Significance
Dicer is a ribonuclease III enzyme in biosynthesis of miRNAs, regulators of gene expression involved in macrophage differentiation. We found a specific truncation of Dicer in monocytic cells resulting from apparently constitutive cleavage by a serine protease. Inhibition of this proteolytic truncation, which occurred during macrophage differentiation in presence of TLR ligands or prostaglandin E2, up-regulates full-length Dicer and promotes miR biosynthesis. Regulation of transcription of pri-miRNA is one mode to regulate biosynthesis of mature miRNA. Inhibition of constitutive proteolysis of Dicer, as described here, provides a second layer of regulation, at the level of miRNA processing. Our data provide insights to Dicer and miRNAs in macrophage polarization/differentiation, a key process in the innate immune response.
Keywords: miRNA, PGE2, macrophage
Abstract
Dicer is a ribonuclease III enzyme in biosynthesis of micro-RNAs (miRNAs). Here we describe a regulation of Dicer expression in monocytic cells, based on proteolysis. In undifferentiated Mono Mac 6 (MM6) cells, full-length Dicer was undetectable; only an ∼50-kDa fragment appeared in Western blots. However, when MM6 cells were treated with zymosan or LPS during differentiation with TGF-β and 1,25diOHvitD3, full-length Dicer became abundant together with varying amounts of ∼170- and ∼50-kDa Dicer fragments. Mass spectrometry identified the Dicer fragments and showed cleavage about 450 residues upstream from the C terminus. Also, PGE2 (prostaglandin E2) added to differentiating MM6 cells up-regulated full-length Dicer, through EP2/EP4 and cAMP. The TLR stimuli strongly induced miR-146a-5p, while PGE2 increased miR-99a-5p and miR-125a-5p, both implicated in down-regulation of TNFα. The Ser protease inhibitor AEBSF (4-[2-aminoethyl] benzene sulfonyl fluoride) up-regulated full-length Dicer, both in MM6 cells and in primary human blood monocytes, indicating a specific proteolytic degradation. However, AEBSF alone did not lead to a general increase in miR expression, indicating that additional mechanisms are required to increase miRNA biosynthesis. Finally, differentiation of monocytes to macrophages with M-CSF or GM-CSF strongly up-regulated full-length Dicer. Our results suggest that differentiation regimens, both in the MM6 cell line and of peripheral blood monocytes, inhibit an apparently constitutive Dicer proteolysis, allowing for increased formation of miRNAs.
Dicer is an RNA endonuclease type III/RNase III enzyme best known for its canonical function in biosynthesis of micro-RNAs (miRNA), which are predicted to control up to 60% of protein-coding genes by targeting specific mRNA for degradation or translation repression. MiRNA are formed from primary transcripts (pri-miRNA) in two stages of processing. Pri-miRNAs, transcribed by RNA polymerase II, are first cleaved by the nuclear microprocessor complex (Drosha and its partner DGCR8) (1). The resulting hairpin structures called pre-miRNAs (precursor-miRNAs) consist of ∼70 nucleotides with two nucleotides overhanging at the 3′ end. Following exportin-5–mediated transport to the cytosol, final processing by Dicer yields ∼22 nucleotide mature double-stranded miRNAs. Only one of the strands (the guide strand) of miRNA is then incorporated to RNA-induced silencing complex (RISC) and directs this complex to 3′ untranslated regions (UTR) of target mRNAs for inhibition of translation (1, 2). Biogenesis of miRNAs occurs under stringent spatial and temporal control, and dysregulation can be associated with human diseases (3). Several recent findings show that miRNAs formed in inflammatory cells play major roles in regulating inflammatory responses (4).
Dicer is a large enzyme (∼220 kDa) with several domains including an N-terminal helicase domain, DUF283 (domain of unknown function), PAZ (Piwi-Argonaute-Zwille) domain, two RNase III domains (RNase IIIa/b), and a dsRNA-binding domain (dsRBD) (5–7). An intramolecular dimerization of the two RNase III domains is suggested to form the active RNase center. Crystal structures of human C-terminal RNase IIIb domain (residues 1660 to 1852), and of mouse Dicer domains RNase IIIb plus dsRBD, revealed homodimerization to form an active site, which is similar to the bacterial RNase III enzyme (8, 9). Both these C terminus homodimers were enzymatically active, cleaving dsRNA substrates in vitro. Recently, a substrate processing mechanism was suggested, based on a cryo-EM structure of human Dicer bound with cofactor protein TRBP and its precursor miRNA substrate (7).
Regulation of Dicer expression shows complexity, mRNA transcripts do not always correlate with protein, and there are also posttranscriptional and posttranslational mechanisms (10). Proteolytic cleavages of Dicer have been described in mouse and human cells, and in Caenorhabditis elegans. Calpain I treatment of mouse brain Dicer gave a 75 kDa fragment, with increased RNaseIII activity (11). Cleavage of Dicer in the RNase IIIa domain by caspase-3 in cancer cells resulted in loss of Dicer activity (12). In C. elegans, cleavage by CED-3 caspase generated a C-terminal fragment with DNase activity which could digest chromosomal DNA and promote apoptosis (13). In HeLa cells, Dicer was down-regulated by caspase cleavage during apoptosis (14). In adult C. elegans, half of the Dicer protein was expressed as a stable truncated C-terminal fragment (small DCR-1) containing the RNaseIII and dsRBD domains (15). In platelets of diabetic mice and patients, cleavage of Dicer by calpain determined the miRNA levels (16). These findings on truncated Dicer isoforms have given more insights and open up new views about Dicer noncanonical functions, for example, in genome regulation and surveillance (17, 18).
Specific miRNAs with proinflammatory and antiinflammatory functions accumulate in macrophages activated with TLR stimuli (4, 19–21). As regulators of gene expression, miRNAs also have roles in differentiation (polarization) of macrophages (20, 22, 23). However, the regulation of miRNA processing enzymes required for miRNA biosynthesis in macrophages has not been extensively studied. Here we identified a specific proteolytic truncation of Dicer in monocytic cells. Our results suggest that Dicer is constitutively cleaved by a serine protease in monocytes and that inhibition of this proteolytic cleavage during macrophage differentiation up-regulates full-length Dicer, allowing for miRNA biosynthesis.
Methods
Cell Culture.
Mono Mac 6 cells (MM6), a human cell line with monocyte characteristics, was obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen. MM6 cells were grown in cell culture and differentiated with transforming growth factor-β (TGF-β; 2 ng/mL) and 1,25diOHvitD3 (50 nM) for 96 h as described (24). Cells were also differentiated in presence of either lipopolysaccharide (LPS, 1 µg/mL), zymosan (25 µg/mL), or PGE2 (Prostaglandin E2) at different concentrations. After 20 to 25 passages, fresh undifferentiated cells were started from frozen stock. Different reagents (protease inhibitors, EP receptor antagonists, 8-Br-cAMP, and forskolin) were obtained from Sigma, if not otherwise stated.
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and Western Blotting.
Cells were harvested, washed twice with phosphate buffered saline (PBS), resuspended in lysis buffer (20 mM Tris⋅HCl, pH 8.0, 1% Triton X-100, 2 mM EDTA, 2 mM PMSF, 100 mM NaCl, 1 mM Na3VO4, 0.1% SDS, and complete protease inhibitor mixture), sonicated 2 × 5 s on ice, and centrifuged (10,000 g, 10 min, 4 °C). Laemmli sample loading buffer was added to supernatant protein samples (typically 50 µg) and heated (90 °C, 5 min). Proteins were separated by SDS-PAGE on 4 to 20% gradient gels (Bio-Rad) and transferred to nitrocellulose membrane (Hybond C, Amersham GE) by electroblot. Membranes were washed 2 × 10 min with TTBS (20 mM Tris⋅HCl, pH 7.4, 100 mM NaCl, and 0.1% Tween 20), blocked with nonfat dry milk powder in TTBS for 1 h at room temperature, and incubated with antibodies at 4 °C overnight. Four Dicer antibodies were used: rabbit polyclonal (N-terminal epitope region 600 to 650, Bethyl Laboratories no. A301-936A) at 1:1,000, mouse monoclonal (epitope region 1239 to 1255, Abcam no. 14601) at 1:1,000, mouse monoclonal (C-terminal epitope 1701 to 1912, Santa Cruz sc-136981) at 1:100, and rabbit polyclonal (C-terminal epitope around residue 1902, BioVision no. 3697-100) at 1:100. The mPGES-1 antibody was from Cayman. Peroxidase-conjugated primary antibody against β-actin (1:2,000 dilution) and peroxidase-conjugated secondary antibodies (anti-rabbit 1:5,000 and anti-mouse 1:5,000) were from Sigma. Protein bands were detected by enhanced chemiluminescence (GE Healthcare) with an Odyssey scanner (LI-COR). Band intensity was calculated with Odyssey Imaging software.
Immunoprecipitation.
Differentiated MM6 cells were suspended in lysis buffer (Tris⋅HCl, pH 8.0, 0.1% Triton X-100, 1 mM PMSF, and complete protease inhibitor mixture), sonicated 2 × 5 s on ice, and centrifuged (10,000 g, 10 min, 4 °C). Cell lysates (600 to 1,000 µg of protein) were precleared by incubation with protein A-Sepharose for 1 h at 4 °C with continuous mixing. The precleared samples were incubated with Dicer antibody (1 to 2 µL) overnight at 4 °C. Protein A-Sepharose was added, and incubations continued for additional 1 h. The immunocomplexes were washed four times with lysis buffer and eluted by heating (at 90 °C) for 5 min in Laemmli sample loading buffer. After centrifugation, the supernatants were analyzed by SDS-PAGE and immunoblotting or Coomassie blue staining of the gel.
Mass Spectrometric Analysis.
Dicer immunoprecipitates were subjected to SDS-PAGE, and the gel was stained with Coomassie brilliant blue. Bands corresponding to the molecular mass of full-length and Dicer fragments were excised and subjected to in-gel trypsinization and mass spectrometric analysis, performed at Proteomics Karolinska (PK/KI).
Analysis of Dicer mRNA and miRNAs.
RNA isolation including DNA digestion was performed using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. RNA was reverse transcribed with the High Capacity RNA-to-cDNA kit (Applied Biosystems). Dicer levels were analyzed using the Power SYBR Green PCR Master Mix or the Fast SYBR Green Master Mix (Applied Biosystems). β-actin or GAPDH served as an endogenous control. For primer sequences, see SI Appendix, Table S1. For qPCR analysis of miRNAs, primers and kits from Qiagen (miScript II RT kit, miScript SYBR Green PCR kit) were used according to the manufacturer’s protocols. The Qiagen miScript Primer Assay utilizes miRNA-specific forward primers and the miScript Universal primer as reverse primer. U6 or miRNA-20a-5p served as an endogenous control. qPCR was carried out on Applied Biosystems 7300 or StepOnePlus machines. Affymetrix screening of miRNAs in MM6 cells was performed at the Bioinformatics and Expression analysis core facility, Novum, Karolinska Institutet.
Generation of mPGES-1 Knockdown MM6 Cells.
Generation of stable knockdown for mPGES-1 (microsomal Prostaglandin E Synthase-1) in MM6 cells was performed as described (24). Lentiviral constructs with shRNAs directed against mPGES-1 (NM_004878.3-306s1c1) was obtained from Sigma Aldrich. Lentivirus was prepared by transient cotransfection of HEK293T cells with shRNA plasmid (pLKO.1-puro) along with third-generation packaging constructs (pMDLg/pRRE + pRSV-Rev + pMD2.G). Control cells were obtained by transfection with lentivirus obtained from the pLKO.1-puro nontarget shRNA control plasmid (Sigma SHC002). Stable knockdown cells were selected by culture with puromycin (10 µg/mL) for 2 wk. The efficiency of knockdown was verified by Western blotting and analyzed each time when cells were used for different experiments.
Preparation and Differentiation of Monocytes.
Monocytes were isolated from buffy coats of 12 healthy human donors (Karolinska Hospital Blood Bank) as described previously (25, 26). Peripheral blood mononuclear cells were isolated by gradient centrifugation with Ficoll-Paque PREMIUM (GE Healthcare) and seeded at 5 × 106 cells per mL for 2 h, allowing monocytes to adhere to plates. Cells were then vigorously washed twice with PBS, to remove lymphocytes. Finally, monocytes were cultured in RPMI (Roswell Park Memorial Institute) medium 1640 with glutamine supplemented with 10% FBS, 100 mg/mL streptomycin, 100 U/mL penicillin, 1× nonessential amino acids, 25 mM Hepes. Cells were differentiated for 7 d in rhGM-CSF (granulocyte-macrophage colony-stimulating factor, 10 ng/mL) and in rhM-CSF (macrophage colony-stimulating factor, 10 ng/mL) to obtain to obtain M1- or M2-primed macrophages, respectively. Cells were supplied with fresh medium and cytokines on days 3 and 6. To yield M1/M2 macrophages, GM-CSF primed cells were treated also with LPS (100 ng/mL) and IFNγ (20 ng/mL), and M-CSF primed cells also with IL-4 (20 ng/mL) for the final 24 h of differentiation. These treatments resulted in increased formation of IL-6 and surface markers CD86 and ICAM-1 for M1 and IL-10 and surface markers CD14 and DC-SIGN for M2, as described in our previous study (26).
Data Availability.
All relevant data, associated protocols, and materials are within the manuscript and its SI Appendix files.
Results
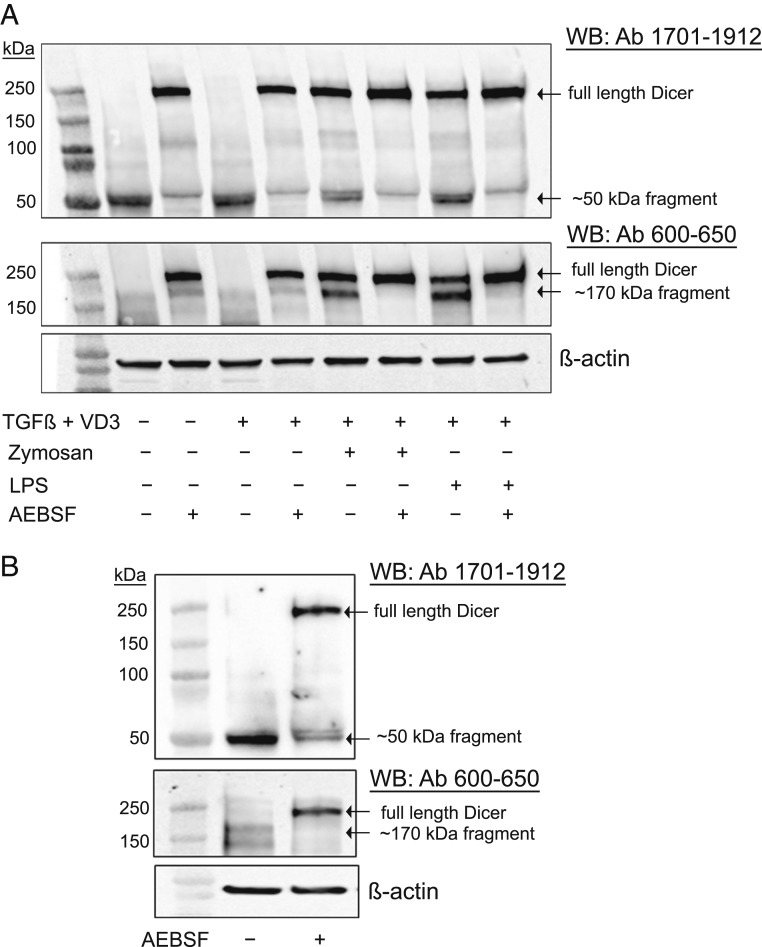
Up-Regulation of Full-Length Dicer and Truncated Fragments in MM6 Cells Differentiated in Presence of LPS and Zymosan.
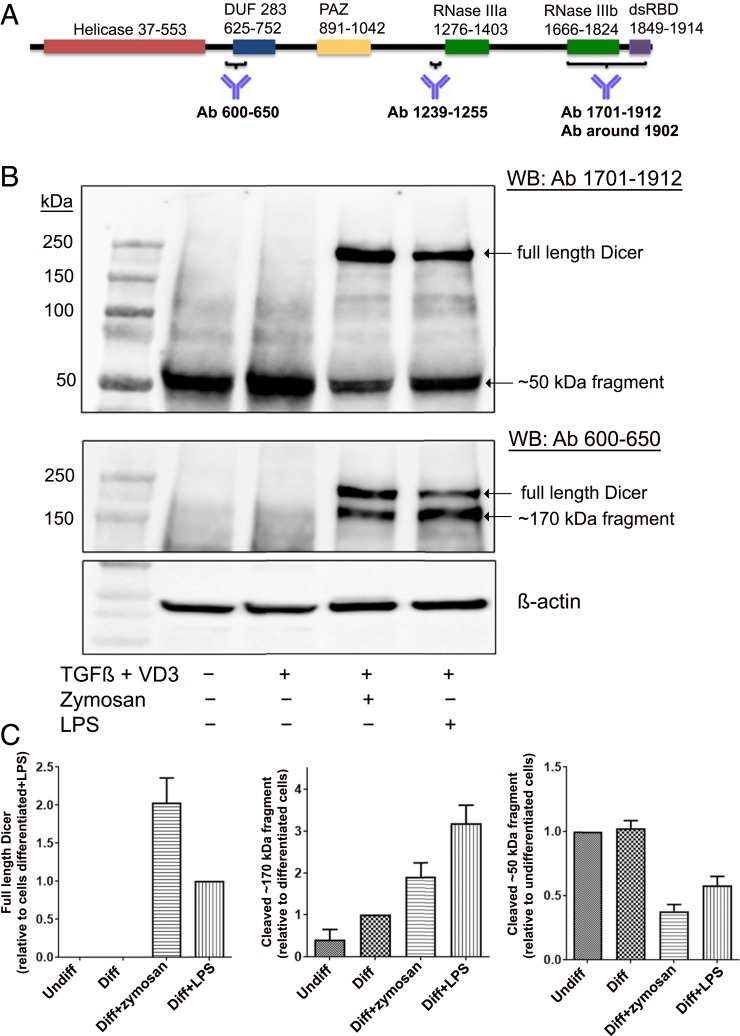
Mono Mac 6 (MM6) is a human cell line with monocytic properties; differentiation with TGF-β and 1,25diOHvitD3 for 96 h leads to macrophage-like cells (24). Screening of Dicer expression by Western blot was routinely performed using two antibodies, raised against N-terminal (epitope 600 to 650) and C-terminal (epitope 1701 to 1912) parts of Dicer (Fig. 1A). In both undifferentiated MM6 and in cells differentiated with TGF-β and 1,25diOHvitD3, truncated fragments (∼50 and ∼170 kDa) appeared, while full-length Dicer (∼220 kDa) was undetectable. However, when cells were differentiated with TGF-β and 1,25diOHvitD3 in presence of LPS or zymosan, full-length Dicer was up-regulated (Fig. 1B). The N-terminal (epitope 600 to 650) antibody detected full-length Dicer and the ∼170-kDa fragment but not the ∼50-kDa fragment. Similar results were obtained with an antibody directed against the middle region of Dicer (epitope 1239 to 1255; SI Appendix, Fig. S1). The antibody with epitope at the C terminus (epitope 1701 to 1912) recognized full-length Dicer as well as the ∼50-kDa fragment but not the ∼170-kDa fragment (Fig. 1B). These results indicated that the small ∼50-kDa fragment should be from the C terminus.
Fig. 1.
Up-regulation of full-length Dicer and Dicer fragments, induced by zymosan or LPS during differentiation of MM6 cells. (A) Scheme of Dicer domains. Epitope regions of antibodies used in this study are indicated. Most data were obtained with antibody (Ab) 600 to 650 and Ab 1701 to 1912. (B) Analysis of Dicer expression with Dicer antibodies 1701 to 1912 and 600 to 650. Lane 1, molecular weight markers; lane 2, undifferentiated MM6 cells; lane 3, MM6 cells differentiated with TGF-β (5 ng/mL) and 1,25diOHvitD3 (50 nM) (VD3) for 96 h; lane 4, zymosan (25 µg/mL) present during 96 h differentiation; and lane 5, LPS (1 µg/mL) present during 96 h differentiation. Whole-cell lysates (∼50 µg protein) were analyzed by Western blot (WB) using Dicer Ab 1701 to 1912. The membrane was reblotted with Dicer Ab 600 to 650. (C) Relative expression levels of full-length Dicer and Dicer fragments (∼50 and ∼170 kDa). In undifferentiated MM6 cells (Undiff), MM6 cells differentiated with TGF-β + 1,25diOHvitD3 (Diff) for 96 h, with zymosan present during differentiation, and with LPS present during differentiation. Band intensities were normalized to β-actin, before comparisons between samples. Data are from B and six additional experiments. Mean ± SE, n = 7.
The relative levels of Dicer and fragments were evaluated in seven independent experiments (Fig. 1C). The C-terminal ∼50-kDa fragment was present in all samples. Full-length Dicer appeared only when cells were differentiated in presence of zymosan (stimulates TLR2) or LPS (stimulates TLR4). In zymosan-treated cells, full-length Dicer dominated over the ∼170-kDa fragment, while the opposite was observed for LPS-treated cells. We also tested to add zymosan late during differentiation (at time 72 h of the 96 h differentiation period). Also in this condition, full-length Dicer was up-regulated but less intense compared to ∼170-kDa fragment (SI Appendix, Fig. S2). In several experiments an inverse relationship between full-length Dicer and the fragments was observed, suggesting a specific proteolytic cleavage.
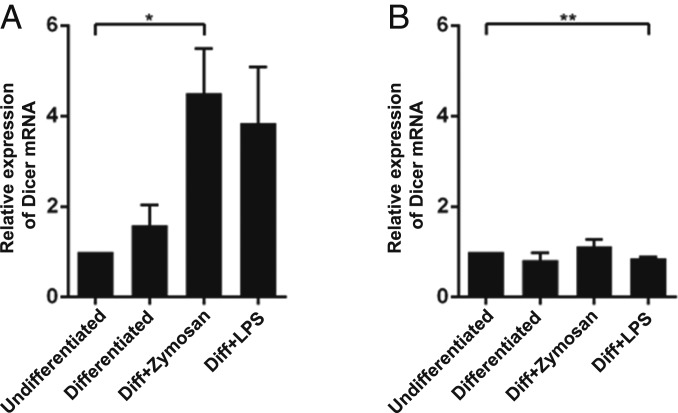
Quantitative analysis of Dicer mRNA in MM6 cells by qPCR showed a modest twofold to threefold up-regulation when also LPS or zymosan was present during the 96 h differentiation with TGF-β and 1,25diOHvitD3 (Fig. 2A). When LPS or zymosan was present only during the final 24 h of differentiation, Dicer mRNA was quite constant (Fig. 2B). The primers used amplified the Dicer mRNA sequence 5662 to 5811, which corresponds to the region of the ∼50-kDa fragment.
Fig. 2.
Dicer mRNA levels in MM6 cells. RNA was prepared from MM6 cells, reverse transcribed, and analyzed by qPCR with β-actin as reference gene. The data are presented relative to undifferentiated cells, set as 1. Two-tailed unpaired t test. **P < 0.01; *P < 0.05. (A) Undifferentiated cells and cells differentiated with TGF-β + 1,25diOHvitD3 (Diff) for 96 h, with LPS or zymosan present during the entire differentiation period (96 h). Mean ± SEM, n = 3. (B) Undifferentiated cells and cells differentiated with TGF-β + 1,25diOHvitD3 for 96 h, with LPS or zymosan present during the final 24 h of the differentiation period (96 h). Mean ± SEM, n = 3.
Mass Spectrometric Analysis of Dicer Immunoprecipitates Validate Truncation in C-Terminal Part.
Dicer was immunoprecipitated (IP) from cells differentiated in presence of zymosan or LPS using the epitope 1239 to 1255 antibody. When the IP was analyzed by Western blot with the same antibody, both full-length Dicer and the ∼170-kDa Dicer fragment appeared (SI Appendix, Fig. S3A). The membrane was reprobed with a C-terminal epitope antibody; then the ∼170-kDa Dicer fragment could not be observed (SI Appendix, Fig. S3B). The same pattern was also obtained when the N-terminal epitope (region 600 to 650) antibody was used for immunoprecipitation and Western blot analysis. These results confirm that the ∼170-kDa fragment lacked the C-terminal region.
Dicer and the fragments were positively identified by mass spectrometry (MS). A Dicer IP from zymosan-treated MM6 cells was divided in two parts. These were resolved on SDS-PAGE and subjected to Coomassie blue staining or Western blot. Western blot again showed full-length Dicer and the ∼170-kDa Dicer fragment, as above. From the Coomassie-stained gel, bands corresponding to full-length Dicer (upper band) and the ∼170 kDa Dicer fragment (lower band) were excised (SI Appendix, Fig. S4A). After trypsinization, peptides were analyzed by MS. Material in the upper band showed peptides corresponding to all parts of Dicer. Material in the lower band showed many similar peptides but none corresponding to the Dicer sequence after Lys-1465 (SI Appendix, Fig. S4B). Similar results were obtained also with samples from MM6 cells differentiated with TGF-β + 1,25diOHvitD3 in presence of LPS for 96 h. Furthermore, analysis of the ∼50-kDa fragment (IP with C-terminal epitope 1701 to 1912 antibody) showed peptides corresponding only to the C-terminal part of Dicer (SI Appendix, Fig. S5). These results clearly confirmed proteolytic truncation in the C-terminal part of Dicer.
PGE2 Present During Differentiation of MM6 Cells Up-Regulates Full-Length Dicer.
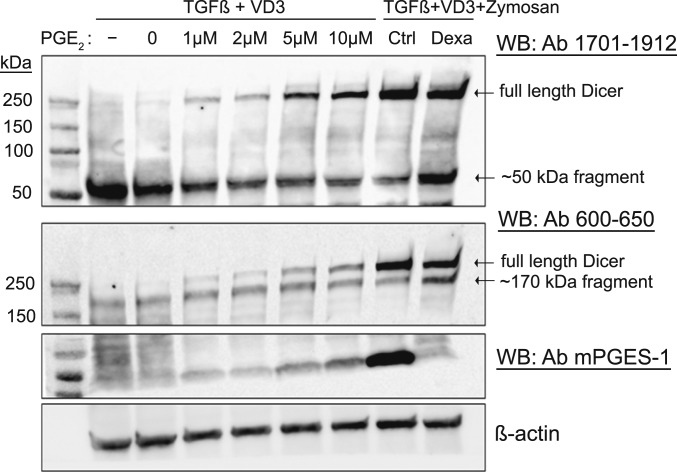
Differentiation of MM6 cells in presence of PGE2 (no TLR stimuli) also up-regulated full-length Dicer, in a dose-dependent fashion (Fig. 3). A dose-dependent up-regulation of mPGES-1 expression was also observed, presumably by the positive feedback mechanism reported earlier (27). Since dexamethasone down-regulates mPGES-1 expression (28), we treated MM6 cells differentiated in presence of zymosan with dexamethasone. This resulted in a slightly attenuated up-regulation of full-length Dicer and a corresponding increase of Dicer fragments (Fig. 3, rightmost lanes).
Fig. 3.
Effect of exogenous PGE2 added to differentiating wild-type MM6 cells. MM6 cells were differentiated with TGF-β + 1,25diOHvitD3 (VD3) and increasing concentrations of PGE2 (1 to 10 µM) present during the 96-h differentiation period. In addition, cells were differentiated with TGF-β, 1,25diOHvitD3, and zymosan, ±dexamethasone (Dexa, 150 nM). Whole-cell lysates were analyzed by Western blot (WB) using Dicer antibody (Ab) 1701 to 1912. The membrane was reblotted with Dicer Ab 600 to 650 and with mPGES-1 Ab. Similar results were obtained in two additional experiments.
The expression level of Dicer in mPGES-1 knockdown MM6 cells further supported a role for PGE2. It was reported before (29) that MM6 cells differentiated in presence of zymosan up-regulate mPGES-1 and release PGE2. The relative amount of full-length Dicer was about half in zymosan-treated mPGES-1 knockdown cells compared to zymosan-treated control cells, in parallel with increased levels of Dicer fragments (SI Appendix, Fig. S6 A and B). Addition of exogenous PGE2 to mPGES-1 knockdown MM6 cells reduced truncation (SI Appendix, Fig. S7).
E-Prostanoid Receptors EP2/EP4 Mediate the PGE2 Effect on Dicer via cAMP.
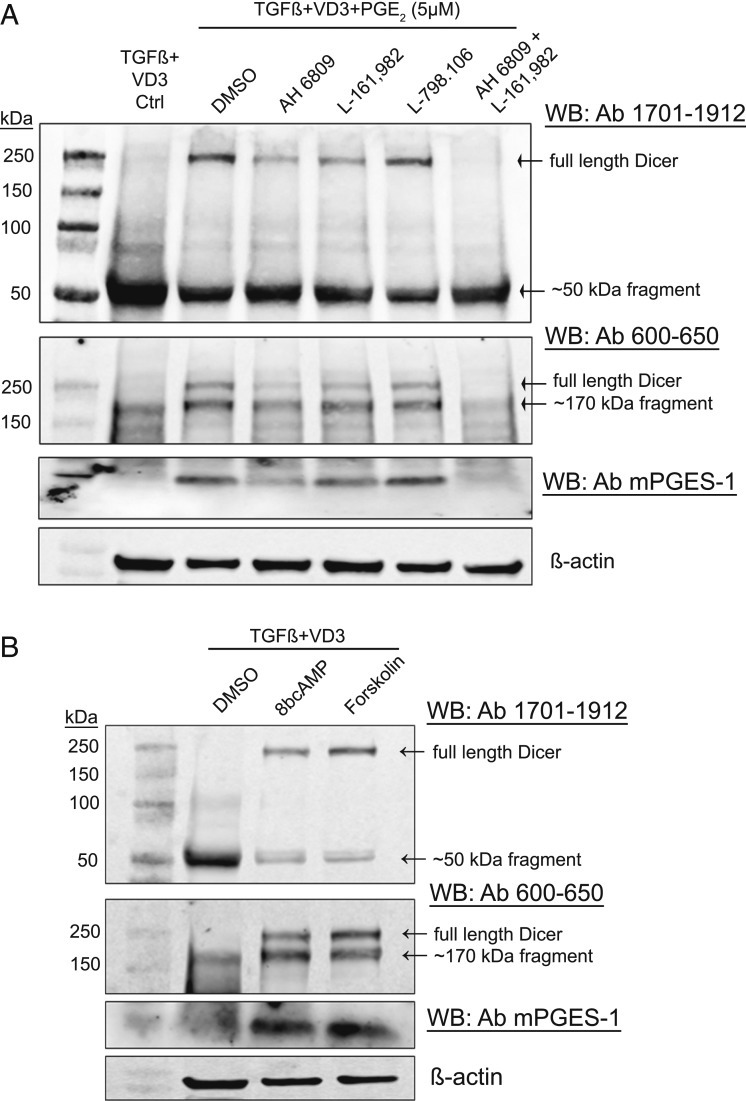
PGE2 signals through four different EP receptors (EP1 to EP4) (30). Using antagonists, we evaluated PGE2 receptors involved in up-regulation of Dicer. MM6 were differentiated (96 h) in presence of PGE2 (5 µM) and treated with inhibitors. AH6809 is known to strongly inhibit EP2, with less affinity for EP1 and EP3. L798.106 is EP3 specific, and L161.982 is specific for EP4. Antagonists to EP2 and EP4 receptors reduced the PGE2-mediated up-regulation of Dicer, and there was a complete block when AH6809 and L161.982 were added together (Fig. 4A). The EP3-specific antagonist had no effect. Also the positive feedback up-regulation of mPGES-1 expression by PGE2 was inhibited by EP2 and EP4 receptor antagonists. PGE2 signaling via EP2 and EP4 receptor increases intracellular cAMP levels by activating adenylate cyclase. Treatment of MM6 cells during differentiation, with cell-permeable 8-Br-cAMP or with adenylate cyclase activator forskolin, also up-regulated Dicer and mPGES-1 expression in MM6 cells (Fig. 4B). Taken together, the results suggest that PGE2-mediated activation of intracellular cAMP signaling may inhibit a protease involved in Dicer cleavage.
Fig. 4.
PGE2-mediated activation of cAMP signaling up-regulates full-length Dicer in MM6 cells. (A) Effect of different EP receptor antagonists on PGE2 up-regulation of Dicer in MM6 cells. MM6 cells were differentiated with TGF-β + 1,25diOHvitD3 (VD3) in presence of PGE2 (5 µM) for 96 h. EP-receptor antagonists (10 µM) were present during the entire differentiation period. For EP1-3 (AH6809), EP3 (L798.106), and EP4 (L161.982). Whole-cell lysates (∼50 µg protein) were analyzed by Western blot (WB) using Dicer antibody (Ab) 1701 to 1912. The membrane was reblotted with Dicer Ab 600 to 650 and with mPGES-1 Ab. Similar results were obtained in two additional experiments. (B) Effect of cell-permeable 8-Br-cAMP and adenylate cyclase activator forskolin on Dicer up-regulation in MM6 cells during differentiation. MM6 cells were differentiated with TGF-β + 1,25diOHvitD3 (VD3), in presence of 8-Br-cAMP (100 μM) or forskolin (20 μM) during the 96-h differentiation period. Cell lysates were analyzed by Western blot (WB) using Dicer antibody (Ab) 1701 to 1912. The membrane was reprobed with Dicer Ab 600 to 650 and with mPGES-1 Ab. Similar results were obtained in two additional experiments.
Serine Protease-Specific Inhibitor AEBSF Prevents Proteolysis of Dicer in MM6 Cells and in Primary Human Monocytes.
To determine the protease involved in the cleavage of Dicer in MM6 cells, we used inhibitors against various classes of proteases. Limited cleavage of Dicer by activated caspases and calpain in different cell types has been reported in previous studies (17). We tested cell-permeable selective inhibitors for caspase-1 (Ac-YVAD-cmk), caspase-3 (Ivachtin), caspase-6 (Ac-VEID-CHO), and calpain-1 (ALLN, PD151746, and PD150606). None of these prevented Dicer cleavage in MM6 cells, when tested at different concentrations and time points during differentiation. Also the proteasome inhibitor MG132 failed to prevent the Dicer cleavage in MM6 cells. However, the serine protease-specific inhibitor AEBSF (4-[2-aminoethyl] benzene sulfonyl fluoride) was effective. Thus, when MM6 cells, undifferentiated or differentiating with or without LPS or zymosan, were treated with AEBSF (150 to 250 μM for the final 18 h of the 96-h differentiation period), Dicer proteolysis was blocked. Both ∼170- and ∼50-kDa fragments were practically absent, with a concomitant increase of full-length Dicer (Fig. 5A). When AEBSF was added just before cell lysis or in the lysis buffer, there was no effect (SI Appendix, Fig. S8). Also, qPCR analysis showed no effect of AEBSF on Dicer mRNA expression levels in MM6 cells. These results suggest a serine protease-specific cleavage of Dicer in MM6 cells.
Fig. 5.
Serine protease inhibitor AEBSF (4-[2-Aminoethyl] benzenesulfonyl fluoride hydrochloride) inhibits proteolytic cleavage of Dicer. (A) MM6 cells (undifferentiated) were treated with AEBSF (250 μM) during final 18 h of 96 h culture. Cells differentiated with TGF-β + 1,25diOHvitD3 (VD3) were treated with AEBSF (200 μM) during final 18 h of 96 h differentiation. Cells differentiated with TGF-β, 1,25diOHvitD3, and zymosan/LPS were treated with AEBSF (150 μM) during final 18 h of 96 h differentiation. Whole-cell lysates (∼50 µg protein) were analyzed by Western blot using Dicer Ab 1701 to 1912. The membrane was reblotted with Dicer Ab 600 to 650. Similar results were obtained in three additional experiments. (B) Freshly isolated human monocytes were cultured overnight without or with AEBSF (250 μM) for 18 h. Whole-cell lysates were analyzed by Western blot using Dicer Ab 1701 to 1912. The membrane was reblotted with Dicer Ab 600 to 650. Similar results were obtained with cells from eight additional donors.
We also evaluated the effect of AEBSF in human blood monocytes. Monocytes, isolated from buffy coats of nine healthy donors, were cultured overnight with or without AEBSF. In untreated control cells, full-length Dicer was absent, but a clear band corresponding to the ∼50-kDa fragment was observed also in these primary cells. Treatment of monocytes with AEBSF (250 μM) resulted in strong up-regulation of full-length Dicer, with reduction of the ∼50-kDa fragment (Fig. 5B). Together, these results indicate a serine protease-specific cleavage of Dicer in monocytic cells.
Up-Regulation of miRNAs in Differentiating MM6 Cells.
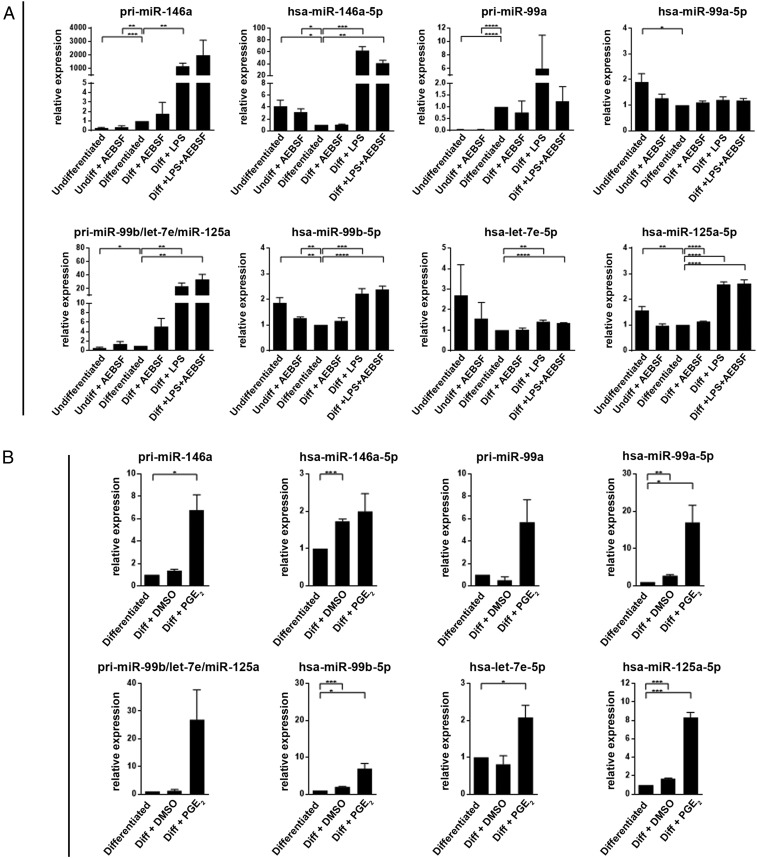
Stimulation of TLRs in monocytic cells leads to up-regulation of certain miRNAs (4, 21). This was confirmed also for MM6 cells differentiated in presence of LPS (TLR4) or zymosan (TLR2) for 96 h. qPCR analysis was performed for seven miRNAs, selected from Affymetrix screening. The results were quite similar for LPS and zymosan; the strongest relative increase (about 400-fold) was observed for miR-146a-5p. Also miR-146b-5p and miR-21-5p were up-regulated, 20- to 40-fold (SI Appendix, Fig. S9). Three clustered miRs (miR-99b-5p, let-7e-5p, and miR-125a-5p) were up-regulated, 40- to 60-fold for miR-99b and miR-125a but less for the third cluster member let-7e (about 15-fold). miR-155-5p was abundant in undifferentiated MM6 cells and increased only twofold. This may be related to the leukemic origin of this cell line; high levels of miR-155 have been reported in B-cell lymphomas and other malignancies (31).
In subsequent experiments, LPS was added to differentiating cells at time point 72 h (present during the final 24 h of the 96-h differentiation period). This late addition of LPS had a less prominent effect on the miRs; again the strongest relative increase (about 60-fold) was observed for miR-146a-5p (Fig. 6A). The corresponding pri-miR increased about 1,000-fold. The cluster miR-99b-5p/let-7e-5p/miR-125a-5p was modestly up-regulated (1.5- to 3-fold), while the corresponding pri-miR increased 20-fold. In these experiments we also treated MM6 cells with AEBSF. qPCR analysis showed quite constant levels for five miRs in the AEBSF-treated samples (Fig. 6A), and also Affymetrix screening did not suggest a generally increased expression of miRNAs.
Fig. 6.
qPCR analysis of miRNAs in MM6 cells. (A) Effects of LPS and AEBSF. MM6 cells were differentiated (Diff) with TGF-β + 1,25diOHvitD3 for 96 h. LPS (1 µg/mL) was added as indicated at time 72 h (present final 24 h). AEBSF (150 to 250 µM) was added at time 80 h (present final 16 h). Performed with the Qiagen miScript Primer Assay (Methods). The data are presented relative to cells differentiated with only TGF-β+ 1,25diOHvitD3, set as 1. For the pri-miRNA, GAPDH was reference gene. For the mature miRNA, U6 was reference gene. Mean ± SEM, n = 3 to 4, two-tailed unpaired t test, ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05. (B) Effect of PGE2. MM6 cells were differentiated with TGF-β + 1,25diOHvitD3 in presence of PGE2 (5 µM) or vehicle (DMSO) added at time 0 of the 96-h differentiation period. The data are presented relative to cells differentiated with only TGF-β + 1,25diOH, set as 1. For the pri-miRNA, GAPDH was reference gene. For the mature miRNA, U6 was reference gene. Mean ± SEM, n = 3, two-tailed unpaired t test, ***P < 0.001; **P < 0.01; *P < 0.05.
When MM6 cells were differentiated in presence of PGE2, the strongest relative up-regulation (17-fold; Fig. 6B) was observed for miR-99a-5p, recently described to target TNFα (32). The corresponding pri-miR increased about sixfold. Also miR-99b and miR-125a were clearly up-regulated (sevenfold to eightfold) but again less for the third cluster member let-7e (twofold). The pri-miR for this cluster was up-regulated about 27-fold. Interestingly, PGE2 had no effect on miR-146a-5p, although the pri-miR was increased (Fig. 6B). Conversely, LPS had no effect on miR-99a-5p, although the pri-miR was increased (Fig. 6A).
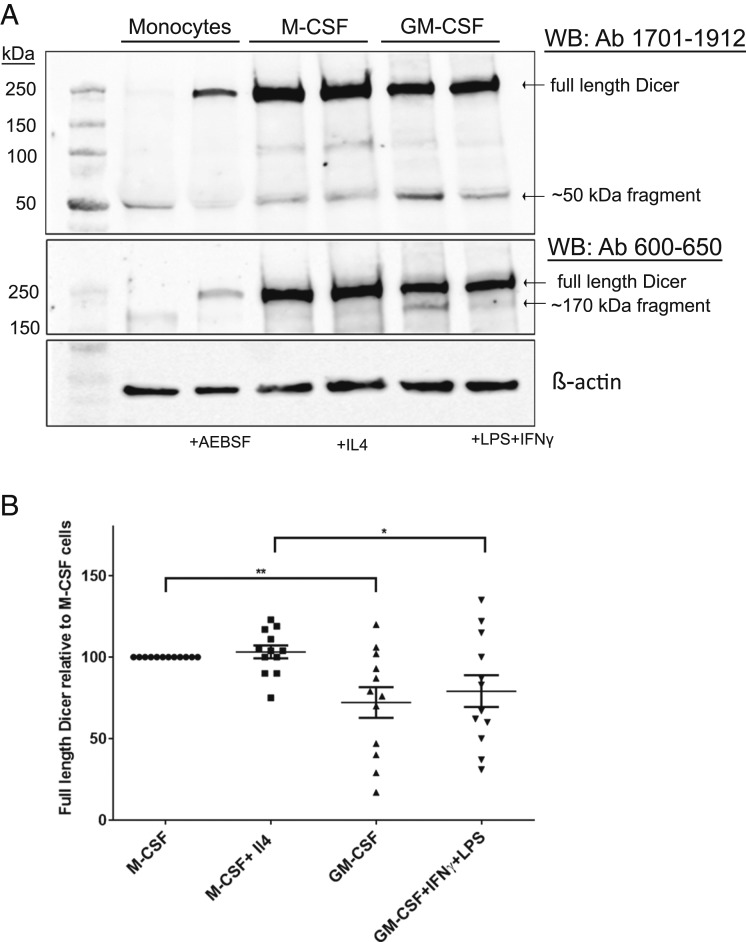
Differentiation of Monocytes in Presence of M-CSF and GM-CSF Up-Regulates Full-Length Dicer, with Relatively More Proteolysis in GM-CSF–Treated Cells.
We next evaluated up-regulation and cleavage pattern of Dicer in human macrophages polarized to different phenotypes. Human monocytes were cultured for 7 d in presence of M-CSF and activated with IL4 (final 24 h) to obtain M2 macrophages. Alternatively, monocytes were cultured for 7 d in presence of GM-CSF and activated (final 24 h) with IFNγ and LPS to obtain an M1 phenotype. Dicer proteins were evaluated in cells from 12 different donors. Our results show profound up-regulation of full-length Dicer in both differentiation regimens, with slightly higher up-regulation in M-CSF–treated macrophages (Fig. 7 A and B). The lower level of full-length Dicer in GM-CSF–treated macrophages was concomitant with increased levels of the fragments. Interestingly, treatment with the CSFs alone gave practically the same results as when also the activating factors (IL-4 or IFNγ + LPS) were added.
Fig. 7.
Dicer up-regulation during differentiation of monocytes to M1/M2 macrophages. (A) Human monocytes were isolated from buffy coats of healthy donors. M2- and M1-primed macrophages were prepared by 7-d differentiation culture with M-CSF or GM-CSF, respectively. Treatment with IL-4 (M2) or IFNγ plus LPS (M1) during the final 24 h leads to M2/M1 phenotypes. For comparison, monocytes were also cultured with or without AEBSF (250 μM) for 18 h. Whole-cell lysates (∼50 µg protein) were analyzed by Western blot (WB) using Dicer antibody (Ab) 1701 to 1912. The membrane was reblotted with Dicer Ab 600 to 650. (B) Relative expression levels of full-length Dicer in human macrophages polarized and activated to different M1/M2 phenotypes. Full-length Dicer in macrophage cell lysates was determined by Western blot. Band intensities were normalized to β-actin, before comparisons between phenotypes. Data from Fig. 7A and 11 additional donors. Mean ± SE, n = 12, two-tailed unpaired t test, **P < 0.01; *P < 0.05.
Discussion
Dicer plays a critical role in generation of miRNAs. In macrophages, specific miRNAs regulate M1/M2 polarization and fine-tune expression of many proteins which are important for both proinflammatory and antiinflammatory macrophage responses (4, 20, 22, 23). The expression levels of enzymes in the miRNA processing machinery should be relevant for miRNA production. Here we describe a mode of Dicer regulation, based on inhibition of proteolysis. In monocytes from peripheral blood, as well as in undifferentiated Mono Mac 6 cells, full-length Dicer was absent. In these cells, only truncated forms were found; a C-terminal ∼50-kDa fragment predominated, and an ∼170-kDa fragment was less abundant. However, during differentiation to macrophages, full-length Dicer became the clearly dominating form. An inverse relationship was observed between full-length Dicer and the Dicer fragments. For example, nearly double amount of full-length Dicer appeared in MM6 cells differentiated in presence of zymosan compared to LPS. This correlated with increased amounts of cleaved fragments in the LPS-treated cells. Previous studies reported the absence of Dicer in human monocytic cell lines (THP1, U937, and U1) and in fresh monocytes from human blood (33). However, the mechanism for up-regulation of full-length Dicer in these cells has not been described.
The cleavage of Dicer, both in MM6 cells and in human blood monocytes, was completely inhibited by the Ser-protease inhibitor AEBSF. Inhibitors of other proteolytic enzymes previously shown to degrade Dicer (Introduction and Results) had no effect. These observations suggest the presence of a Ser-protease in monocytic cells which cleaves Dicer at one specific site, apparently within residues 1465 to 1480. The quite constant expression of Dicer mRNA in undifferentiated and differentiated MM6 cells suggest a constitutive expression and cleavage of Dicer protein in the undifferentiated monocytic cells and that full-length Dicer is up-regulated by inhibition or down-regulation of the Ser-protease during differentiation to macrophages. This mode of Dicer regulation may be specific to monocytic cells. A recent report suggested that degradation of Dicer in C57BL/6 mouse microglia involved JNK-mediated phosphorylation of a particular Ser residue (34). Interestingly, the corresponding residue in human Dicer is Ser-1470, within the apparent cleavage site observed here (1465 to 1480). The calculated molecular mass of Dicer sequence 1470 to 1922 is 51.4 kDa. Splice variants of Dicer have been described in cancer cells, encoding proteins of 93 and 113 kDa (35) different from the Dicer fragments found here in monocytic cells.
Intracellular serine protease activities in monocytes and macrophages have been described previously. A Ser-protease activity in human monocyte microsomes was responsible for cleavage of HIV reverse transcriptase, and this protease activity was down-regulated upon cell activation or differentiation (36). The p65 subunit of NFκB in myelo-monocytic cells was cleaved by a Ser-protease, and inhibition of this activity was implicated in HIV replication (37). In another study, Ser-protease activity in the crude membrane fractions from monocytic cells processed TNF-α precursor (26 kDa) to its active form (17 kDa) (38). A Ser-protease on the cytoplasmic surface of an organelle or vesicle from undifferentiated monocytes was responsible for cleavage of IFN regulatory factor 1 (IRF-1); inhibition of this activity is implicated in Mycobacterium tuberculosis infection (39). The Ser-protease activities which were down-regulated when monocytes were differentiated (36, 39) could be related to the Ser-protease cleaving Dicer. However, to our knowledge these Ser-proteases have not been cloned or otherwise identified.
Recombinant Dicer constructs containing both RNaseIII domains have been found to cleave pre-let-7a more rapidly compared to a dsRNA substrate. When the N-terminal helicase domain was deleted, the conversion of a dsRNA substrate became more efficient, while the effect with pre-let-7a as substrate was more subtle (40). We and others found that shorter C-terminal Dicer fragments containing only the RNaseIIIb domain digested let-7a pre-miRNA but to a variety of fragments rather than to mature let-7a (8, 9, 41). During this study we attempted to determine the enzymatic activity for a larger expressed 1465 to 1912 fragment containing the RNaseIIIb domain (corresponding to the ∼50-kDa fragment). However, no reproducible formation of 21- to 23-nt miRNA species was found for this fragment. In MM6 cells, the increased formation of miRNAs was connected with increased full-length Dicer and concomitant decreased amounts of the ∼50- and ∼170-kDa fragments. Thus, it appears that intact Dicer, rather than the fragments, was the active species in the increased cellular formation of miRNAs. However, it has been published that when N- and C-terminal parts of Dicer were expressed separately and mixed, the resulting complex could form 22-nt products (6). It may be speculated that Dicer fragments could combine also in cells, leading to activity producing miRNAs. Possibly, such combination of fragments could explain formation of miRNAs in monocytic cells lacking intact Dicer (33).
Full-length Dicer protein was strongly up-regulated when monocytic cells were subjected to differentiating factors, MM6 cells with TGF-β and 1,25diOHvitD3 plus LPS/zymosan or PGE2, and blood monocytes with M-CSF or GM-CSF. Thus, in addition to cAMP signal transduction pathways (downstream EP2/4), MAP kinases and NFkB (downstream TLR2/4, GM-CSF receptor) and Tyr phosphorylation (downstream M-CSF receptor) also are implicated. Dicer expression in GM-CSF–treated macrophages (M1 primed) was about 70% compared to M-CSF–treated cells (M2 primed). This may be related to the recent observation that deletion of Dicer in mouse macrophages impeded alternative activation (M2) and accelerated atherosclerosis. This effect, connected with promoted mitochondrial oxidative metabolism, was due to impaired formation of certain miRNAs and indicated an antiinflammatory role for Dicer (42). Also, a disease-promoting phenotype of alveolar macrophages from smokers was linked to down-regulation of several miRNAs due to impaired Dicer activity linked with SUMOylation (43). In a mouse model of Parkinson’s disease, activation of the inflammatory process conferred down-regulation of Dicer in microglia by a JNK-mediated pathway (34). Thus, decreased expression of Dicer in macrophages may be connected with a more proinflammatory phenotype.
Tumor-associated macrophages (TAM) are M2-like, and PGE2 acts as an immunosuppressive factor suggested to promote M2 phenotype of TAMs in cancer tissues (44). We found that differentiation of MM6 cells in presence of exogenous PGE2 up-regulated full-length Dicer, indicating a role for PGE2 in preventing proteolysis. Signaling via EP2/EP4 receptors and intracellular cAMP mediated this PGE2 effect, apparently leading to down-regulation of the unknown Ser-protease. This is reminiscent to the immunosuppressive and antiinflammatory effects of PGE2, also mediated through EP2/EP4 receptors, activating the cAMP/PKA/CREB pathway (45, 46). The effect of PGE2 to up-regulate full-length Dicer could have relevance for the tumor microenvironment. We speculate that up-regulation of full-length Dicer may contribute to promote the TAM phenotype. This is in line with the finding that conditional deletion of Dicer in mouse TAMs could reprogram the macrophages into an antitumor mode (M1-like phenotype), allowing an enhanced immunotherapy response and inhibition of tumor progression (47).
When differentiating MM6 cells were treated with zymosan/LPS or with PGE2, different miRNAs were produced. TLR stimulation of monocytic cells is well known to induce miR146a which down-regulates mRNAs for IRAK1 and TRAF6; this is an established regulation of the LPS response (48). This was found also for MM6 cells; differentiation in presence of zymosan or LPS strongly up-regulated miR-146a-5p. PGE2 induced miR-99a-5p and miR-125a-5p in MM6 cells. Effects of PGE2 in monocytic cell functions are mostly inhibitory (49); 25 y ago it was published that PGE2 inhibits LPS-induced formation of TNFα in murine peritoneal macrophages, by a mechanism involving IL-10 (50). Proinflammatory effects also have been observed; in mouse BMDMs and in human monocytes, PGE2 boosted LPS-induced IL-1β while production of TNFα was inhibited (51). It was recently published that miR-99a-5p targets TNFα in mouse BMDMs (32), and we found that PGE2 induced miR-99a-5p. This suggests a mechanism for the effect of PGE2 on TNFα in monocytic cells, in addition to the previously described involvement of protein kinase A anchoring protein 8 (52). Furthermore, it was recently published that miR-125a-5p and let-7e down-regulate TNFα (and other cytokines) in THP-1 cells (21), and we found here that PGE2 up-regulated the cluster miR-99b-5p/let-7e-5p/miR-125a-5p. This cluster was suggested to be part of the late-induced IL-10 effects functioning as negative regulator of the LPS response (21). Bacterial components such as LPS induce formation of PGE2 in monocytic cells (53); thus, it appears reasonable that this, in turn, could induce formation of miR-99a and miR-125a, contributing to negative feedback control of TNFα formation.
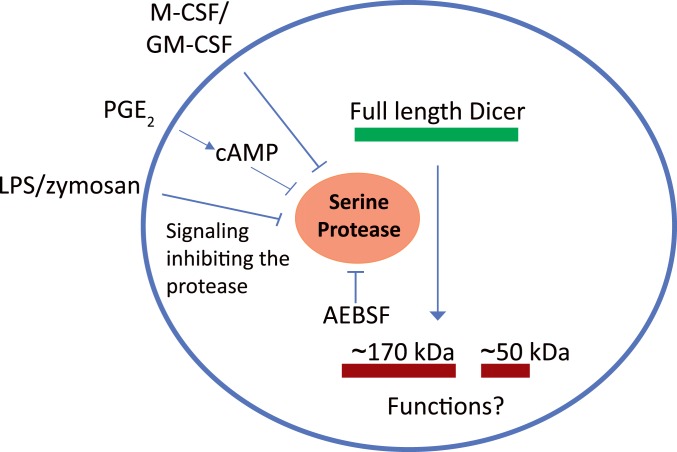
In view of the effect of AEBSF on full-length Dicer, we examined if AEBSF itself could lead to expression of miRNAs in MM6 cells. Affymetrix screening did not suggest a generally increased miRNA expression, and qPCR analysis showed quite constant levels for five miRs. However, when MM6 cells were treated with LPS or PGE2, full-length Dicer was up-regulated, and certain miRNAs were produced. The degree of miR up-regulation varied considerably, both between miRs and between stimuli (LPS or PGE2). Furthermore, the degree of up-regulation of pri-miRNAs was quite different compared to the corresponding mature miRNA. These observations together show that in addition to presence of active full-length Dicer, activation of additional mechanisms is required for cellular production of mature miRNAs. These additional mechanisms, including pri-miR transcription, processing by Drosha, export of pre-miR to cytosol, and miRNA stability, should lead to diverse formation of miRNAs, depending on the cell status and surrounding milieu. In a report discussing biosynthesis of miR-21 in macrophages, induction of primary transcripts was described as an immediate early response, followed by accumulation of mature functional miRNAs as a late response (54). These two processes, separated in space and time, would provide two layers of regulation, one at the transcriptional level and another at the miRNA processing level. Regulation of Dicer as described here, by inhibition of an apparent constitutive proteolysis during differentiation of monocytic cells to macrophages (Fig. 8), may be part of cell type-specific regulation at the miRNA processing level.
Fig. 8.
Schematic representation of Dicer proteolysis in monocytic cells. Dicer is cleaved by a constitutively activate serine protease (inhibited by AEBSF) in monocytes or undifferentiated MM6 cells. Dicer up-regulation occurs during differentiation to macrophages, by down-regulation of the serine protease activity. In MM6 cells when zymosan, LPS, or PGE2 were added together with TGF-β + 1,25diOHvitD3. In monocytes differentiated with M-CSF or GM-CSF.
Supplementary Material
Acknowledgments
This work was supported by Else Kröner-Fresenius-Stiftung (Else Kröner-Fresenius-Graduiertenkolleg) and by the Karolinska Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916249117/-/DCSupplemental.
References
- 1.Ha M., Kim V. N., Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Gregory R. I., Chendrimada T. P., Cooch N., Shiekhattar R., Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Paul P., et al. , Interplay between miRNAs and human diseases. J. Cell. Physiol. 233, 2007–2018 (2018). [DOI] [PubMed] [Google Scholar]
- 4.O’Connell R. M., Rao D. S., Baltimore D., microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 30, 295–312 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Provost P., et al. , Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 21, 5864–5874 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma E., Zhou K., Kidwell M. A., Doudna J. A., Coordinated activities of human dicer domains in regulatory RNA processing. J. Mol. Biol. 422, 466–476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z., et al. , Cryo-EM structure of human dicer and its complexes with a pre-miRNA substrate. Cell 173, 1191–1203.e12 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Takeshita D., et al. , Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer. J. Mol. Biol. 374, 106–120 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Du Z., Lee J. K., Tjhen R., Stroud R. M., James T. L., Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage. Proc. Natl. Acad. Sci. U.S.A. 105, 2391–2396 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurzynska-Kokorniak A., et al. , The many faces of Dicer: The complexity of the mechanisms regulating Dicer gene expression and enzyme activities. Nucleic Acids Res. 43, 4365–4380 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lugli G., Larson J., Martone M. E., Jones Y., Smalheiser N. R., Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J. Neurochem. 94, 896–905 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Ghodgaonkar M. M., et al. , Abrogation of DNA vector-based RNAi during apoptosis in mammalian cells due to caspase-mediated cleavage and inactivation of Dicer-1. Cell Death Differ. 16, 858–868 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa A., Shi Y., Kage-Nakadai E., Mitani S., Xue D., Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science 328, 327–334 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matskevich A. A., Moelling K., Stimuli-dependent cleavage of Dicer during apoptosis. Biochem. J. 412, 527–534 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Sawh A. N., Duchaine T. F., A truncated form of dicer tilts the balance of RNA interference pathways. Cell Rep. 4, 454–463 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Elgheznawy A., et al. , Dicer cleavage by calpain determines platelet microRNA levels and function in diabetes. Circ. Res. 117, 157–165 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Burger K., Gullerova M., Swiss army knives: Non-canonical functions of nuclear Drosha and Dicer. Nat. Rev. Mol. Cell Biol. 16, 417–430 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Johanson T. M., Lew A. M., Chong M. M., MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biol. 3, 130144 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Neill L. A., Sheedy F. J., McCoy C. E., MicroRNAs: The fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 11, 163–175 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Liu G., Abraham E., MicroRNAs in immune response and macrophage polarization. Arterioscler. Thromb. Vasc. Biol. 33, 170–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtale G., et al. , Multi-step regulation of the TLR4 pathway by the miR-125a∼99b∼let-7e cluster. Front. Immunol. 9, 2037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X. Q., et al. , Emerging role of microRNAs in regulating macrophage activation and polarization in immune response and inflammation. Immunology 148, 237–248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curtale G., MiRNAs at the crossroads between innate immunity and cancer: Focus on macrophages. Cells 7, E12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basavarajappa D., et al. , Roles of coactosin-like protein (CLP) and 5-lipoxygenase-activating protein (FLAP) in cellular leukotriene biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 111, 11371–11376 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray P. J., et al. , Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 41, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukic A., et al. , GM-CSF- and M-CSF-primed macrophages present similar resolving but distinct inflammatory lipid mediator signatures. FASEB J. 31, 4370–4381 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Díaz-Muñoz M. D., Osma-García I. C., Fresno M., Iñiguez M. A., Involvement of PGE2 and the cAMP signalling pathway in the up-regulation of COX-2 and mPGES-1 expression in LPS-activated macrophages. Biochem. J. 443, 451–461 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Thorén S., Jakobsson P. J., Coordinate up- and down-regulation of glutathione-dependent prostaglandin E synthase and cyclooxygenase-2 in A549 cells. Inhibition by NS-398 and leukotriene C4. Eur. J. Biochem. 267, 6428–6434 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Esser J., et al. , Zymosan suppresses leukotriene C4 synthase activity in differentiating monocytes: Antagonism by aspirin and protein kinase inhibitors. FASEB J. 25, 1417–1427 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Hirata T., Narumiya S., Prostanoids as regulators of innate and adaptive immunity. Adv. Immunol. 116, 143–174 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Mashima R., Physiological roles of miR-155. Immunology 145, 323–333 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaiswal A., Reddy S. S., Maurya M., Maurya P., Barthwal M. K., MicroRNA-99a mimics inhibit M1 macrophage phenotype and adipose tissue inflammation by targeting TNFα. Cell. Mol. Immunol. 16, 495–507 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coley W., et al. , Absence of DICER in monocytes and its regulation by HIV-1. J. Biol. Chem. 285, 31930–31943 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q., et al. , JNK-mediated microglial DICER degradation potentiates inflammatory responses to induce dopaminergic neuron loss. J. Neuroinflammation 15, 184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinkal G. W., Grelier G., Puisieux A., Moyret-Lalle C., Complexity in the regulation of Dicer expression: Dicer variant proteins are differentially expressed in epithelial and mesenchymal breast cancer cells and decreased during EMT. Br. J. Cancer 104, 387–388 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Château M. T., et al. , Human monocytes possess a serine protease activity capable of degrading HIV-1 reverse transcriptase in vitro. Biochem. Biophys. Res. Commun. 285, 863–872 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Franzoso G., et al. , A family of serine proteases expressed exclusively in myelo-monocytic cells specifically processes the nuclear factor-kappa B subunit p65 in vitro and may impair human immunodeficiency virus replication in these cells. J. Exp. Med. 180, 1445–1456 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robache-Gallea S., et al. , In vitro processing of human tumor necrosis factor-alpha. J. Biol. Chem. 270, 23688–23692 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Qiao Y., et al. , Host defense responses to infection by Mycobacterium tuberculosis. Induction of IRF-1 and a serine protease inhibitor. J. Biol. Chem. 277, 22377–22385 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Ma E., MacRae I. J., Kirsch J. F., Doudna J. A., Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 380, 237–243 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dincbas-Renqvist V., et al. , Human Dicer C-terminus functions as a 5-lipoxygenase binding domain. Biochim. Biophys. Acta 1789, 99–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei Y., et al. , Dicer in macrophages prevents atherosclerosis by promoting mitochondrial oxidative metabolism. Circulation 138, 2007–2020 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Gross T. J., et al. , A microRNA processing defect in smokers’ macrophages is linked to SUMOylation of the endonuclease DICER. J. Biol. Chem. 289, 12823–12834 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalinski P., Regulation of immune responses by prostaglandin E2. J. Immunol. 188, 21–28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luan B., et al. , CREB pathway links PGE2 signaling with macrophage polarization. Proc. Natl. Acad. Sci. U.S.A. 112, 15642–15647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaslona Z., Serezani C. H., Okunishi K., Aronoff D. M., Peters-Golden M., Prostaglandin E2 restrains macrophage maturation via E prostanoid receptor 2/protein kinase A signaling. Blood 119, 2358–2367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baer C., et al. , Suppression of microRNA activity amplifies IFN-γ-induced macrophage activation and promotes anti-tumour immunity. Nat. Cell Biol. 18, 790–802 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Mehta A., Baltimore D., MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 16, 279–294 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Aronoff D. M., Canetti C., Serezani C. H., Luo M., Peters-Golden M., Cutting edge: Macrophage inhibition by cyclic AMP (cAMP): Differential roles of protein kinase A and exchange protein directly activated by cAMP-1. J. Immunol. 174, 595–599 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Strassmann G., Patil-Koota V., Finkelman F., Fong M., Kambayashi T., Evidence for the involvement of interleukin 10 in the differential deactivation of murine peritoneal macrophages by prostaglandin E2. J. Exp. Med. 180, 2365–2370 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zasłona Z., et al. , The induction of pro-IL-1β by lipopolysaccharide requires endogenous prostaglandin E2 production. J. Immunol. 198, 3558–3564 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Kim S. H., et al. , Distinct protein kinase A anchoring proteins direct prostaglandin E2 modulation of Toll-like receptor signaling in alveolar macrophages. J. Biol. Chem. 286, 8875–8883 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hempel S. L., Monick M. M., Hunninghake G. W., Lipopolysaccharide induces prostaglandin H synthase-2 protein and mRNA in human alveolar macrophages and blood monocytes. J. Clin. Invest. 93, 391–396 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheedy F. J., Turning 21: Induction of miR-21 as a key switch in the inflammatory response. Front. Immunol. 6, 19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data, associated protocols, and materials are within the manuscript and its SI Appendix files.