The human genome contains more than 3 billion base pairs, and some estimates suggest that nearly 75% of the genome may be transcribed (1), yet only a small fraction (1 to 2%) of the genome that encodes for protein coding regions has been systematically probed for function. The transcribed genome consists of both short noncoding RNAs (ncRNAs) and long ncRNAs (lncRNAs). The lncRNAs are defined as being greater than 200 nucleotides in length and may be capped, polyadenylated, and spliced like protein-coding genes. By definition, lncRNAs do not contain open reading frames of greater than 50 amino acids, but recent work has uncovered a new world of smaller “microproteins” that are encoded by lncRNAs (2). Other functions of lncRNAs include their ability to form aptamers, which bind and regulate protein complexes (3). The lncRNAs can modulate gene expression by enhancer trapping (4), and by recruiting histone modifiers to chromatin (5, 6). These examples notwithstanding, the function of most lncRNAs is unknown. In PNAS, Raffeiner et al. (7) develop CRISPR-based technology to silence the expression of lncRNAs, and use their technology to identify functional lncRNAs that are regulated by the oncogene MYC.
Recent efforts to identify functional lncRNAs have relied on CRISPR technology, which can exquisitely target CRISPR-associated (Cas) proteins to specific genomic locations. This technology has ushered in a new era of functional genomics in which large-scale screens have exponentially increased our understanding of complex genotype−phenotype relationships (8–10). CRISPR-Cas9 pooled screens have become a mainstay of cancer research (11, 12), revealing gene essentiality, synthetic lethality, and mechanisms of resistance to targeted therapy and immunotherapy, among countless other phenotypes. Most of these screens have relied on Cas9-induced DNA double-stranded breaks that, when repaired, introduce small insertions or deletions, ultimately producing either mutant or truncated proteins. Cas9-induced genome editing has also been used to alter gene expression by disrupting noncoding regulatory elements (13). However, CRISPR screens with Cas9 are less able to reliably elucidate lncRNA function, as focal indels may not perturb lncRNA function, and the functional domains of most lncRNAs have not been mapped.
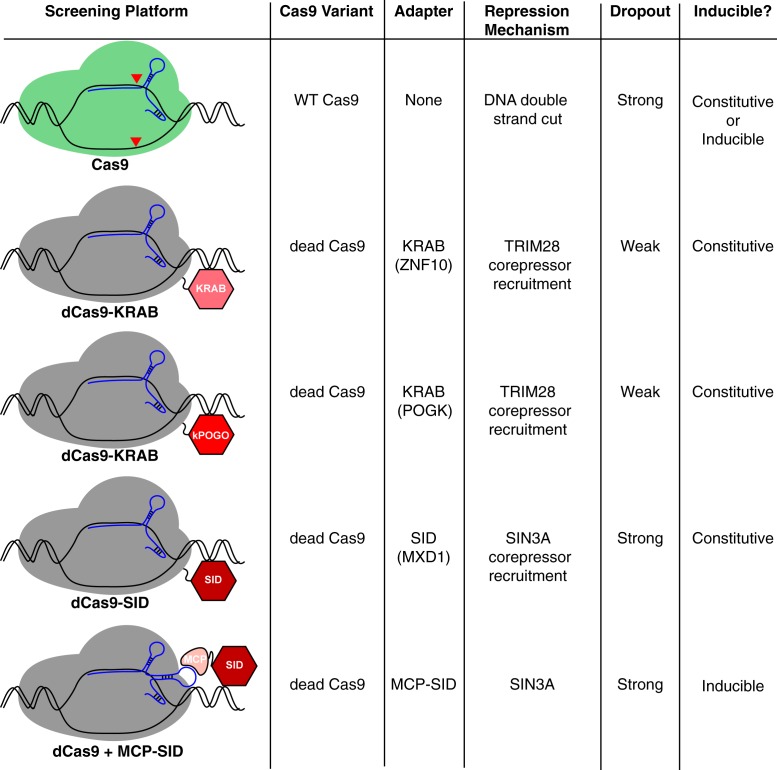
Previous studies have shown that Cas9 can be engineered to silence gene expression (rather than induce double-stranded breaks) by fusing transcriptional repressor domains to an enzymatically inactive “dead” Cas9 (dCas9) isoform (14). In these CRISPR interference (CRISPRi) systems, a single guide RNA (sgRNA) is used to recruit dCas9 repressor domain fusions to a specific genomic location where they can repress the transcription. Raffeiner et al. (7) turn to this technology to probe the function of lncRNAs that are induced by MYC in B cell malignances such as Burkitt lymphoma (BL). They began by fusing the Krüppel-associated box (KRAB) repression domain of ZNF10 to dCas9 and screened BL cell lines with an sgRNA library of 5,708 sgRNAs targeting 508 MYC-regulated lncRNAs, 100 MYC-regulated protein-coding genes, and 14 known panessential protein-coding genes as a positive control. Unexpectedly, only two of the positive control genes appeared essential in this screen, and subsequent experiments using dCas9 fused to a different KRAB domain yielded similarly suboptimal results (Fig. 1, kPOGO). However, the authors did not test the KRAB domain of Kox1, which has been shown to be effective in other CRISPRi screens (14).
Fig. 1.
Summary of CRISPRi screens performed by Raffeiner et al. (7) using variants of catalytically dead Cas9 (dCas9, gray) fused to various domains of adapter proteins that recruit transcriptional repressors. For comparison, wild-type Cas9 (green) is also displayed, but CRISPR screens were not performed. KRAB domain of POGK, kPOGO.
The authors (7) hypothesize that their approach might have been more successful if they had used a repression domain that was highly active in MYC-transformed lymphomas. MYC target gene activation is normally opposed by complexes that include MXD1 (MAD), which competes with MYC for binding to MAX and recruits the transcriptional corepressor SIN3A (15). Raffeiner et al. therefore fused the SIN3-interacting domain (SID) from MXD1 to dCas9 (dCas9-SID) and tested whether this bespoke repressor would outperform dCas9-KRAB in BL cells. As they predicted, 12 of 14 panessential genes scored as essential in BL cells using dCas9-SID, and this fusion protein revealed the essentiality of several noncoding RNAs (Fig. 1).
Given the apparent efficacy of the SID domain, Raffeiner et al. (7) created an inducible system for SID-mediated gene repression. They built on previous work showing that MS2 coat protein (MCP) can be recruited to dCas9 using sgRNAs containing MS2 RNA aptamers to which MCP can bind (16) (Fig. 1). The authors fused MCP to the SID domain and used a Tet-On cassette to inducibly express this fusion protein in BL cells. Using a library of sgRNAs containing the MS2 aptamer sequence, they showed that this system could efficiently identify essential genes.
This system has several technical advantages. Large Cas9 fusion proteins can be challenging to express using lentiviral vectors and could be toxic to some cells due to their interaction with corepressors. By contrast, Raffeiner et al. (7) only require the stable expression of dCas9, which should be functionally inert in the absence of an sgRNA. The MCP-SID fusion protein is relatively small and thus can be readily expressed using a lentiviral vector that coexpresses an sgRNA, enabling efficient inducible gene repression. An intriguing possibility afforded by this modular design is that cells could be engineered to express two different MCP fusion proteins, each coupled to a different repression domain. Raffeiner et al. observe that the SID domain was able to repress the transcription of most lncRNAs more efficiently than the ZNF10 KRAB domain, but the opposite was true for one well-known lncRNA, MALAT1. Thus, it is conceivable that genome-wide CRISPRi screens could silence target genes more effectively if distinct repression domains could be recruited simultaneously to a promoter or enhancer, perhaps producing stronger phenotypes and identifying novel essential genes.
All this technological wizardly is in the service of identifying functional important lncRNAs that may play a role in MYC-driven B cell lymphomas. Importantly, the screen performed by Raffeiner et al. (7) rediscovers the importance of MIR17HG, which is a MYC-regulated lncRNA that encodes several microRNAs that target the tumor suppressor PTEN (17). Among the novel lncRNAs that are essential in BL cells, Raffeiner et al. validate two—SNHG17 and SNHG26—as bona fide MYC target genes using chromatin immunoprecipitation assays. Interestingly, SNHG17 has been reported to be necessary to maintain phosphorylation of PI3 kinase (PI3K) and AKT in cell line models of melanoma (18). PI3K activation is triggered by the constitutive and oncogenic activity of the B cell receptor (BCR) in BL, which has been dubbed “toncogenic” BCR signaling (19–21). Moreover, MIR17HG also activates PI3K by downmodulating PTEN, a negative regulator of PI3K. Thus, it is attractive to speculate that two critical oncogenic pathways in BL—MYC and PI3K—are functionally interconnected by two MYC-regulated lncRNAs. More generally, the large number of essential lncRNAs in BL uncovered by Raffeiner et al. may reveal mechanisms that modulate other important BL attributes, such as their explosive and clinically aggressive proliferation, and their distinctive morphology that reflects their derivation from the centroblast subpopulation of germinal center B cells (20).
Inducible CRISPRi screens could reveal phenotypes that are missed in CRISPR screen using catalytically active Cas9. Since gene inactivation by Cas9 is stochastic, the timing of gene inactivation will be variable within a population of sgRNA-expressing cells. By contrast, gene silencing by inducible CRISPRi will be more synchronous, which is a decided advantage when conducting combinatorial genetic screens that aim to knock down two genes at the same time. Further, for some genes, a complete knockout could be cell lethal, but a partial knockdown by CRISPRi could reveal a role in nonlethal phenotypes. Indeed, technologies that partially knock down genes may better model the effect of drugs, which rarely completely inactivate their targets in vivo. And lastly, as demonstrated by Raffeiner et al. (7), CRISPRi technologies are effective tools to probe the noncoding genome. From these perspectives, it will be important to start building a public database of CRISPRi results so that metaanalyses can be performed, which can highlight cell type- and context-specific effects of lncRNAs. The value of such public repositories is evident from the widespread usage of a similar database (Depmap) for Cas9 loss-of-function screens targeting the coding genome (22).
Footnotes
The authors declare no competing interest.
See companion article, “An MXD1-derived repressor peptide identifies noncoding mediators of MYC-driven cell proliferation,” 10.1073/pnas.1921786117.
References
- 1.Atianand M. K., Caffrey D. R., Fitzgerald K. A., Immunobiology of long noncoding RNAs. Annu. Rev. Immunol. 35, 177–198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J., et al. , Pervasive functional translation of noncanonical human open reading frames. Science 367, 1140–1146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willingham A. T., et al. , A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309, 1570–1573 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Cho S. W., et al. , Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell 173, 1398–1412.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao J., Sun B. K., Erwin J. A., Song J. J., Lee J. T., Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai M. C., et al. , Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raffeiner P., et al. , An MXD1-derived repressor peptide identifies noncoding mediators of MYC-driven cell proliferation. Proc. Natl. Acad. Sci. U.S.A. 117, 6571–6579 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M., et al. , A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L., et al. , Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P., et al. , RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang T., Wei J. J., Sabatini D. M., Lander E. S., Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T., et al. , Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanjana N. E., et al. , High-resolution interrogation of functional elements in the noncoding genome. Science 353, 1545–1549 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert L. A., et al. , CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayer D. E., Lawrence Q. A., Eisenman R. N., Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80, 767–776 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Konermann S., et al. , Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiao C., et al. , Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 9, 405–414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao H., Liu R., Sun X., STAT3-induced upregulation of lncRNA SNHG17 predicts a poor prognosis of melanoma and promotes cell proliferation and metastasis through regulating PI3K-AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 8000–8010 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Sander S., et al. , Synergy between PI3K signaling and MYC in Burkitt lymphomagenesis. Cancer Cell 22, 167–179 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitz R., et al. , Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 490, 116–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young R. M., Phelan J. D., Wilson W. H., Staudt L. M., Pathogenic B-cell receptor signaling in lymphoid malignancies: New insights to improve treatment. Immunol. Rev. 291, 190–213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsherniak A., et al. , Defining a cancer dependency map. Cell 170, 564–576.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]