Significance
Epithelial cellular activating molecule (EpCAM) is vastly overexpressed in tumor cell membranes of certain carcinomas, and thus represents an auspicious clinical target for cancer therapies. To date, therapeutic targeting of EpCAM has principally involved antibodies; smaller, nucleic-acid based aptamers have shown great promise, but have not yet reached clinical application. Here, using experimentally validated in silico techniques, we have designed two RNA aptamers that bind EpCAM with improved affinity over existing aptamers. We confirm that these aptamers bind the EpCAM dimer, a complex distinct from and more physiologically relevant than the oft-targeted EpCAM monomer. This successful structure-based in silico approach advocates for the broad use of such methods in RNA aptamer design.
Keywords: RNA aptamer, rational design, molecular dynamics, isothermal titration calorimetry, epithelial cellular adhesion molecule
Abstract
Nucleic acid aptamers hold great promise for therapeutic applications due to their favorable intrinsic properties, as well as high-throughput experimental selection techniques. Despite the utility of the systematic evolution of ligands by the exponential enrichment (SELEX) method for aptamer determination, complementary in silico aptamer design is highly sought after to facilitate virtual screening and increased understanding of important nucleic acid–protein interactions. Here, with a combined experimental and theoretical approach, we have developed two optimal epithelial cellular adhesion molecule (EpCAM) aptamers. Our structure-based in silico method first predicts their binding modes and then optimizes them for EpCAM with molecular dynamics simulations, docking, and free energy calculations. Our isothermal titration calorimetry experiments further confirm that the EpCAM aptamers indeed exhibit enhanced affinity over a previously patented nanomolar aptamer, EP23. Moreover, our study suggests that EP23 and the de novo designed aptamers primarily bind to EpCAM dimers (and not monomers, as hypothesized in previous published works), suggesting a paradigm for developing EpCAM-targeted therapies.
Since the inception of the systematic evolution of ligands by exponential enrichment (SELEX) technique for identifying nucleic acid aptamers (1, 2), major efforts have been undertaken to create aptamer-based therapeutics for a wide range of diseases (3, 4). During this time, the field of RNA biology has exploded in discovery and elucidation of RNA physiological processes (5, 6). Each discovery in RNA biology [e.g., primer in CRISPR (7, 8), siRNA in RNAi (9)], presents opportunities for designed RNA therapeutics, explaining why RNA aptamers have remained a popular area of research (4, 10).
One active area of aptamer research is the in silico design and development of RNA aptamers complementing the voluminous SELEX experimental technique (11). Despite its strengths, the SELEX experiment is time and resource intensive and suffers from several disadvantages, such as only allowing for the blind selection and enrichment of a few candidates, when numerous additional sequences in the library may bind the target with comparable or higher affinity. Adding to these concerns is the general desire to increase our understanding of RNA design and structure. Unfortunately, creation of an in silico design methodology for RNA aptamers has remained a desirable yet daunting task. One challenge in structure-based in silico RNA aptamer design is the accurate generation of RNA 3D structure from the primary sequence. Numerous RNA structure prediction models exist (12, 13), but their accuracies vary [as determined by the RNA-Puzzles (14) competition]. Further, these models have not yet reached the scale to predict plausible 3D aptamer structures from a sequence library, although more scalable 2D models are available (15). Another barrier to structure-based in silico aptamer design is the prediction of aptamer binding modes to the target biomolecule. Although several groups have sought to improve protein–RNA docking (16, 17), whole-molecule docking approaches are still mostly focused on proteins (18). Combining the techniques of RNA structure prediction, docking, and molecular modeling, several works have sought to design RNA aptamers in silico, using a structure-based procedure (19, 20); however, these methodologies have produced varied results. In this work, we propose an aptamer design framework composed of three major phases, including constructing the aptamer structures, predicting their binding conformations, and optimizing their binding through mutations. During each step of the workflow, both system physiology and biochemistry knowledge as well as the desired therapeutic action are taken into careful account for appropriate aptamer design.
Human epithelial cellular activating molecule (or epithelial cellular adhesion molecule; EpCAM) is a 314-amino acid transmembrane protein located on the cell surface that features a large, 242-residue extracellular domain. Despite recent questions surrounding its self-adhesion mechanism (21), EpCAM is known to perform several prominent physiological roles in cellular signaling (22, 23). Most notably, EpCAM is overexpressed, up to 1,000-fold, in many tumor cells of epithelial origin (carcinomas), and is a prominent prognostic indicator for specific types of cancer (24). Due to its abundance on tumor cells, numerous antibodies have been created to target EpCAM (25) for cancer therapy, some of which [e.g., catumaxomab (26)] have reached the clinics. Given the advantages of aptamers for targeted therapy, as well as the overexpression of EpCAM in cancer, EpCAM presents an auspicious opportunity for aptamer design.
Here, we computationally design and experimentally validate two high-affinity aptamer sequences targeting EpCAM. This represents a successful attempt at using rational, structure-based design to improve aptamer binding affinity in a targeted molecular fashion with blind experimental confirmation. An RNA sequence derived from SELEX experiments termed EP23, which was known to bind EpCAM, was the initial input of our work. RNA structure and binding were predicted using a combination of equilibrium and steered molecular dynamics (MD) simulations, free energy perturbation (FEP) calculations, Dot2.0 docking (27), and Mfold (28) secondary structure prediction. The output of our work consists of two designed RNA aptamers with putative EpCAM binding conformations and mutational free energy data. Subsequent isothermal titration calorimetry (ITC) experiments verified the predicted mutational affinity changes of the aptamer, suggesting the binding conformation found in silico is indeed representative of the true structure. Further experiments confirm our simulation-based hypothesis that these aptamers bind to EpCAM dimers, demonstrating a mechanism underlying target–aptamer interaction.
Results
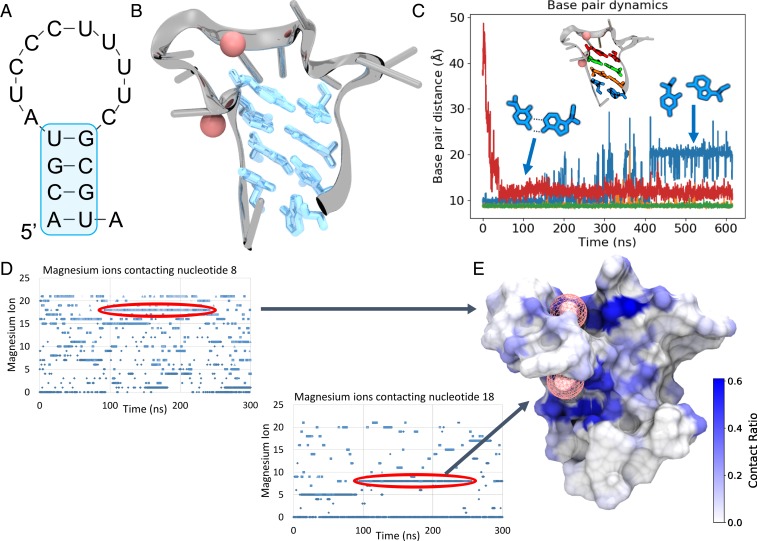
The predicted RNA aptamer structure and Mg2+ binding sites are shown in Fig. 1. The initial input for this work was an RNA sequence (ACGUAUCCCU UUUCGCGUA), termed EP23, that was known to bind EpCAM, as demonstrated in our previous works (29, 30). RNA secondary structure (Fig. 1A) was predicted using Mfold (28). Secondary structure constraints were initially applied to the 3D RNA structure during MD simulations. Once the constraints were removed (150 ns), the MFold-predicted base pairs remained paired, as shown in Fig. 1B, even after 450 ns simulation, despite normal fluctuations of the terminal base pair. Experimentally, the EP23 aptamer was determined to require a moderate Mg2+ concentration for proper folding. As such, the Mg2+ concentration was kept artificially high (0.15 M) during MD simulations to mimic a locally high Mg2+ concentration surrounding the RNA. Mg2+ concentration dependence on RNA structure is well-studied (31, 32), and is likewise found to be an important factor in the EP23 aptamer structure, as observed here. By studying the Mg2+ binding plots (Fig. 1C), we identified and selected an RNA structure (Fig. 1D) with two long-lived bound Mg2+ ions for advancement to EpCAM binding.
Fig. 1.
EP23 Structure and Mg2+. (A) The Mfold-predicted secondary structure of EP23. (B) The 3D folded structure of EP23 determined from MD simulations, with the 4 base pairs predicted from Mfold highlighted in blue. (C) Base pair dynamics of the Mfold predicted secondary structure. The terminal base pair (blue) changed conformations slightly from a Watson-Crick to a Hoogsteen-like configuration. (D) Mg2+ ions binding nucleotides 8 (top) and 18 (bottom) over the course of the MD simulations. Two long-lived bound ions are circled in red. (E) Mg2+ ion contact ratio mapped onto the EP23 aptamer surface. The two bound Mg2+ ions correspond to the long-lived bound ions in D.
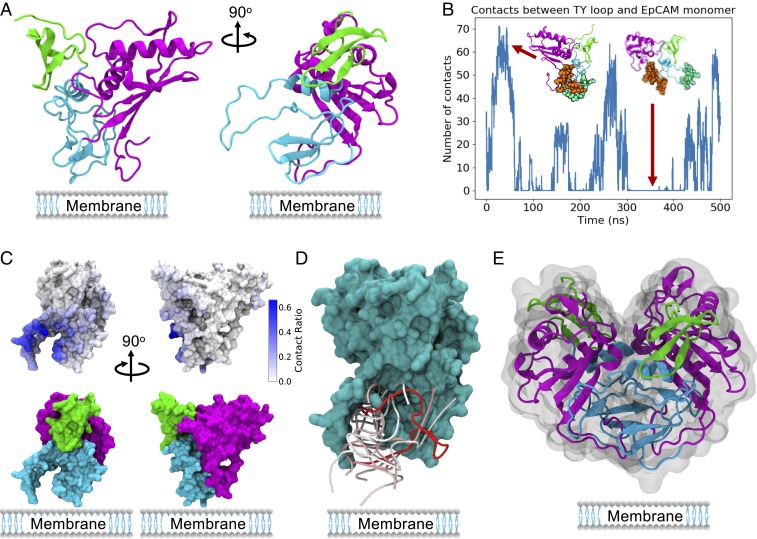
The crystal structure of EpCAM (33), as presented in Fig. 2A, includes 3 distinct domains: a short N-terminal domain (ND), a thyroglobulin (TY) loop, and a large C-terminal domain (CD). Notably, EpCAM exists as a dimer in vivo (21), was crystallized as a dimer (33), and was observed to form dimers in solution (21, 34). However, to be thorough, we first studied aptamer binding to the EpCAM monomer. In vivo, this would be the special case where the EP23 aptamer binds EpCAM immediately after translation and before dimerization. Interestingly, in the control EpCAM monomer MD simulations, we observed extensive flexibility of the TY loop with reversible opening and closing behavior (Fig. 2B). Aptamer docking simulations using Dot2.0 (27) revealed a highly localized preference for the TY loop (Fig. 2C), binding between the TY loop and the rest of the protein (Fig. 2D). The TY loop is the interface for EpCAM dimerization (33) (Fig. 2E), and aptamer binding the TY loop at the precise time directly after translation could at least theoretically inhibit EpCAM dimerization. However, this binding behavior seems unlikely, for several reasons.
Fig. 2.
EP23-EpCAM monomer binding results. (A) EpCAM monomer showing the N-terminal domain in green, TY loop in blue, and C-terminal domain in magenta. (B) EpCAM TY loop fluctuations observed in control EpCAM explicit solvent MD simulations. The residues used for contact analysis are shown as spheres in green (TY loop) and brown (core EpCAM). (C) EpCAM mapped contacts from the top 200 Dot2.0 docking conformations, showing the strong preference for TY loop binding conformations. (D, Top) Ten docking conformations predicted by Dot2.0, all targeting the TY loop region at the dimer interface and the membrane side of the protein. The RNA backbone is shown, colored by energy from strong (red) to weak (blue). (E) EpCAM dimer crystal structure, showing dimerization using the TY loop.
First, the predicted monomer binding modes place the aptamer close to the cell membrane surface that would be present in vivo, a relationship that could result in a significant steric conflict with putative aptamer interactions. Although soluble extracellular EpCAM (EpEX) has been found in sera (35) and ascites (36), most clinical and therapeutic targeting of EpCAM (including this work) is focused on the most-studied, membrane-bound form. Second, EpCAM has two c-terminal transmembrane and intracellular domains that were not crystallized but would be located in proximity to the TY loop and could, along with the membrane surface itself, obscure binding. Third, the EpCAM dimerization dissociation constant (kd) has been estimated at 10 nM (37), revealing a strong preference for intracellular dimerization. Last, given the transient nature of the TY loop opening and closing behavior, as well as the prevalence of intracellular dimerization, the aptamer is restricted to a very narrow monomer-binding window even after endocytosis. Considering the above arguments (including experimental observations that EpCAM dimerizes in vivo using the TY loop), we argue that the monomer aptamer binding conformations proposed in Fig. 2 and found in the literature (38, 39), which bind EpCAM at the TY loop dimer interface, should be discounted as not physiologically relevant. Although the experiments performed in both papers indicate the aptamers do bind to EpCAM (38, 39), the proposed binding conformations are inconsistent with solved crystal structures of EpCAM (33, 40). These conclusions lead us to include the background sections of physiology and biochemistry and of therapeutic action in our aptamer design protocol presented in SI Appendix, Fig. S1. Based on our findings, we hypothesize that the EpCAM dimer presents a more appropriate aptamer binding target, as confirmed in simulations and experiments described here.
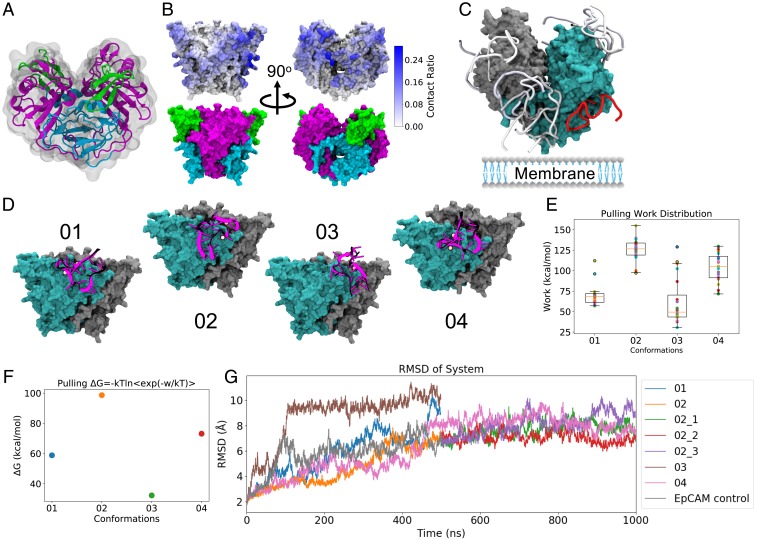
EpCAM dimerizes at an interface including the TY loop, leaving its N-terminal (ND) and C-terminal (CD) domains solvent-exposed (Fig. 3A). Accordingly, predicted aptamer binding modes [generated with the Dot2.0 (27) docking software] reveal a much different binding preference for the EpCAM dimer than for monomer. As shown in Fig. 3B, docked modes show a preference for EP23 binding to the exposed CD top and ND and CD sides. From the docking computations, four unique EP23 binding conformations (Fig. 3D) were selected for advanced interrogation of EpCAM binding. From these conformations, steered MD pulling simulations (Fig. 3 E and F) and additional MD simulations (Fig. 3G) revealed one of them as an exceptionally strong binder. This conformation (conformation 02) targeted the CD of EpCAM, making important electrostatic interactions with Lys155, Lys160, Arg163, and Lys168. Given that the physiological roles of EpCAM structure remain unknown, it is difficult to speculate whether binding at this location will have any therapeutic action on EpCAM itself. In the crystal structure paper (33), the authors hypothesized that intercellular EpCAM-EpCAM binding occurred as a tetramer, using docking software to find the plausible tetrameric binding conformations along the CD. However, the same authors later presented compelling evidence disproving this intercellular EpCAM oligomerization, although maintaining solution and in vivo EpCAM cis-dimerization (21). The authors posited that EpCAM could bind another unknown ligand, resulting in intercellular adhesion, or that EpCAM could solely function in other roles including its known signaling roles. The majority of EpCAM antibodies bind to the ND domain of EpCAM (33); however, the ND is not required for intercellular adhesion (41). Therapeutically efficacious antibodies, including catumaxomab (26) and mm-131 (40), bind multiple proteins and only use EpCAM binding to target tumor cells, localizing the action of other therapeutic protein effectors. Ultimately, using EpCAM binding to target particular cells may also be the therapeutic fate of this aptamer, as previous experimental work has shown that attachment of another aptamer allows for dual binding of both EpCAM and the transferrin receptor (42). However, the RNA aptamer-derived direct inhibition of interactions between EpCAM and other cellular ligands should not be discounted without further study.
Fig. 3.
EP23 binding EpCAM dimer. (A) EpCAM dimer, showing dimerization along the TY loop (blue) and exposure of the N and C-terminal domains (green and magenta, respectively). (B) Aptamer EP23 contacts mapped onto the EpCAM dimer from the top 200 Dot2.0 docking conformations. For the dimer system, docked conformations are spread across the protein, but show higher population for the exposed C-terminal domain. (C, Top) 10 Dot2.0 docking conformations for the EpCAM dimer, colored by energy from strong (red) to weak (blue). EpCAM dimer is shown in a surface representation and colored by protein monomer: cyan and gray. (D) Four unique docking conformations from the top 10 docking conformations selected for further binding assessment. EpCAM dimer is shown in a surface representation and colored by protein monomer: cyan and gray, while the EP23 aptamer is illustrated as a cartoon in magenta, with bound Mg2+ ions shown as pink spheres. (E) The work required to pull the EP23 conformation off of EpCAM dimer, with larger work showing stronger binding. The orange line is the median, boxes extend form lower to upper quartiles, whiskers show range of nonoutlier data. (F) Jarzynski equality averages computed from pulling EP23 off of the EpCAM dimer. Values shown are the mean, SE bars are shown but within the data points. (G) The root-mean-square deviation (RMSD) of atomic positions for the selected EP23 binding conformations + EpCAM dimer configurations across the MD simulations. Note: 02_1, 02_2, and 02_3 are 3 replicas of 500-ns MD extension starting from the 500-ns structure of run 02.
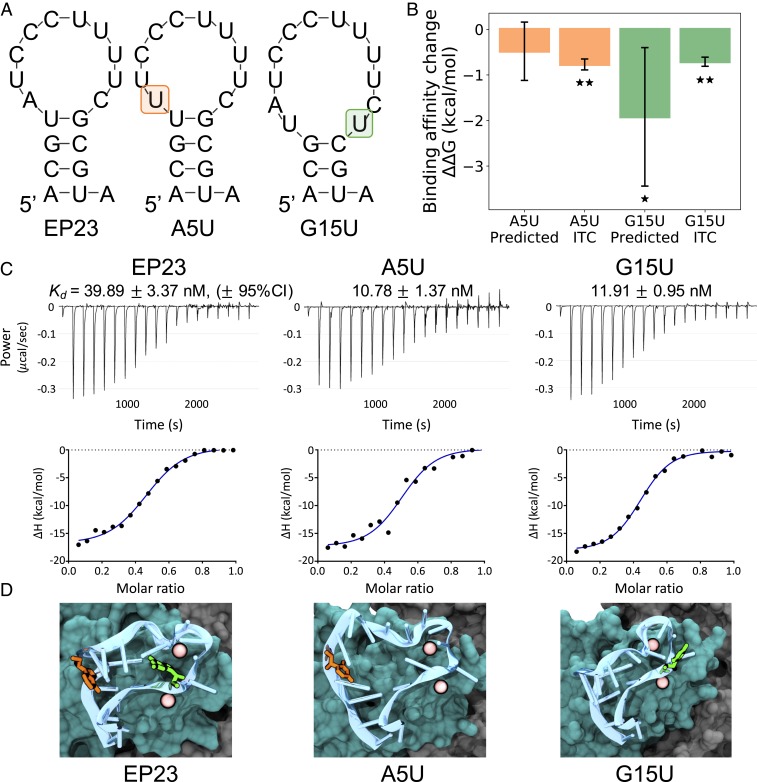
To explore the design of more potent aptamers for EpCAM binding, we identified two sites, A5 and G15, that (if mutated) would likely retain a simple hairpin folding yet result in different binding outcomes. Here, we first analyzed the dominant binding conformation (02) found from MD simulations and identified plausible aptamer mutations that might affect the EpCAM binding. For the largest effect, we decided to mutate A5 and G15 (both purines) to the pyrimidine uracil. Secondary structures for these sequence mutations [again computed using Mfold (27)] did not reveal any major changes from the EP23 fold (Fig. 4A), although the G15U mutant did lose one base pair. Notably, even in proteins, single point mutations can substantially change structure (43, 44). Given RNA structure’s notoriously dynamic conformations, understanding mutational effects on structure is imperative, and a quick check of secondary structure will give a reasonable first approximation of any changes. The mutational change in binding affinity was computed in silico, using the free energy perturbation technique, which we have used with previous success (45–48). Both A5U and G15U mutations resulted in favorable binding affinity changes for the predicted dominant binding conformation. These results and their verification by isothermal titration calorimetry are shown in Fig. 4 B and C. Confirming our conjecture that RNA structure would not be adversely affected by the A5U and G15U mutations, we observed only minor changes in aptamer structure during the FEP calculation (Fig. 4D).
Fig. 4.
EP23 mutation binding affinity. (A) Comparison of Mfold-predicted secondary structures for the original EP23 sequence, the A5U mutation, and the G15U mutation. (B) The change in binding affinity for two mutations, both as predicted by FEP simulations and determined by ITC experiments. Error bars represent 95% CIs. Starred values (*) indicate P < 0.05, and double-starred (**) experimental values indicate P < 0.0001 for a one-sided t test. (C) Isothermal titration calorimetric analysis of the interaction of the 50-μM aptamer solutions from EP23, A5U, or G15U, with EpCAM protein (5 μM) in ITC buffer at 25 °C. (Top and Bottom) Raw data and binding isotherm obtained over a series of injections of aptamer into EpCAM protein. (Top) Differential power (μcal/sec) versus time is presented in the form of integrated heat values. The data were fitted using a one binding site model. BSA control data are presented in SI Appendix, Fig. S2. (D) Structural comparison of bound EP23 (blue cartoon) with the A5U mutation (orange) and the G15U mutation (green). Bound Mg2+ ions shown as pink spheres; EpCAM dimer is shown in a surface representation and colored cyan or gray for different monomers.
Experimental ITC measurements confirmed that both A5U and G15U mutations produced favorable binding affinity changes and also validated our hypothesis that all three aptamers studied in this work bind to the EpCAM dimer. Fig. 4 and SI Appendix, Fig. S2 show typical aptamer titration profiles for the EpCAM solution (Fig. 4) and the control BSA solution(SI Appendix, Fig. S2). Corresponding thermodynamic parameters are listed in SI Appendix, Fig. S2. The quality of the ITC data is described by the Wiseman c value, with a c value of 10 to 500 designating optimal curve fitting (49). In the current experiment, the c value ranges from 55.16 to 278.09, indicating that the ITC assay is reliable. It is noteworthy that the equilibrium dissociation constant kd of EP23 binding to EpCAM obtained from the current ITC experiment (39.89 ± 3.37 nM, 95% CI) is similar to that measured previously using flow cytometry (∼37 nM) (50), further supporting the robustness of the ITC assay.
Enthalpy changes (ΔH) reflect the change in number and strength of noncovalent bonding and can be measured by ITC. In addition, the change in entropy (ΔS) from desolvation and conformational changes upon binding can be calculated as ΔS = (ΔH – ΔG)/T, where ΔG is the Gibbs free energy change. Our ITC results reveal that aptamer–EpCAM protein binding is mainly driven by enthalpy contributions, ΔH. The stoichiometry N for the binding of EP23, A5U, or G15U to EpCAM protein ranges from 0.458 to 0.474, close to 0.5, indicating one aptamer binds to approximately two EpCAM proteins. The reason that the N value is lower than 0.5 may be due to the accuracy of the aptamer and/or EpCAM protein concentrations or inactive EpCAM protein in the solution. The N values obtained from the ITC experiment are in accordance with published data that the EpCAM proteins form a dimer in solution (34), and our computational modeling also indicates that one aptamer binds to one EpCAM dimer.
Our experimental results overlap well within our predicted affinity values (A5U ΔΔG: −0.48 ± 0.64 vs. −0.77 ± 0.12 kcal/mol computed vs. experiment, and G15U ΔΔG: −1.92 ± 1.52 vs. −0.71 ± 0.10 kcal/mol computed vs. experiment; 95% CI), providing additional evidence that the predicted dominant binding conformation is indeed representative of the experimental binding structure. The relatively large error of the predicted G15U mutation is a result of the broken G15-U4 base pair, resulting in larger structural fluctuations than the A5U mutant. We also computed mutational binding affinity changes for other binding conformations (01 and 04; SI Appendix, Fig. S3); associated results were inconsistent with experiments, further supporting conformation 02 as the dominant binding mode.
Discussion
Our in silico design of RNA aptamers for EpCAM binding is exciting for several reasons. Starting from an RNA sequence, we constructed a stable, folded RNA structure using only MD simulations and secondary structure-predicted constraints. At this time, numerous RNA structure prediction models exist, but with varied accuracies and shortcomings (14). Accommodating the dynamic nature of RNA structure, our physics-based RNA structure-building framework allowed for appropriate fluctuations in the RNA structure while still identifying stable, robust RNA conformations. Given the importance of Mg2+ in RNA folding, we selected folded RNA structures for advancement based on the presence of long-lived Mg2+ binding sites rather than more global metrics such as root-mean-square deviation. This in silico selection criterion worked well to identify stable RNA structures for our system, and future work will be necessary to determine its utility for other RNAs.
We further identified how the designed RNA aptamer binds to the in vivo-relevant and in vitro-relevant form of EpCAM: the EpCAM dimer. Similar to other studies (38, 39), we used docking to predict RNA binding conformations onto the EpCAM monomer. However, the predicted docking conformations showed strong binding preference for the TY loop of the EpCAM monomer. This result led us to conclude that aptamer binding to the EpCAM monomer TY loop [as found by others (38, 39)] is most likely not physiologically relevant, considering the TY loop’s close proximity to the cell membrane and its location at a strong [kd of 10 nM (37)] intracellular dimerization interface. Hence, we conducted RNA aptamer docking onto the EpCAM dimer instead. From EpCAM dimer docking, we found a considerable preference for binding to the top and sides of the EpCAM C-terminal domain, with some crossover onto the N-terminal domain. These observed binding modes stood in stark contrast to most antibody binding sites, which often bind the N-terminal domain (33). The sides and top of the C-terminal domain have numerous positively charged residues that form favorable electrostatic interactions with the negatively charged RNA aptamer backbone; furthermore, the C-terminal domain is solvent exposed, rather than being buried by the membrane similar to the EpCAM TY loop. This domain thus represents a strong target for EpCAM-focused aptamer binding (as confirmed by our free energy simulations and experiments on aptamer mutants).
Last, it is significant that we discovered two RNA sequences that bound more strongly to EpCAM than the initial EP23 molecule identified via SELEX experiments. As Fig. 4C indicates, the A5U and G15U mutants both bind to EpCAM at approximately four times the potency of EP23. While factors other than affinity (e.g., binding specificity, toxicity, and deliverability) need to be considered in evaluating aptamer drugs, the two sequences presented in this work should certainly be explored as EpCAM-targeted therapeutics in their own right. More generally, we illustrated that the SELEX procedure, while effective, is not completely exhaustive in exploring RNA sequence space for particular targets, and can be further optimized with in silico techniques such as those applied in this work.
Conclusions
In this work, we used a combined experimental and theoretical approach to investigate the EP23 aptamer binding EpCAM and discovered multiple aptamer sequences with increased binding affinity. Aptamer binding conformations and favorable binding affinity mutations were predicted in silico, using structure-based techniques including MD simulations and FEP calculations. Starting from a known aptamer sequence, we built the RNA structure and used docking to predict EpCAM binding. Both monomer and dimer forms of EpCAM were investigated, but the EpCAM dimer was selected for further investigation because of extensive experimental evidence of EpCAM existing in vivo and in vitro as a dimer. MD simulations including nonequilibrium steered MD simulations revealed one strong binding conformation, binding to the C-terminal domain of EpCAM. From FEP calculations of EP23 aptamer mutations, we found that two mutations resulted in stronger EpCAM binding affinity. The binding affinity of these mutations was investigated experimentally, using isothermal titration calorimetry, and was found to agree with the in silico predictions, resulting in stronger binding affinity with the mutated residues. This work confirms the utility of using in silico structure-based techniques to design and determine favorable RNA aptamers for physiological targets.
Materials and Methods
MD Simulations.
All MD simulations were conducted using NAMD2 (51), specifically configured for IBM Power systems. The RNA structure was initially built as a single-stranded A-form helix. Mfold (28) base pair constraints were implemented, and then the RNA was slowly heated up to 310 K, using Generalized Born implicit solvent. Then, solvent and ions (0.15 M Mg2+) were added and MD was performed for 450 ns, using a 2 fs time step, with the first 150 ns maintaining base pair constraints. The Amberff14SB force field (52) was used for RNA parameters. The EpCAM protein structure was taken from PDB ID: 4MZV (33). All EpCAM simulations (both monomer and dimer, with and without bound aptamer conformations) were run for 500 ns. The top binding conformation for EpCAM dimer was further simulated for an additional 500 ns; in total, more than 7 µs of MD were conducted. The CHARMM36 force field (53) was used for protein parameters. MD simulations with protein were conducted in the NPT ensemble, with a pressure of 1 atm and temperature of 310 K. Van der Waals interactions were cutoff at 12 Å, while long-range electrostatics were treated with the Particle Mesh Ewald method.
FEP Calculations.
FEP calculations were computed using NAMD2, as described previously (54). Although dual topologies exist for amino acids, nucleotide dual topologies had to be created using existing AMBERff14SB force field (52) parameters. All FEP calculations were computed with at least 12 replicas for each structure (up to 100+ replicas for each mutation), using 34 windows with at least 400 ps/window.
DOT2 Docking.
Dot2.0 (27) software was downloaded and run locally on IBM Power systems. The top 200 conformations were saved and used to generate the heat maps in Figs. 2 and 3. Three aptamer conformations (RNA + bound Mg2+) were selected for docking. Ultimately, only one aptamer conformation resulted in plausible binding conformations and was selected for analysis. Only unique EpCAM binding conformations were selected for advancement, and MD simulations were performed on the top 10 Dot2.0 predicted conformations.
EpCAM Reagents.
Human EpCAM protein was purchased from Sino Biological Inc. Australia (catalog number: 10694-H08H).
Aptamer Sequences.
The original EpCAM aptamer EP23 and the mutant aptamers A5U and G15U were synthesized by Suzhou Genepharma (Suzhou, China), followed by HPLC purification. The sequences of EP23, A5U, and G15U are shown here:
EP23 aptamer: 5′-A (2′-F-C) G (2′-F-U) A (2′-F-U) (2′-F-C) (2′-F-C) (2′-F-C) (2′-F-U) (2′-F-U) (2′-F-U) (2′-F-U) (2′-F-C) G (2′-F-C) G (2′-F-U) A-3′
A5U aptamer: 5′-A (2′-F-C) G (2′-F-U) (2′-F-U) (2′-F-U) (2′-F-C) (2′-F-C) (2′-F-C) (2′-F-U) (2′-F-U) (2′-F-U) (2′-F-U) (2′-F-C) G (2′-F-C) G (2′-F-U) A-3′
G15U aptamer: 5′-A (2′-F-C) G (2′-F-U) A (2′-F-U) (2′-F-C) (2′-F-C) (2′-F-C) (2′-F-U) (2′-F-U) (2′-F-U) (2′-F-U) (2′-F-C) (2′-F-U) (2′-F-C) G (2′-F-U) A-3′
In these sequences, the uppercase letters A, U, G and C indicate RNAs and 2′-F represents a substitution of 2′-fluoro for 2′-hydroxyl group in an RNA monomer. For all of the aptamers, an amino-modifier-C6-TFA phosphoramidite group (6-[trifluoroacetylamino]hexyl-[(2-cyanoethyl)-(N,N-diisopropyl)]- phosphoramidite) and an inverted deoxythymidine group were added to the 5′- and 3′-end of the aptamers, respectively, for enhanced nuclease resistance.
The aptamers were prepared using the dialysis buffer in which the major components are PBS containing 5 mM MgCl2, as described in ITC, and then folded by denaturation at 85 °C for 5 min, followed by 10 min incubation at room temperature and 37 °C for at least 15 min.
ITC.
ITC experiments were performed at 25 °C with a Microcal PEAQ-ITC instrument (Malvern). The EpCAM proteins were dissolved in PBS containing 5 mM MgCl2 to achieve a final concentration of 5 µM. To minimize buffer mismatch and remove small molecules including mannitol and trehalose, which exist in the protein solution, the EpCAM protein solution was extensively dialyzed against 1 L PBS containing 5 mM MgCl2, using a dialysis cassette with a 10-kDa molecular weight cutoff (Thermo). Briefly, the samples were dialyzed at 4 °C for 2 h and the buffer was refreshed with 1 L PBS containing 5 mM MgCl2. Then the protein samples were dialyzed against the dialysis buffer at 4 °C overnight.
An aptamer solution (50 μM) in a syringe was injected into 300 μL of 5 μM EpCAM protein in a sample cell. The volume of aptamer solution in each injection was 2 μL, except for the first injection, which was 0.4 µL. The time interval between injections was 150 s, and the syringe stirring speed was 750 rpm. The reference power was 10.0 μcal/s. As a control, the aptamer samples were injected into the ITC buffer alone to subtract the heat of dilution of the samples. Stoichiometry of binding (N), dissociation constant (Kd), and thermodynamic parameters including enthalpy change (ΔH), entropy change (ΔS), and Gibbs free energy change (ΔG) were obtained according to a single-site binding model using the MicroCal PEAQ-ITC analysis software (Malvern). Data were from three independent measurements.
ITC Data Analysis.
All data and results were analyzed using the Affinimeter software (AFFINImeter, Spain) and are shown as mean ± SD (mean ± S. D.) unless otherwise stated. The differences among multiple groups (n ≥ 3) and between two specific groups were analyzed by one-way ANOVA and t test, using GraphPad Prism 8.0.1, respectively. In all tests, P < 0.05 was considered as statistically significant.
Data Availability.
All data and analyses tools/scripts are available upon request.
Supplementary Material
Acknowledgments
The authors wish to acknowledge insightful discussions with Hongsuk Kang, Binquan Luan, Serena H. Chen, Leili Zhang, Francisco X. Vasquez, Zhifeng Francis Jing, and Jose Aguilar. R.Z. gratefully acknowledges the financial support from W. M. Keck Foundation (2019-2022) and the IBM Bluegene Science Program (grant numbers: W125859, W1464125, and W1464164).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913242117/-/DCSupplemental.
References
- 1.Ellington A. D., Szostak J. W., In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C., Gold L., Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Keefe A. D., Pai S., Ellington A., Aptamers as therapeutics. Nat. Rev. Drug Discov. 9, 537–550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J., Rossi J., Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 16, 181–202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carthew R. W., Sontheimer E. J., Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson R. C., Doudna J. A., Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 42, 217–239 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doudna J. A., Charpentier E., Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Cong L., et al. , Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Setten R. L., Rossi J. J., Han S. P., The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 18, 421–446 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Nimjee S. M., White R. R., Becker R. C., Sullenger B. A., Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol. 57, 61–79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Presnell K. V., Alper H. S., Thermodynamic and first-principles biomolecular simulations applied to synthetic biology: Promoter and aptamer designs. Mol. Syst. Des. Eng. 3, 19–37 (2018). [Google Scholar]
- 12.Laing C., Schlick T., Computational approaches to RNA structure prediction, analysis, and design. Curr. Opin. Struct. Biol. 21, 306–318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell D. R., Cheng S. Y., Salazar H., Ren P., Capturing RNA folding free energy with coarse-grained molecular dynamics simulations. Sci. Rep. 7, 45812 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao Z., et al. , RNA-puzzles round III: 3D RNA structure prediction of five riboswitches and one ribozyme. RNA 23, 655–672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim N., Izzo J. A., Elmetwaly S., Gan H. H., Schlick T., Computational generation and screening of RNA motifs in large nucleotide sequence pools. Nucleic Acids Res. 38, e139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Y., Zhang D., Zhou P., Li B., Huang S.-Y., HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 45, W365–W373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwakiri J., Hamada M., Asai K., Kameda T., Improved accuracy in RNA-protein rigid body docking by incorporating force field for molecular dynamics simulation into the scoring function. J. Chem. Theory Comput. 12, 4688–4697 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Pagadala N. S., Syed K., Tuszynski J., Software for molecular docking: A review. Biophys. Rev. 9, 91–102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisold A., Labudde D., Detailed analysis of 17β-estradiol-aptamer interactions: A molecular dynamics simulation study. Molecules 23, E1690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.La Penna G., Chelli R., Structural insights into the osteopontin-aptamer complex by molecular dynamics simulations. Front Chem. 6, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaber A., et al. , EpCAM homo-oligomerization is not the basis for its role in cell-cell adhesion. Sci. Rep. 8, 13269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trzpis M., McLaughlin P. M. J., de Leij L. M. F. H., Harmsen M. C., Epithelial cell adhesion molecule: More than a carcinoma marker and adhesion molecule. Am. J. Pathol. 171, 386–395 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnell U., Cirulli V., Giepmans B. N. G., EpCAM: Structure and function in health and disease. Biochim. Biophys. Acta. 1828, 1989–2001 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Imrich S., Hachmeister M., Gires O., EpCAM and its potential role in tumor-initiating cells. Cell Adhes. Migr. 6, 30–38 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon M., Stefan N., Plückthun A., Zangemeister-Wittke U., Epithelial cell adhesion molecule-targeted drug delivery for cancer therapy. Expert Opin. Drug Deliv. 10, 451–468 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Heiss M. M., et al. , The trifunctional antibody catumaxomab for the treatment of malignant ascites due to epithelial cancer: Results of a prospective randomized phase II/III trial. Int. J. Cancer 127, 2209–2221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts V. A., Thompson E. E., Pique M. E., Perez M. S., Ten Eyck L. F., DOT2: Macromolecular docking with improved biophysical models. J. Comput. Chem. 34, 1743–1758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuker M., Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shigdar S., et al. , RNA aptamer against a cancer stem cell marker epithelial cell adhesion molecule. Cancer Sci. 102, 991–998 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Xiang D., et al. , Superior performance of aptamer in tumor penetration over antibody: Implication of aptamer-based theranostics in solid tumors. Theranostics 5, 1083–1097 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng H., Shabalin I. G., Handing K. B., Bujnicki J. M., Minor W., Magnesium-binding architectures in RNA crystal structures: Validation, binding preferences, classification and motif detection. Nucleic Acids Res. 43, 3789–3801 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cunha R. A., Bussi G., Unraveling Mg2+-RNA binding with atomistic molecular dynamics. RNA 23, 628–638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavšič M., Gunčar G., Djinović-Carugo K., Lenarčič B., Crystal structure and its bearing towards an understanding of key biological functions of EpCAM. Nat. Commun. 5, 4764 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Pavšič M., Lenarčič B., Expression, crystallization and preliminary x-ray characterization of the human epithelial cell-adhesion molecule ectodomain. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1363–1366 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petsch S., et al. , Concentrations of EpCAM ectodomain as found in sera of cancer patients do not significantly impact redirected lysis and T-cell activation by EpCAM/CD3-bispecific BiTE antibody MT110. MAbs 3, 31–37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seeber A., et al. , Soluble EpCAM levels in ascites correlate with positive cytology and neutralize catumaxomab activity in vitro. BMC Cancer 15, 372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trebak M., et al. , Oligomeric state of the colon carcinoma-associated glycoprotein GA733-2 (Ep-CAM/EGP40) and its role in GA733-mediated homotypic cell-cell adhesion. J. Biol. Chem. 276, 2299–2309 (2001). [DOI] [PubMed] [Google Scholar]
- 38.Alshaer W., et al. , Selection and targeting of EpCAM protein by ssDNA aptamer. PLoS One 12, e0189558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bavi R., Liu Z., Han Z., Zhang H., Gu Y., In silico designed RNA aptamer against epithelial cell adhesion molecule for cancer cell imaging. Biochem. Biophys. Res. Commun. 509, 937–942 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Casaletto J. B., et al. , MM-131, a bispecific anti-Met/EpCAM mAb, inhibits HGF-dependent and HGF-independent Met signaling through concurrent binding to EpCAM. Proc. Natl. Acad. Sci. U.S.A. 116, 7533–7542 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balzar M., et al. , Epidermal growth factor-like repeats mediate lateral and reciprocal interactions of Ep-CAM molecules in homophilic adhesions. Mol. Cell. Biol. 21, 2570–2580 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macdonald J., et al. , Development of a bifunctional aptamer targeting the transferrin receptor and epithelial cell adhesion molecule (EpCAM) for the treatment of brain cancer metastases. ACS Chem. Neurosci. 8, 777–784 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Chen S.-H., Elber R., The energy landscape of a protein switch. Phys. Chem. Chem. Phys. 16, 6407–6421 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Chen S.-H., Meller J., Elber R., Comprehensive analysis of sequences of a protein switch. Protein Sci. 25, 135–146 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luan B., Xu G., Feng M., Cong L., Zhou R., Combined computational-experimental approach to explore the molecular mechanism of SaCas9 with a broadened DNA targeting range. J. Am. Chem. Soc. 141, 6545–6552 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Joglekar A. V., et al. , T cell receptors for the HIV KK10 epitope from patients with differential immunologic control are functionally indistinguishable. Proc. Natl. Acad. Sci. U.S.A. 115, 1877–1882 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowell D., et al. , Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359, 582–587 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S. H., Kang S. G., Luo J., Zhou R., Charging nanoparticles: Increased binding of Gd@C82(OH)22 derivatives to human MMP-9. Nanoscale 10, 5667–5677 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Turnbull W. B., Daranas A. H., On the value of c: Can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 125, 14859–14866 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Shigdar S., et al. , The use of sensitive chemical antibodies for diagnosis: Detection of low levels of EpCAM in breast cancer. PLoS One 8, e57613 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheatham T. E. 3rd, Case D. A., Twenty-five years of nucleic acid simulations. Biopolymers 99, 969–977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Best R. B., et al. , Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia Z., et al. , The complex and specific pMHC interactions with diverse HIV-1 TCR clonotypes reveal a structural basis for alterations in CTL function. Sci. Rep. 4, 4087 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and analyses tools/scripts are available upon request.