GTP)/guanosine diphosphate (GDP) binding motifs present in Rab family proteins, designated G1–G5, are well conserved in RABL3 while the C-terminal prenylation site is absent, and ∼20 C-terminal amino acids serve as the membrane-targeting motif (SI Appendix, Fig. S6). Phylogenetic studies suggest that RABL3 is the founder of an independent subfamily of the Ras superfamily (3, 4). Given its divergence from other Rab proteins, we were interested in the physical and structural properties of RABL3 and the effect of the xm mutation upon them.
Both WT and mutant mouse RABL3 were strongly stabilized by nucleotide binding in thermal shift assays (SI Appendix, Fig. S7). WT RABL3 (mouse protein unless otherwise specified) displayed a higher melting temperature (Tm) when bound to GTPγS than when bound to GDP (Tm = 51 °C vs. Tm = 48 °C), suggestive of a structural difference between 5’-O-[gamma-thio]triphosphate (GTPγS)- and GDP-bound WT RABL3. The Tm of RABL3xm was similar for GTPγS- and GDP-bound forms (Tm = 39 °C) and was lower than that of either GTPγS- or GDP-bound WT RABL3, indicating that the 4-aa deletion caused by the xm mutation reduces the thermal stability of RABL3.
Size exclusion chromatography showed that WT RABL3, alone or in the presence of GTPγS or GDP, and RABL3xm alone eluted in the same position, which corresponds to a dimer (SI Appendix, Fig. S8). By analytical ultracentrifugation, WT RABL3 at 24 μM had a molar mass of 41.7 kilodaltons (kDa), slightly lower than the dimeric molar mass (48.212 kDa) but consistent with a dimer (SI Appendix, Fig. S9). At a lower concentration (3.6 μM), WT RABL3 had a decidedly lower and more disperse distribution, likely indicating a dynamic equilibrium between the monomeric and dimeric forms (SI Appendix, Fig. S9). These data suggest that the dissociation constant of the WT RABL3 dimer is in the low micromolar range.
We determined the crystal structure of the G-domain of WT RABL3, alone (apo) and in complex with GTPγS or GDP (SI Appendix, Fig. S10 and Table S1). In all three structures, RABL3 formed a similar symmetrical dimer (SI Appendix, Fig. S11) (rmsd <0.5). Analysis of RABL3 monomers showed a typical small G protein fold consisting of six β-strands flanked by five α-helices (Fig. 4A). Nucleotides and magnesium bound to a pocket that is similar to canonical Rab nucleotide binding pockets (Fig. 4 B and C). The Rab family consensus sequences for G1–G4 are conserved in RABL3 (SI Appendix, Fig. S6) and made canonical interactions with GTPγS/GDP (Fig. 4 B and C and SI Appendix, Fig. S12A). The G5 motif of RABL3 does not conform to the consensus sequence [E(T/C/G)SA(K/L/A); RABL3 sequence: 177NLDCT181], with Asp-179 and Cys-180 in RABL3 substituting functionally for canonical Ser and Ala (SI Appendix, Figs. S6 and S12A). Mg2+ is coordinated tightly via water molecules, Ser-20, Thr-38, and β/γ-phosphate groups (SI Appendix, Fig. S12 B and C). The dimerization interface of RABL3 is composed of residues from the interswitch and switch II regions and is stabilized by two hydrogen bonds, two ionic interactions, and several hydrophobic interactions, with xm-deleted residues 43 to 46 located within the interswitch region (SI Appendix, Fig. S11D).
Fig. 4.
Crystal structures of WT RABL3 and RABL3xm monomers. (A) Ribbon diagram of the crystal structure of RABL3WT apo monomer. Secondary structures are labeled. (B and C) Ribbon diagrams of the crystal structures of RABL3WT:GTPγS:Mg2+ (B) and RABL3WT:GDP:Mg2+ (C) monomers. Nucleotide and magnesium binding motifs are highlighted in cyan and labeled G1 to G5. GTPγS and GDP are shown in yellow sticks. (D) Structural comparison of the monomers of RABL3WT apo, RABL3WT:GTPγS:Mg2+, and RABL3WT:GDP:Mg2+. GTPγS and GDP are shown in gray sticks. The disordered switch I of the apo structure is shown in cyan. GTPγS-bound switch II showed a large conformational difference compared to the apo and GDP-bound switch II. (E) Ribbon diagram of the crystal structure of RABL3xm:GTPγS:Mg2+. GTPγS is shown in yellow sticks. The GTPγS binding pocket is more open, and Mg2+ coordination is weaker than in the WT RABL3. (F) Structural comparison of the monomers of RABL3WT:GTPγS:Mg2+ and RABL3xm:GTPγS:Mg2+. GTPγS is shown in yellow sticks. RABL3xm switch I adopted a dramatically different conformation than in RABL3WT. RABL3xm switch II showed a large conformational change. Switch regions are highlighted by discrete colors. The magnesium ion is shown as a black sphere. Water molecules are shown as brown spheres. Red dotted lines show the magnesium ion coordination.
The apo and GTPγS- and GDP-bound WT RABL3 monomers were similar (rmsd <0.5), except for some conformational differences in switch I and switch II (Fig. 4D). GTPγS or GDP binding stabilized switch I, which had a well-defined conformation with these ligands but was disordered in the apo structure. The switch II helix is shorter, and the loop is more flexible in GTPγS-bound RABL3 than the GDP-bound or apo forms (Fig. 4D). This behavior contrasts with that of other Rab proteins, in which switch II is more flexible in the GDP-bound inactive form than in the GTP-bound active form (5).
We determined the crystal structure of RABL3xm in complex with GTPγS (SI Appendix, Table S1). RABL3xm formed a symmetrical dimer through hydrogen bonds and ionic and hydrophobic interactions (SI Appendix, Fig. S11 E and F), but its conformation was different from that of the WT dimer, with the two monomers adopting a different relative orientation (SI Appendix, Fig. S11E). The 4-aa deletion (residues 43 to 46) in the interswitch region of RABL3xm caused major conformational changes in switch I, which converted from a loop to two short antiparallel β-strands, one of which (residues 38 to 40) replaced the β-strand structure missing due to deletion of residues 43 to 46 (Fig. 4E). This alternate interswitch formed by residues 38 to 40 adopted a slightly different conformation from that of the WT GTPγS-bound RABL3 (Fig. 4 E and F). As a result of the change in switch I, the RABL3xm nucleotide binding pocket was more open and Mg2+ coordination was weaker than in WT RABL3 (Fig. 4E and SI Appendix, Fig. S12 B and D). Switch II also showed large conformational differences in the loop region that likely contributed to the weaker Mg2+ binding (Fig. 4F and SI Appendix, Fig. S12D). Altogether, the effect of the 4-aa deletion in RABL3xm was to impair nucleotide binding, consistent with the diminished thermal stability of nucleotide-bound RABL3xm compared to WT RABL3 (SI Appendix, Fig. S7).
The crystal structures of WT RABL3 and RABL3xm showed the formation of dimers mediated by residues in switch II and interswitch. Other Ras superfamily proteins also form dimers, an interaction that regulates their function (6–10). Because Rab effector binding is typically mediated by the dimerization interface, especially hydrophobic residues Tyr-61, Trp-65, and Phe-84 (5), we hypothesized that RABL3 dimerization may regulate effector binding.
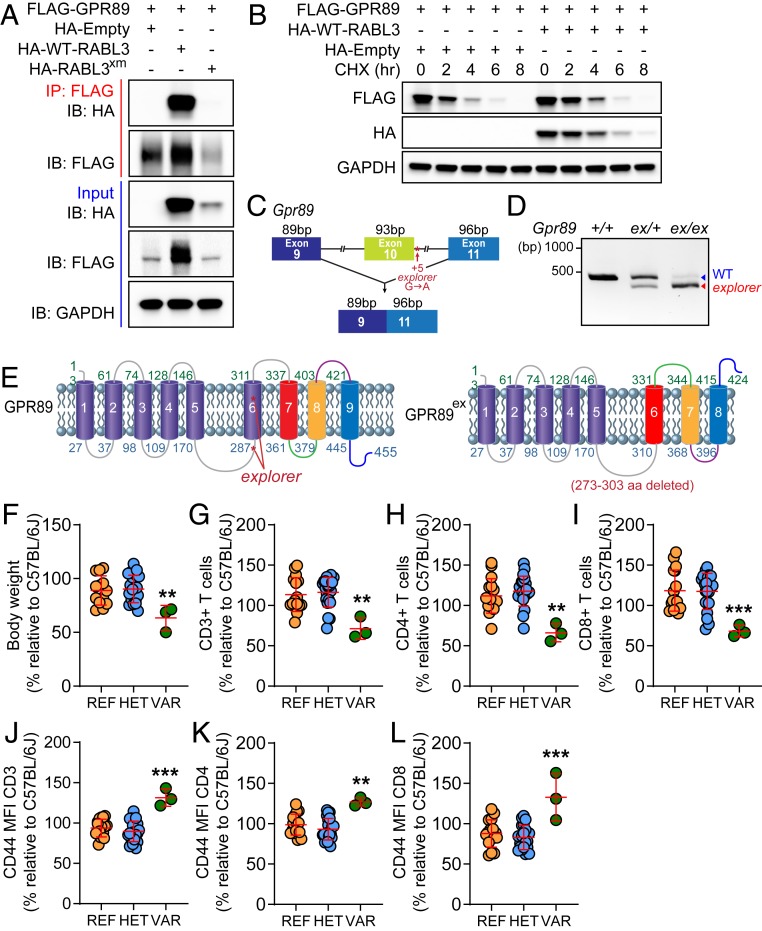
Through mass spectrometry analysis of FLAG-tagged RABL3 interacting proteins in EL4 cells (Dataset S1), we identified GPR89 as a putative interactor, which we confirmed by coimmunoprecipitation (co-IP) in human embryonic kidney 293T (HEK293T) cells after transient transfection (Fig. 5A). In contrast, RABL3xm displayed diminished interaction with GPR89 (Fig. 5A), a nine transmembrane-spanning protein with topology similar to GPCRs. GPR89 was also reported to function as an anion channel regulating Golgi pH (11). We noted higher expression levels of GPR89 when it was coexpressed with WT RABL3 than when it was expressed by itself or with RABL3xm. Supporting GPR89 stabilization by RABL3, FLAG-tagged GPR89 had slower decay kinetics when coexpressed with hemagglutinin (HA)-tagged RABL3 than when expressed alone after treatment with the translation inhibitor cycloheximide (Fig. 5B). Through forward genetic screening, we identified a pedigree of G3 mice carrying an ENU-induced germline mutation in Gpr89 that had a peripheral T cell deficiency. This pedigree was named explorer. The explorer mutation resulted in a G-to-A substitution in the fifth nucleotide of Gpr89 intron 10 and was predicted to impair the donor splice site, resulting in skipping of exon 10 (Fig. 5C). RT-PCR and complementary DNA (cDNA) sequencing verified that exon skipping occurred in explorer homozygotes but also showed the presence of a small amount of full-length Gpr89 messenger RNA (mRNA) (Fig. 5D), indicating that the mutated splice site supported normal splicing with very low efficiency. The predicted protein product contains an in-frame deletion of 31 amino acids from 273 to 303 of GPR89 (Fig. 5E). GPR89 is a putative GPCR with nine predicted transmembrane domains (TDs) and a long intracellular loop between the fifth and sixth TDs (170 to 287 aa). In canonical GPCRs, this intracellular loop is critical for coupling the receptor to its cognate G protein (12). The 31-aa deletion resulting from the explorer mutation was predicted to delete TD6 and reverse the orientation of TD7, TD8, and TD9 (Fig. 5E). Altogether, our analysis suggests that the explorer allele is a hypomorphic variant of GPR89. Many of the phenotypes found in Rabl3xm/xm mice were also found in Gpr89ex/ex mice (Fig. 5 F–L). Gpr89ex/ex mice had no obvious external deformities but were ∼40% smaller than their WT littermates (Fig. 5F). They also showed reduced frequencies of CD3+ T cells (Fig. 5G), CD4+ T cells (Fig. 5H), and CD8+ T cells (Fig. 5I) that displayed elevated CD44 expression (Fig. 5 J–L) compared to WT mice. These data strongly suggest that GPR89 interacts with RABL3 and may function as a RABL3 effector in hematopoietic cells.
Fig. 5.
Identification of GPR89 as a RABL3 interacting protein and the explorer phenotype. (A and B) HEK293T cells were transfected with FLAG-tagged GPR89 and either HA-tagged WT-RABL3, RABL3xm, or empty vector. (A) Total cell lysates (TCLs) were immunoprecipitated by using anti-FLAG M2 agarose beads (IP). Immunoblot (IB) analysis of IPs or TCLs with antibodies against HA or FLAG. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. (B) Cells were treated with cycloheximide (CHX) 14 h after transfection and harvested after treatment for the indicated times. TCLs were immunoblotted with the indicated antibodies. (C) Schematic of the explorer (ex) mutation, a G-to-A transition of the fifth nucleotide of intron 10 predicted to damage the intron 10 donor splice site leading to skipping of exon 10. (D) RT-PCR amplification across exon 10 of Gpr89 mRNA from WBC of the indicated genotypes. (E) Predicted membrane topology of GPR89ex. The explorer mutation is predicted to cause in-frame deletion of 31 aa (indicated in red). (F–L) The explorer phenotype. Mice from a single pedigree with REF (+/+), HET (ex/+), or VAR (ex/ex) genotypes for the Gpr89ex allele were analyzed and compared to C57BL/6J (WT) mice. (F) Body weights of 6-wk-old mice (n = 3–21 mice per genotype). (G–I) Frequency of CD3+ T cells (G), CD4+ T cells (H), CD8+ T cells (I) in the peripheral blood expressed as a percentage relative to the average frequency for 30 C57BL/6J (WT) mice. (J–L) Mean fluorescence intensity (MFI) of CD44 expression on peripheral blood CD3+ T cells (J), CD4+ T cells (K), or CD8+ T cells (L) expressed as a percentage relative to the average CD44 MFI in the corresponding cells from 30 C57BL/6J (WT) mice. Data are representative of one experiment with three to 21 mice per genotype (F–L), or one (B), two (D), or three (A) independent experiments. (F–L) Data points represent individual mice. Error bars indicate SD. **P < 0.01; ***P < 0.001. bp, base pairs.
Previous reports have provided evidence that human RABL3 (hRABL3) is an oncogene that predisposes individuals to pancreatic and other types of cancer (2, 13–16). A truncated form of hRABL3 resulting from a premature stop codon (Ser36*) was reported to interact with SmgGDS to promote KRAS prenylation, resulting in KRAS activation and hereditary human pancreatic ductal adenocarcinoma (13). Our data indicate that this mutation terminates the protein within switch I and may therefore impair dimerization and consequently dysregulate the binding of effectors such as SmgGDS. However, the physiological function of RABL3, beyond suppression of cancer, was an open question. Based on the studies reported here, RABL3 is essential in mice as a null mutation was embryonic lethal in the homozygous state. The viable hypomorphic allele xm, discovered through random germline mutagenesis, revealed that mouse RABL3 also acts in the hematopoietic compartment where it is necessary to support lymphocyte development. RABL3 dimerization, discovered in biophysical and crystallography studies, may regulate the binding of GPR89 or other proteins that mediate survival or differentiation signals in lymphocyte precursors.
Materials and Methods
Mice.
C57BL/6J male mice (8- to 10-wk-old) were purchased from The Jackson Laboratory and mutagenized with N-ethyl-N-nitrosourea (ENU) as described previously (1). Mutagenized G0 males were bred for three generations as described previously (1) to yield male and female G3 mice carrying heterozygous and homozygous ENU-induced mutations. G3 mice were screened for phenotypes. Whole-exome sequencing and mapping were performed following previously described protocols (1).
To generate mice carrying a Rabl3-null allele, female C57BL/6J mice were superovulated by injection with 6.5 units (U) of pregnant mare serum gonadotropin (PMSG) (Millipore) and then 6.5 U of human chorionic gonadotropin (hCG) (Sigma) 48 h later. The superovulated mice were subsequently mated with C57BL/6J male mice overnight. The following day, fertilized eggs were collected from the oviducts, and in vitro-transcribed Cas9 mRNA (50 ng/μL) and Rabl3 small base-pairing guide RNA (50 ng/μL; 5′-GATTTCCTAACACTTGATTG-3′) were injected into the cytoplasm or pronucleus of the embryos. The injected embryos were cultured in M16 medium (Sigma) at 37 °C and 95% air/5% CO2. For the production of mutant mice, two-cell stage embryos were transferred into the ampulla of the oviduct (10 to 20 embryos per oviduct) of pseudopregnant Hsd:ICR (CD-1) (Harlan Laboratories) females. Mice were housed in specific pathogen-free conditions at the University of Texas Southwestern Medical Center, and all experimental procedures were performed in accordance with institutionally approved protocols.
Flow Cytometry.
Peripheral blood was collected from G3 mice >6 wk old by cheek bleeding. Red blood cells (RBCs) were lysed with hypotonic buffer (eBosceince). Samples were washed with fluorescence-activated cell sorter (FACS) staining buffer (phosphate-buffered saline [PBS] with 1% [wt/vol] bovine serum albumin [BSA]) one time at 500 × g for 6 min. The RBC-depleted samples were stained for 1 h at 4 °C, in a 100-μL (1:200 dilution) mixture of fluorescence-conjugated antibodies to 15 cell surface markers encompassing the major immune lineages B220 (clone RA3-6B2; BD), CD19 (clone 1D3; BD), IgM (clone R6-60.2; BD), IgD (clone 11-26c.2a; Biolegend), CD3ε (clone 145-2C11; BD), CD4 (clone RM4-5; BD), CD8α (clone 53-6.7; Biolegend), CD11b (clone M1/70; Biolegend), CD11c (clone HL3; BD), F4/80 (clone BM8.1; Tonbo), CD44 (clone 1M7; BD), CD62L (clone MEL-14; Tonbo), CD5 (clone 53-7.3; BD), CD43 (clone S7; BD), NK 1.1 (clone PK136; Biolegend), and Fc shield (clone 2.4G2; Tonbo). Data were collected on a BD LSR Fortessa, and the proportions of immune cell populations in each G3 mouse were analyzed with FlowJo software. The resulting screening data were uploaded to Mutagenetix for automated mapping of causative alleles.
Hematopoietic stem and progenitors, B and T cell subsets were stained using antibodies listed below: anti-mouse lineage mixture (clones 17A2/RB6-8C5/RA3-6B2/Ter-119/M1/70; Biolegend), CD135 (clone A2F10; Biolegend), CD16/CD32 (clone 93; eBioscience), CD34 (clone RAM34; eBioscience), CD127 (clone SB/199; BD), Ly-51 (clone BP-1; BD), CD93 (AA4.1; BD), Ly-6A/E (Sca-1) (clone D7; Biolegend), CD117 (c-kit) (clone 2B8; Biolegend), CD4 (clone RM4-5; BD), CD8α (clone 53-6.7; Biolegend), CD25 (clone 3C7; Biolegend), and CD44 (clone 1M7; BD), CD23 (clone B3B4; BD), CD43 (clone S7; BD), CD5 (clone 53-7.3; BD), B220 (clone RA3-6B2; BD), CD19 (clone 1D3; BD), IgM (clone R6-60.2; BD), IgD (clone 11-26c.2a; Biolegend), and CD21/CD35 (clone 7E9; Biolegend).
Bone Marrow Chimeras.
Recipient mice were given a 7 Gy exposure by X-ray irradiator twice at a 5-h interval. Antibiotic water was provided to the recipients after irradiation. Femurs from donor WT (CD45.1), Rabl3xm/- (CD45.2), or Rabl3+/+ (CD45.2) mice were placed in a small dish containing medium (RPMI-1640 medium; Life Technologies) supplemented with 10% (vol/vol) fetal bovine serum (FBS) (Life Technologies), 10 units/mL penicillin, and streptomycin (Life Technologies). The femurs were flushed with this medium using a 25G needle. To remove bits of bone, the marrow was homogenized, and the solution was put through a sterile 40-µm nylon cell strainer (BD) and collected in a 50-mL tube. The volume was brought to 50 mL with medium and then centrifuged at 700 × g for 5 min at 4 °C. The cells were then resuspended in 5 mL of red blood cell lysing buffer (Sigma) and incubated for 1 to 2 min. Five milliliters of sterile PBS was added to the tube, and 10 μL of cells and 10 μL of Trypan Blue were mixed to count the total cells. The cells were centrifuged at 700 × g for 5 min, and cells were resuspended in 1 mL of PBS and transferred into 1.5-mL Eppendorf tubes and kept on ice. Bone marrow cells from each donor or 1:1 mixture were transferred into the indicated recipient mice through retroorbital injection.
Peripheral blood was sampled at time points posttransplantation, and the fraction of donor chimeras within the major immune cell populations was assessed with flow cytometry using fluorescence-conjugated antibodies against the CD45 congenic markers (CD45.1, clone A20 [Biolegend]; CD45.2, clone 104 [Biolegend]) and a mixture of fluorescence-conjugated antibodies to 15 cell surface markers encompassing the major immune lineages as described above.
Immunization and Enzyme-Linked Immunosorbent Assay Analysis.
Eight- to 12-wk-old mice were immunized with T cell-dependent antigen rSFV-βGal (2 × 106 IU) (17) on day 0 and the T cell-independent antigen NP-Ficoll (50 μg; Biosearch Technologies) on day 8 (intraperitoneally [i.p.]) as previously described (18). Six days after NP-Ficoll immunization, blood was collected in minicollect tubes (Mercedes Medical) and centrifuged at 700 × g to separate serum for the antigen-specific IgG or IgM concentration analysis.
For enzyme-linked immunosorbent assay (ELISA) analysis of antibody responses, Nunc MaxiSorp flat-bottom 96-well microplates (Thermo) were coated with 5 μg of antigens and incubated at 4 °C overnight. Plates were washed four times with washing buffer (0.1% [vol/vol] Tween 20 in PBS) using a BioTek microplate washer and then blocked with 1% (vol/vol) BSA in PBS for 1 h at room temperature.
Serum samples were serially diluted in blocking buffer, and then the 1:150 dilutions were added to the prepared ELISA plates. After a 2-h incubation, plates were washed eight times with washing buffer and then incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG or IgM for 1 h at room temperature. Plates were washed eight times with washing buffer and then developed with 3,3′,5,5′-tetramethylbenzidine (TMB) peroxidase substrate and stop solution (KPL). Absorbance was measured at 450 nm on a Synergy Neo Multi-Mode Microplate Reader (BioTek).
In Vivo CTL and NK Cytotoxicity.
CD8+ T cell cytolytic effector function was measured using a standard in vivo cytotoxic T lymphocyte (CTL) assay (19, 20). Briefly, splenocytes from naive mice were collected, and half were stained with 5 µM carboxyfluorescein succinimidyl ester (CFSEhi) and the other half with 0.5 µM CFSE (CFSElo). The CFSEhi cells were pulsed with peptide (5 μM) containing the Escherichia coli β-galactosidase major histocompatibility complex (MHC) I epitope for mice with the H-2b haplotype (ICPMYARV; New England Peptide). CFSElo cells were not stimulated. Then 2 × 106 cells consisting of CFSEhi and CFSElo cells mixed 1:1 were injected retroorbitally into naive mice, and the mice were immunized with rSFV-βgal. Twenty-four hours after adoptive transfer, blood was collected, and CFSE intensities were assessed by flow cytometry. Lysis of target (CFSEhi) cells was calculated as follows: percent lysis = [1 – (ratio control mice/ratio vaccinated mice)] × 100; ratio = percent CFSElo/percent CFSEhi.
NK cell-mediated killing was measured as previously described (20). CellTrace Violet was used to stain splenocytes from C57BL/6J mice (0.5 μM Violet; Violetlo) and B2m−/− mice (5 μM Violet; Violethi). Recipient mice were injected retroorbitally with equal numbers of Violethi and Violetlo cells. After 24 h, blood was collected for measurement of the number of remaining Violethi and Violetlo cells by flow cytometry as follows: percent lysis = [1 – (target cells/control cells)/(target cells/control cells in B2m−/−)] × 100.
Homeostatic Proliferation of T Cells.
Isolated pure WT CD45.1 or Rabl3xm/− CD45.2 pan T cells were isolated by using the EasySep Mouse Pan T Cell Isolation Kit (StemCell Technologies) and labeled with 5 µM CellTrace Far Red (Life Technologies). The labeled cells were transferred to mice that had been sublethally irradiated (8 Gy) 6 h earlier or into unirradiated controls by retroorbital injection. Seven days after adoptive transfer, splenocytes were prepared, surface stained for CD45.1, CD45.2, together with CD3, CD4, and CD8, and then analyzed by flow cytometry for Far Red dye dilution.
MCMV Challenge and Quantification.
MCMV (Smith strain) was administered i.p. at 105 plaque-forming units (PFU) per 20 g of body weight as described previously (21). After 5 d, total genomic DNA (including host and virus DNA) was extracted from ∼30 mg of spleen tissue from each infected mouse using 200 μL of PBND buffer (10 mM Tris⋅HCl, pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 0.1 mg/mL gelatin, 0.45% [vol/vol] IGEPAL CA-630, and 0.45% [vol/vol] Tween 20) supplemented with 100 μg/mL proteinase K at 56 °C overnight. The next day, the supernatant containing the DNA was collected, and proteinase K was inactivated by incubation at 95 °C for 15 min. A 1:100 dilution of the supernatant was used for real-time quantitative PCR (RT-qPCR) to measure copy numbers of mouse genome (represented by Actb) and MCMV genome (represented by MCMV immediate-early gene IE1). The ratio of MCMV-IE1 to Actb represented the viral titer (shown in log10). RT-qPCR primer sequences were as follows: MCMV-IE1 (5′-GCATCGAAAGACAACGCAAG-3′ [fwd] and 5′- ACGTAGCTCATCCAGACTCTC-3′ [rev]); Actb (5′-AGCTCACCATTCACCATCTTG -3′ [fwd] and 5′-GACTCATCGTACTCCTGCTTG -3′ [rev]).
Detection of Apoptosis.
Annexin V/PI labeling and detection were performed with the fluorescein isothiocyanate (FITC)-Annexin V Apoptosis Detection Kit I (BD) according to the manufacturer’s instructions.
Detection of Antigen-Specific CD8+ T Cells.
Mice were immunized with aluminum hydroxide precipitated ovalbumin (OVA/alum) (200 μg; Invivogen) at day 0, and the frequency of OVA-specific CD8+ T cells was analyzed at day 14 by flow cytometry. Phycoerythrin (PE)-conjugated Kb/SIINFEKL tetramer, a reagent specific for the ovalbumin epitope peptide SIINFEKL presented by H-2 Kb (MHC Tetramer Core at Baylor College of Medicine) was used to detect antigen-specific CD8+ T cells.
Plasmids.
Full-length or mutant mouse Rabl3 was tagged with N-terminal HA epitope in pCMV6 vector. Full-length mouse Gpr89a with FLAG epitope was purchased from Origene. For liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis, full-length mouse Rabl3 was cloned with C-terminal FLAG epitope in pMSCV-IRES-GFP vector (22). Plasmids were sequenced to confirm the absence of undesirable mutations.
Generation of Stable Cell Lines for LC-MS/MS Analysis.
PT67 (ATCC) cells were transfected with FLAG-tagged Rabl3-pMSCV-IRES-GFP or empty vector (pMSCV-IRES-GFP) using Lipofectamine 2000 (Life Technologies). Green fluorescent protein (GFP)-positive PT67 cells were sorted twice by Aria II SORP (BD). Retroviruses recovered from the media of PT67 cells were concentrated by Retro-X concentrator kit (Takara) according to the manufacturer’s instructions and infected to WT EL-4 cells (ATCC) with polybrene (4 µg/mL). Forty-eight hours after retrovirus infection, GFP expressing EL-4 cells were sorted as described above. Immunoprecipitation was performed using anti-FLAG M2 agarose beads (Sigma) for 4 h at 4 °C, and beads were washed five times in Nonidet P-40 lysis buffer. The proteins were eluted with 100 μg/mL 3× Flag at 4 °C for 30 min. Lysates were loaded onto a 12% (wt/vol) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) gel and run ∼1 cm into separation gel. The gel was stained with Coomassie blue (Thermo Fisher), and whole stained lanes were subjected to mass spectrometry analysis (LC-MS/MS) as described previously (20).
Coimmunoprecipitation and Western Blot.
The HEK293T cells were grown at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies)/10% (vol/vol) FBS (Gibco)/1% antibiotics (Life Technologies) in 5% CO2. Transfection of plasmids was carried out using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were harvested in Nonidet P-40 lysis buffer for 45 min at 4 °C. For the cycloheximide (CHX) chase assay, cells were treated with CHX 14 h after transfection and harvested at various times posttreatment. Immunoprecipitation was performed using anti-FLAG M2 agarose beads (Sigma). Whole-cell lysates and immunoprecipitates were analyzed using anti-FLAG M2 antibody (Sigma) and anti-HA antibody (Cell Signaling Technology) using standard procedures for Western blot analysis.
Detection of Cytokine Secretion by Peritoneal Macrophages.
Brewer’s thioglycolate medium was used to boost peritoneal exudate cells (PECs). The concentration of each cell sample was adjusted to 5 × 105 cells per milliliter, and 200 μL of each sample was added to duplicate columns of a tissue culture-treated 96-well flat-bottomed plate. The NLRP3, NLRC4 inflammasome, and TLR (TLR4, TLR1/2, TLR7, TLR3, and TLR9) screenings were performed as described previously (23). IL-1β ELISA was used to determine the activity of NLRP3 and NLRC4. Tumor necrosis factor (TNF) ELISA was performed to investigate the response of TLR ligands.
Cloning, Protein Expression, and Purification.
The coding region of mouse RABL3 truncated at the C terminus (2-216) was cloned into pHis-parallel vector. The xm deletion mutation of RABL3 (Δ43–46) and a D44G point mutation were generated by standard site-directed mutagenesis following the QuikChange II site-directed mutagenesis (Agilent Technologies) protocol. The WT, xm, and D44G RABL3 proteins were expressed in bacteria (Rosetta DE3) and induced at 0.6 to ∼0.8 A600nm with 200 μM isopropyl-β-d-thiogalactopyranoside (IPTG) in the presence of 1 mM MgCl2 for 20 h at 16 °C. The proteins were purified from the clarified lysate with an Ni2+-Sepharose fast flow (GE Healthcare) gravity affinity column at 4 °C, followed by ion exchange purification (HiTrap Q HP 5 mL; GE Healthcare). The peak fractions were collected and incubated with TEV protease overnight at 4 °C to remove 6×His tag. The protein was further purified by a second Ni2+-Sepharose affinity purification to remove 6×His-TEV and uncleaved 6×His-RABL3. The tag-free protein was incubated with 10 mM ethylenediaminetetraacetic acid (EDTA) for 2 h and purified by a HiLoad 16/600 Superdex 75 gel filtration column (GE Healthcare) in a buffer containing 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), pH 7.5, 150 mM NaCl. The fully purified RABL3 was concentrated to 8 mg/mL in the presence of 1 mM Tris(2-carboxyethyl)phosphine (TCEP).
Crystallization.
WT RABL3 was incubated either with 10 times molar excess GTPγS in the presence of 5 mM MgCl2 or with five times molar excess GDP in the presence of 3 mM MgCl2 before crystallization. xm and D44G RABL3 were incubated with 10 times molar excess GTPγS in the presence of 10 mM MgCl2. RABL3WT apo, RABL3WT:GTPγS, RABL3WT:GDP, and RABL3xm:GTPγS crystals were grown with a hanging-drop vapor diffusion method by mixing 1 μL of protein with 1 μL of crystallization solution. RABL3D44G:GTPγS crystals were grown with a sitting-drop vapor diffusion method by mixing 1 μL of protein with 1 μL of crystallization solution. The optimized crystallization conditions for each complex are summarized in SI Appendix, Table S2. The crystals were flash-frozen in liquid nitrogen in cryoprotectant (SI Appendix, Table S2). For phase determination, the RABL3D44G:GTPγS crystals were soaked in 10 mM thimerosal (Sigma-Aldrich) for 10 min before being flash-frozen.
Data Collection and Structure Determination.
RABL3WT:GTPγS native crystals diffracted to a minimum Bragg spacing (dmin) of 2.0 Å and exhibited the symmetry of space group P212121 with cell dimensions of a = 59.5 Å, b = 81.2 Å, c = 109.2 Å, and contained two RABL3WT per asymmetric unit. Crystals of RABL3WT apo, and complexed to GDP, and crystals of RABL3D44G complexed to GTPγS were crystallized in the same space group and similar lattice constants. Crystals of RABL3xm complexed to GTPγS exhibited the symmetry of P6522 and contained one RABL3xm per asymmetric unit. All diffraction data were collected at beamline 19-ID (Structural Biology Center Collaborative Access Team [SBC-CAT]) at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL) and processed in the program HKL-3000 (24) with applied corrections for effects resulting from absorption in a crystal and for radiation damage (25, 26), the calculation of an optimal error model, and corrections to compensate the phasing signal for a radiation-induced increase of nonisomorphism within the crystal (27, 28). These corrections were crucial for successful phasing. Phases were obtained from a combined molecular replacement plus single wavelength anomalous dispersion (MR-SAD) experiment using thimerosal derivatized RABL3D44G complexed to GTPγS. The MR-SAD phasing was performed using Phaser (29) as implemented in the CCP4 suite of programs (30) and human Rab8a (PDB ID code 5SZI) (31) as a molecular replacement search model (sequence identity 20.1%) (SI Appendix, Fig. S6) and experimental phases from four mercury heavy atom sites. Phase improvement via density modification was performed in the program Parrot (32), and an initial model containing 84% of all RABL3D44G residues was automatically built and sequenced in the program Buccaneer (33). Completion of this model was performed by cycles of manual rebuilding in the program Coot (34) and model refinement in the program Phenix (35). The resulting RABL3D44G:GTPγS model was used as an isomorphous or molecular replacement model for all other structures presented herein. For all refined models, positional and isotropic atomic displacement parameter (ADP) as well as Translation/Libration/Screw (TLS) ADP refinement was performed using the program Phenix with a random small percentage of all data set aside for an Rfree calculation. Data collection and structure refinement statistics are summarized in SI Appendix, Table S1.
Thermal Stability Assay.
The thermal stability of the WT and xm RABL3 alone, and with nucleotides, was analyzed using a SYPRO orange thermal shift assay following the manufacturer’s protocol (Life Technologies). Briefly, 5 μL of 100× SYPRO orange were added into a 45-μL protein sample containing WT or xm RABL3 without or with nucleotide and magnesium. The final concentration of each component was as follows: 50 μM protein, 500 μM nucleotide, 10 mM magnesium, and 10× SYPRO orange. The melting profile of each protein sample was monitored using a qPCR instrument reading the fluorescence of SYPRO orange. The temperature was increased from 25 °C to 95 °C. The derivatives of each melting cure were plotted against temperature.
Size Exclusion Chromatography.
Each protein of mRal3 WT alone, RABL3WT:GTPγS, RABL3WT:GDP, and RABL3xm alone was loaded onto a Superdex 200 Increase 10/300 GL column to generate a gel filtration curve. To compare the molecular weight of each protein sample, the three gel filtration curves of the WT RABL3 without and with nucleotides were overlaid, and the gel filtration curves of the WT RABL3 alone and the xm RABL3 alone were overlaid. RABL3WT alone and RABL3xm alone were run in a buffer containing 25 mM Hepes, pH 7.5, 150 mM NaCl. RABL3WT:GTPγS and RABL3WT:GDP were incubated on ice for 1 h with 10:1 molar ratio GTPγS or GDP in the presence of 5 mM MgCl2 and subjected to the gel filtration column in a buffer containing 25 mM Hepes, pH 7.5, 150 mM NaCl, and 5 mM MgCl2.
Analytical Ultracentrifugation.
Two samples of WT RABL3 (optical density [OD] = 0.08 and OD = 0.8) were prepared in a buffer containing 25 mM Hepes, pH7.5, 150 mM NaCl, and 1 mM TCEP. All handling and centrifugation were performed at 4 °C. The samples were introduced into standard centrifugation cells containing 1.2 cm of charcoal-filled Epon centerpieces sandwiched between sapphire windows. The volume used was 400 μL. The cells were placed in an An50-Ti rotor which was then incubated under vacuum in the centrifuge at the experimental temperature for 17 h prior to commencing centrifugation at 50,000 rpm. Laser interferometric and absorbance data at 280 nm were acquired. All data were corrected for time-stamp errors using REDATE. The data were analyzed using the c(s) methodology in SEDFIT (36). The molar-mass values were calculated using the Svedberg equation, capitalizing on the availability of the sedimentation and diffusion coefficients estimated from the analyses. GUSSI (37) was used to transform the distributions to standard conditions (water, 20 °C) and to render the figures.
Data Deposition and Availability.
The coordinates and structure factors of mouse RABL3WT apo, RABL3WT in complex with GTPγS and GDP, and RABL3xm in complex with GTPγS have been deposited with the Protein Data Bank (PDB) under accession codes 6VIH, 6VII, 6VIJ, and 6VIK, respectively. All other data are contained in the manuscript and SI Appendix.
Statistical Analysis.
The statistical significance of differences between two groups was analyzed using Student’s t test in GraphPad Prism. The statistical significance of differences between three or more groups was analyzed by one-way ANOVA with post-hoc Dunnett’s multiple comparisons test (to WT or REF genotype). Differences were considered statistically significant when P < 0.05. P values are denoted as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant with P > 0.05.
Supplementary Material
Acknowledgments
We thank Xuewu Zhang (University of Texas Southwestern Medical Center) for helpful discussions and Diantha La Vine (University of Texas Southwestern Medical Center) for assistance with illustrations. This work was supported by NIH Grants R01 AI125581 and U19 AI100627 (to B.B.). Results shown in this report are derived from work performed at Argonne National Laboratory (ANL), Structural Biology Center (SBC) at the Advanced Photon Source (APS), under US Department of Energy, Office of Biological and Environmental Research Contract DE-AC02-06CH11357. Research reported in this publication was supported by the Office of the Director, NIH under Award S10OD025018.
Footnotes
The authors declare no competing interest.
Data deposition: The coordinates and structure factors of mouse RABL3WT apo, RABL3WT in complex with 5’-O-[gamma-thio]triphosphate (GTPγS), RABL3WT in complex with guanosine diphosphate (GDP), and RABL3xm in complex with GTPγS have been deposited in the Protein Data Bank (PDB), www.wwpdb.org (PDB ID codes 6VIH, 6VII, 6VIJ, and 6VIK, respectively).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000703117/-/DCSupplemental.
References
- 1.Wang T., et al. , Real-time resolution of point mutations that cause phenovariance in mice. Proc. Natl. Acad. Sci. U.S.A. 112, E440–E449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., et al. , Evaluation of the novel gene Rabl3 in the regulation of proliferation and motility in human cancer cells. Oncol. Rep. 24, 433–440 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Rojas A. M., Fuentes G., Rausell A., Valencia A., The Ras protein superfamily: Evolutionary tree and role of conserved amino acids. J. Cell Biol. 196, 189–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wennerberg K., Rossman K. L., Der C. J., The Ras superfamily at a glance. J. Cell Sci. 118, 843–846 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Pylypenko O., Hammich H., Yu I. M., Houdusse A., Rab GTPases and their interacting protein partners: Structural insights into Rab functional diversity. Small GTPases 9, 22–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavas L. M., et al. , Structure of the small GTPase Rab27b shows an unexpected swapped dimer. Acta Crystallogr. D Biol. Crystallogr. 63, 769–779 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Pasqualato S., et al. , The structural GDP/GTP cycle of Rab11 reveals a novel interface involved in the dynamics of recycling endosomes. J. Biol. Chem. 279, 11480–11488 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Wittmann J. G., Rudolph M. G., Crystal structure of Rab9 complexed to GDP reveals a dimer with an active conformation of switch II. FEBS Lett. 568, 23–29 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Inouye K., Mizutani S., Koide H., Kaziro Y., Formation of the Ras dimer is essential for Raf-1 activation. J. Biol. Chem. 275, 3737–3740 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Zhang B., Zheng Y., Negative regulation of Rho family GTPases Cdc42 and Rac2 by homodimer formation. J. Biol. Chem. 273, 25728–25733 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Maeda Y., Ide T., Koike M., Uchiyama Y., Kinoshita T., GPHR is a novel anion channel critical for acidification and functions of the Golgi apparatus. Nat. Cell Biol. 10, 1135–1145 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Rosenbaum D. M., Rasmussen S. G., Kobilka B. K., The structure and function of G-protein-coupled receptors. Nature 459, 356–363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nissim S., et al. , Mutations in RABL3 alter KRAS prenylation and are associated with hereditary pancreatic cancer. Nat. Genet. 51, 1308–1314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An J., et al. , Overexpression of Rabl3 and Cullin7 is associated with pathogenesis and poor prognosis in hepatocellular carcinoma. Hum. Pathol. 67, 146–151 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Pan Y., et al. , The overexpression of Rabl3 is associated with pathogenesis and clinicopathologic variables in hepatocellular carcinoma. Tumour Biol. 39, 1010428317696230 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Zhang W., Sun J., Luo J., High expression of Rab-like 3 (Rabl3) is associated with poor survival of patients with non-small cell lung cancer via repression of MAPK8/9/10-mediated autophagy. Med. Sci. Monit. 22, 1582–1588 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hidmark A. S., et al. , Humoral responses against coimmunized protein antigen but not against alphavirus-encoded antigens require alpha/beta interferon signaling. J. Virol. 80, 7100–7110 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnold C. N., et al. , A forward genetic screen reveals roles for Nfkbid, Zeb1, and Ruvbl2 in humoral immunity. Proc. Natl. Acad. Sci. U.S.A. 109, 12286–12293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi J. H., et al. , A single sublingual dose of an adenovirus-based vaccine protects against lethal Ebola challenge in mice and guinea pigs. Mol. Pharm. 9, 156–167 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi J. H., et al. , LMBR1L regulates lymphopoiesis through Wnt/β-catenin signaling. Science 364, eaau0812 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang D., et al. , Syndromic immune disorder caused by a viable hypomorphic allele of spliceosome component Snrnp40. Nat. Immunol. 20, 1322–1334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holst J., et al. , Generation of T-cell receptor retrogenic mice. Nat. Protoc. 1, 406–417 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Shi H., et al. , NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 17, 250–258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M., HKL-3000: The integration of data reduction and structure solutionFrom diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Borek D., Minor W., Otwinowski Z., Measurement errors and their consequences in protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 59, 2031–2038 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z., Borek D., Majewski W., Minor W., Multiparametric scaling of diffraction intensities. Acta Crystallogr. A 59, 228–234 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Borek D., Cymborowski M., Machius M., Minor W., Otwinowski Z., Diffraction data analysis in the presence of radiation damage. Acta Crystallogr. D Biol. Crystallogr. 66, 426–436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borek D., Dauter Z., Otwinowski Z., Identification of patterns in diffraction intensities affected by radiation exposure. J. Synchrotron Radiat. 20, 37–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winn M. D., et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rai A., et al. , bMERB domains are bivalent Rab8 family effectors evolved by gene duplication. eLife 5, e18675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowtan K., Recent developments in classical density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 470–478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowtan K., The buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuck P., Sedimentation Velocity Analytical Ultracentrifugation: Discrete Species and Size-Distributions of Macromolecules and Particles (CRC Press, Boca Raton, FL, 2016). [Google Scholar]
- 37.Brautigam C. A., Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.