Significance
Evolution of various wings plays an important role in the success of insects in environmental adaptation. During evolution, aphids deploy a wing dimorphism strategy to escape unfavorable conditions. This strategy also poses high risks to crops by distant transmission of viral diseases vectored by aphids. Thus, it is important to understand how aphids perceive environmental cues in triggering wing dimorphism and exploit specific approaches to target this process. Here, we report a microRNA (miR-9b)-mediated signal cascade that controls high-population-density–induced wing dimorphism in aphids. This finding highlights that small RNAs are important in regulating signal transduction of insect phenotypic plasticity when insects face environmental challenges, which may provide an alternative for the development of dispersal restriction-based pest control.
Keywords: microRNA, ABC transporter, trehalose, insulin, population density
Abstract
Wing dimorphism is a phenomenon of phenotypic plasticity in aphid dispersal. However, the signal transduction for perceiving environmental cues (e.g., crowding) and the regulation mechanism remain elusive. Here, we found that aci-miR-9b was the only down-regulated microRNA (miRNA) in both crowding-induced wing dimorphism and during wing development in the brown citrus aphid Aphis citricidus. We determined a targeted regulatory relationship between aci-miR-9b and an ABC transporter (AcABCG4). Inhibition of aci-miR-9b increased the proportion of winged offspring under normal conditions. Overexpression of aci-miR-9b resulted in decline of the proportion of winged offspring under crowding conditions. In addition, overexpression of aci-miR-9b also resulted in malformed wings during wing development. This role of aci-miR-9b mediating wing dimorphism and development was also confirmed in the pea aphid Acyrthosiphon pisum. The downstream action of aci-miR-9b-AcABCG4 was based on the interaction with the insulin and insulin-like signaling pathway. A model for aphid wing dimorphism and development was demonstrated as the following: maternal aphids experience crowding, which results in the decrease of aci-miR-9b. This is followed by the increase of ABCG4, which then activates the insulin and insulin-like signaling pathway, thereby causing a high proportion of winged offspring. Later, the same cascade, “miR-9b-ABCG4-insulin signaling,” is again involved in wing development. Taken together, our results reveal that a signal transduction cascade mediates both wing dimorphism and development in aphids via miRNA. These findings would be useful in developing potential strategies for blocking the aphid dispersal and reducing viral transmission.
Evolution of wing-facilitated flying ability shapes the environmental adaptation behavior of insects, and wing diversity is one of the most fascinating of phenomena. Among them, wing dimorphism functions as an energy trade-off between migration and reproduction as an adaptive switch to environmental changes. Flightless morphs (short-winged or wingless [apterous] morphs) allocate energy into offspring production while winged morphs typically produce fewer offspring but can migrate long distances in search of suitable habitats (1, 2). Factors contributing to insect wing dimorphism include population density, microorganisms, temperature, photoperiod, and host quality (1–3). Ecdysteroids, juvenile hormone, c-Jun NH2-terminal kinases, and insulin/insulin-like growth factor signaling (IIS) pathways contribute to the regulation of wing dimorphism. These pathways have been studied in aphids and planthoppers using RNA interference (RNAi)-based approaches (4–8). Two laterally transferred viral genes (Apns-1 and Apns-2) have been associated with crowding-induced wing dimorphism in the pea aphid Acyrthosiphon pisum (9). In aphids, wing dimorphism is the process that determines wing morphs (proportion), and then wing development is the process that develops wing buds (wing growth). Thus, wing dimorphism and wing development are indispensable continuous processes for aphid wing formation in order to successfully escape unfavorable conditions and to locate new habitats. Therefore, it is important to understand how aphids perceive environmental cues in triggering wing dimorphism and exploit specific approaches to target this process for aphid pest control. However, the signal transduction of this aspect still remains unclear.
microRNAs (miRNAs) are critical components of posttranscriptional gene expression regulation (10, 11). The abundance of miRNAs can be altered by environmental stressors such as insecticides, plant defenses, parasites, and extreme temperatures (10). Thus, miRNAs may facilitate the adaptation of insects to changing environmental conditions. We used the brown citrus aphid Aphis citricidus (SI Appendix, Fig. S1), the main vector of Citrus tristeza virus (one of the most widely distributed pathogens that causes destructive losses in the citrus industry worldwide), as a case study. Here, we report that the miR-9b miRNA plays a key role in the regulation of wing dimorphism and wing development in aphids. Our work promotes the understanding of molecular mechanisms for phenotypic plasticity in aphids as well as the regulation of insect adaptation to the changing environment. The finding of this study might also be helpful in identifying potential targets in pest control through blocking dispersal of flying insect pests.
Results
The aci-miR-9b Emerged as the Only Candidate miRNA Associated with Wing Dimorphism and Wing Development in Aphids.
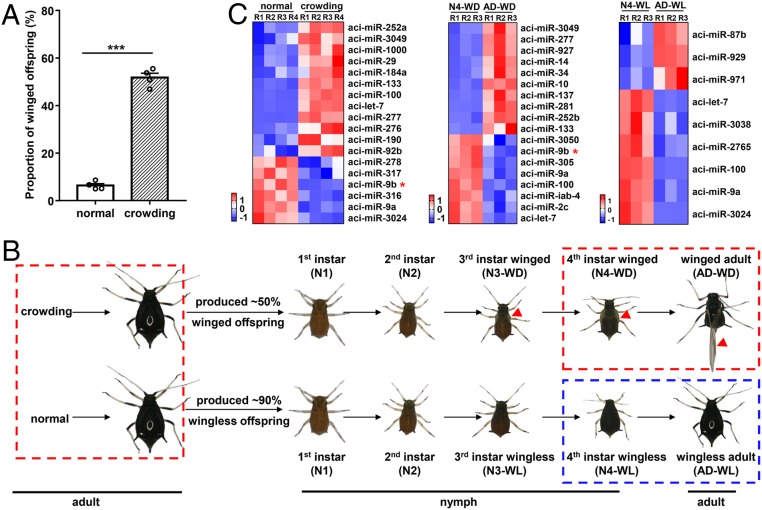
Transgenerational wing dimorphism was observed in A. citricidus in which crowding of the parent (wingless adult) had increased the winged offspring proportion to ∼50% (Fig. 1A). In addition, it was observed that the wing buds developed slowly in nymphal instars until the fully formed wings unfolded after adult emergence in A. citricidus (Fig. 1B). Thus, to identify the potential miRNAs mediating wing dimorphism and wing development, we performed small RNA sequencing for three comparisons: normal vs. crowding (for exploration of the potential miRNAs involved in wing dimorphism), fourth instar winged nymphs (N4-WD) vs. winged adults (AD-WD) (for exploration of the potential miRNAs involved in wing development), and fourth instar wingless nymphs (N4-WL) vs. wingless adults (AD-WL) (as a control for excluding the miRNAs involved in aphid development but not specifically for wing development) (Fig. 1B). Among these three comparisons, only one miRNA (aci-miR-9b) was down-regulated in both wing dimorphism (normal vs. crowding) and wing development (N4-WD vs. AD-WD) but had no change in the control (N4-WL vs. AD-WL) (Fig. 1C, SI Appendix, Fig. S2A, and Datasets S1 and S2). Additionally, the expression level of aci-miR-9b was lower in the winged morphs than in the wingless morphs (N4-WD vs. N4-WL and AD-WD vs. AD-WL) (SI Appendix, Fig. S3). Taken together, the RNA sequencing (RNA-seq) results suggested that aci-miR-9b might act as a key regulator in wing dimorphism and development in aphids.
Fig. 1.
RNA-seq reveals that aci-miR-9b is involved in wing dimorphism and wing development in A. citricidus. (A) The proportion of winged offspring under crowding. normal, 10 adults in a stem-leaf device; crowding, 80 adults in a stem-leaf device. Mean (±SE) is based on four biological replicates. The significant difference between crowding and normal is indicated by asterisks (***P < 0.001). (B) Schematic diagram of wing dimorphism and wing development and strategy for RNA-seq in A. citricidus. The red triangles represent the location of the wing (wing bud) in winged morphs. The comparisons inside the dashed box indicate the strategy of RNA-seq. Red-dashed boxes indicate the potential miRNAs mediating wing dimorphism (normal vs. crowding) and wing development (N4-WD vs. AD-WD) while the blue-dashed box is used to exclude the miRNAs in aphid development but not specifically for wing development (N4-WL vs. AD-WL). (C) The differentially expressed miRNAs among crowding vs. normal, AD-WD vs. N4-WD, and AD-WL vs. N4-WL. miRNAs with an adjusted P value < 0.01 and the absolute value of a fold change > 1.25 found by DESeq were assigned as differentially expressed. In each comparison, the relative expression of miRNAs was clustered based on z-scores from low to high value (with a scale from −1 to 1) among all of the biological replicates. Up-regulation is represented by red shading and down-regulation is represented by blue shading. R1 to R4 represent biological replicates; normal, wingless adult under normal condition; crowding, wingless adult under crowding; N1, first instar nymph; N2, second instar nymph; N3-WD, third instar winged nymph; N3-WL, third instar wingless nymph; N4-WD, fourth instar winged nymph; N4-WL, fourth instar wingless nymph; AD-WD, winged adult; AD-WL, wingless adult.
The aci-miR-9b Targets AcABCG4.
To explore the targets of aci-miR-9b, four miRNA-messenger RNA (mRNA) target prediction programs were performed (SI Appendix, Fig. S4A), and three targets (EVM0006751 [uncharacterized protein LOC111032307], EVM0005413 [phosphoenolpyruvate carboxykinase (GTP)-like], and EVM00015677 [ATP-binding cassette subfamily G member 4, AcABCG4]) were predicted to fit the “down-up” miRNA-target regulation pattern (SI Appendix, Figs. S2B and S4B and Datasets S3–S5). Of these three candidate targets, only the expression levels of AcABCG4 altered accordingly in the aphids treated with aci-miR-9b mimic or inhibitor (Fig. 2 and SI Appendix, Fig. S4 C and D), indicating that AcABCG4 is the target of aci-miR-9b.
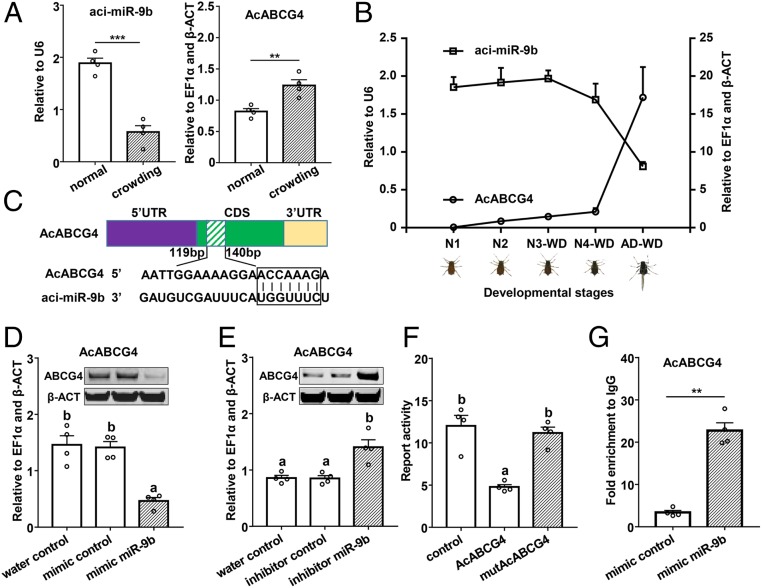
Fig. 2.
aci-miR-9b targets AcABCG4. (A) The expression profiles of aci-miR-9b and AcABCG4 for the wingless adult upon crowding after 24 h by RT-qPCR. normal, 10 adults in a stem-leaf device; crowding, 80 adults in a stem-leaf device. (B) aci-miR-9b and AcABCG4 present the opposite trend in different developmental stages during wing development. N1, first instar nymph; N2, second instar nymph; N3-WD, third instar winged nymph; N4-WD, fourth instar winged nymph; AD-WD, winged adult. (C) The putative aci-miR-9b–binding sites in AcABCG4 were predicted by miRanda, PITA, RIsearch, and RNAhybrid. (D) The mRNA level (RT-qPCR) of ABCG4 in N4-WD treated with miR-9b mimic after 24 h, and the protein level (Western blot) of ABCG4 after 48 h. (E) The mRNA level (RT-qPCR) of ABCG4 in a wingless adult under normal conditions treated with miR-9b mimic after 24 h and the protein level (Western blot) of ABCG4 after 48 h. (Inset) Protein level for ABCG4 after aphids treated with miR-9b mimic (D) or inhibitor (E) through Western blot. U6, small nuclear RNA U6; EF1α, elongation factor-1; β-ACT, beta-actin. (F) aci-miR-9b directly targets AcABCG4 in vitro by using a luciferase reporter assay. (G) aci-miR-9b targets AcABCG4 in vivo demonstrated by RNA immunoprecipitation assay. All of the mean (±SE) is based on four biological replicates. The significant differences among different treatments are indicated by lowercase letters above each bar (one-way ANOVA followed by the least significant difference (LSD) test, P < 0.05) for D, E, and F. The significant differences between treatment and control are indicated by asterisks in A and G (Student’s t test, **P < 0.01; ***P < 0.001).
To determine if aci-miR-9b specifically targets AcABCG4, we used several experimental approaches as follows: 1) aci-miR-9b was observed to have an opposite expression trend with AcABCG4 under crowding treatment (Fig. 2A) and during wing development by quantitative reverse transcription PCR (RT-qPCR) (Fig. 2B). The coding sequencing (CDS) of AcABCG4 exhibited the potential target site of aci-miR-9b (Fig. 2C). 2) The protein level of AcABCG4 decreased or increased (based on Western blot analysis) after the aphids fed on the aci-miR-9b mimic or aci-miR-9b inhibitor, respectively (Fig. 2 D and E and SI Appendix, Fig. S5). 3) To validate the binding activity of aci-miR-9b to AcABCG4, a dual-luciferase reporter assay was performed by cloning an ∼400-bp CDS fragment of AcABCG4 containing the target sequences of the miRNA. The luciferase activities declined when the aci-miR-9b mimic and AcABCG4 target were cotransfected, while the reporter activity recovered when the construct containing the mutated AcABCG4 sequences (Fig. 2F). 4) An RNA immunoprecipitation assay showed that the abundance of AcABCG4 mRNA was 5.77-fold up-regulated when the aci-miR-9b mimic was supplied in the Ago-1 antibody-mediated RNA complex compared to an IgG control (Fig. 2G and SI Appendix, Fig. S6). These data showed that AcABCG4 was targeted by aci-miR-9b.
The aci-miR-9b Is Involved in Wing Dimorphism and Wing Development.
To determine the role of aci-miR-9b and its target in wing dimorphism and wing development, we combined different treatments by feeding aphids with the aci-miR-9b inhibitor dsAcABCG4 and the aci-miR-9b mimic (SI Appendix, Fig. S7).
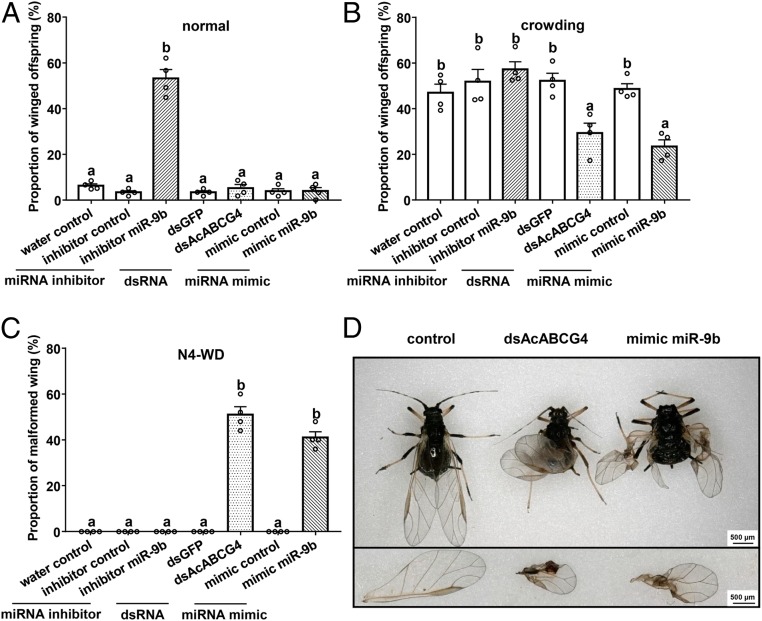
In the wing dimorphism experiment, two population-density—normal (10 adults per stem-leaf device) and crowding (80 adults per stem-leaf device)—were used (SI Appendix, Fig. S7A). Under normal conditions, 53% of offspring were winged morphs when aci-miR-9b was inhibited, while the proportion of winged morphs did not change (<7%) in the dsAcABCG4-fed group, the aci-miR-9b mimic-fed group, and the controls (Fig. 3A and SI Appendix, Figs. S8 and S9). However, under crowding, the proportions of winged offspring in the dsAcABCG4-fed group (29%) and the aci-miR-9b mimic-fed group (23%) were significantly lower than those in the controls (50% in average) and the aci-miR-9b inhibitor-fed group (57%) (Fig. 3B, SI Appendix, Fig. S8, and Dataset S6). The results indicated that aci-miR-9b and its target gene AcABCG4 are involved in crowding-induced wing dimorphism in A. citricidus.
Fig. 3.
aci-miR-9b mediates wing dimorphism and wing development in A. citricidus. (A) The proportion of winged offspring of A. citricidus after a wingless adult was treated by aci-miR-9b inhibitor, dsAcABCG4, and aci-miR-9b mimic under normal. (B) The proportion of winged offspring of A. citricidus after wingless adult treated by aci-miR-9b inhibitor, dsAcABCG4, and aci-miR-9b mimic under crowding. normal, 10 adults in a stem-leaf device; crowding, 80 adults in a stem-leaf device. (C) The proportion of A. citricidus adults with malformed wings after N4-WD treated by aci-miR-9b inhibitor, dsAcABCG4, and aci-miR-9b mimic. (D) Wing-defect phenotypes of A. citricidus under dsAcABCG4 and aci-miR-9b mimic treatment. All of the mean (±SE) is based on four biological replicates. The significant differences among different treatments are indicated by lowercase letters above each bar (one-way ANOVA followed by the LSD test, P < 0.05) for A, B, and C.
In the wing development experiment, 51% of adults had malformed wings after depletion of AcABCG4 (Fig. 3 C and D and SI Appendix, Fig. S8). A high percentage of aphids with malformed wings (41%) occurred after aci-miR-9b mimic feeding, while the malformed wing phenotype did not occur with inhibition of aci-miR-9b (Fig. 3 C and D and SI Appendix, Fig. S8). The aphid forewing size of dsAcABCG4 and aci-miR-9b mimic-fed groups declined compared to controls (declined 48 and 39%, respectively) (SI Appendix, Fig. S10). However, no other phenotypic differences (e.g., adult emergence and body size) were observed in A. citricidus upon miRNA inhibitor/double-stranded RNA (dsRNA)/miRNA mimic treatments (SI Appendix, Figs. S10 and S11). The results suggest that aci-miR-9b and its target gene AcABCG4 play a specific role in wing development.
Taken together, these results demonstrate that aci-miR-9b negatively regulates the expression of AcABCG4 to modulate wing plasticity in both wing dimorphism and wing development. This mechanism was also tested in A. pisum: inhibition of api-miR-9b increased the proportion of winged A. pisum morphs under normal conditions, whereas the silencing of ApABCG4 or the oversupply of api-miR-9b reduced the winged offspring proportion under crowding conditions. During wing development, ∼50% of the aphids showed malformed wings due to silencing of ApABCG4 or an oversupply of api-miR-9b (SI Appendix, Figs. S12 and S13). Furthermore, silencing of ABCG4 decreased the expression of 20 wing-patterning network genes, including decapentaplegic, notch, apterous, wingless, nubbin, and ultrablthorax (SI Appendix, Fig. S14), indicating that the canonical insect wing development pathway was also conserved in aphids (12–14).
Downstream Action of aci-miR-9b.
To clarify the downstream pathway of aci-miR-9b in aphids, we analyzed the tissue-specific expression pattern of aci-miR-9b and its target gene. The results showed that both aci-miR-9b and AcABCG4 are highly expressed in the fat body (SI Appendix, Fig. S15). It is possible that the fat body could regulate insulin-like peptide (ILP) secretion and influence IIS pathway activity in response to the nutritional environment (15). The IIS pathway is a conserved nutrient-sensing pathway that is influenced by carbohydrate content (16). Therefore, we hypothesized that the perception of environmental stressors (e.g., crowding) by aphids triggers the regulation of aci-miR-9b, which negatively regulates the expression of AcABCG4 and might consequently act on the IIS pathway in aphids, ultimately determining wing dimorphism and wing development.
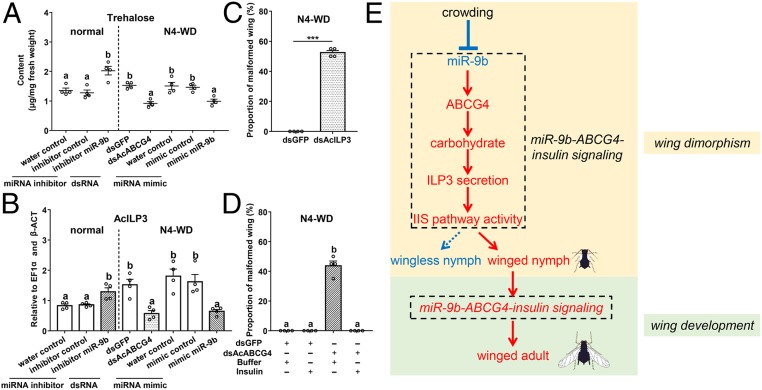
To test this hypothesis, we first measured the level of the main carbohydrate, trehalose. During wing dimorphism, inhibition of aci-miR-9b increased the level of trehalose under normal conditions (Fig. 4A), while during wing development, the silencing of AcABCG4 or feeding with the aci-miR-9b mimic decreased the level of trehalose (Fig. 4A). These data revealed the association of aci-miR-9b with trehalose level. We then tested whether aci-miR-9b/AcABCG4 interacts with the IIS pathway activity. The expression levels of IIS pathway genes were consistent with the trends of AcABCG4 expression dynamics (Fig. 4B and SI Appendix, Fig. S16). The upstream component of the IIS pathway is ILPs and three ILPs (AcILP1, AcILP3, and AcILP5) were identified based on the genome sequence of A. citricidus (SI Appendix, Fig. S17A). The tissue-specific expression patterns showed that AcILP3 and AcILP5 were highly expressed in the fat body, consistent with the expression of aci-miR-9b and AcABCG4, while AcILP1 was specifically expressed in the brain (SI Appendix, Fig. S17B). Silencing of AcILP3 increased the expression of AcABCG4, indicating that AcABCG4 and AcILP3 may be associated (SI Appendix, Fig. S17C). Silencing of AcILP3 resulted in 53% of aphids with malformed wings while silencing of AcILP1 resulted in 48% of the aphids with inhibited adult emergence (Fig. 4C and SI Appendix, Fig. S17 D and E). A rescue experiment was conducted using feeding with bovine insulin. The proportion of malformed wings was 44% under dsAcABCG4 combined with HEPES buffer treatment, while no adults had malformed wings when fed dsAcBCG4 combined with bovine insulin (Fig. 4D). These results indicate that insulin rescued the wing defects caused by the silencing of AcABCG4 and confirmed that the downstream action of aci-miR-9b/AcABCG4 is through the IIS pathway.
Fig. 4.
Mechanism of aci-miR-9b in the regulation of wing dimorphism and wing development in A. citricidus. (A) Trehalose content of AD-WL after aci-miR-9b inhibitor feeding under normal and N4-WD after dsAcABCG4 and aci-miR-9b mimic feeding. (B) The expression level of AcILP3 of AD-WL after aci-miR-9b inhibitor feeding and N4-WD after dsAcABCG4 and aci-miR-9b mimic feeding. (C) The proportion of A. citricidus adults with malformed wings under dsAcILP3 treatment. (D) Cofeeding bovine insulin rescued the wing defects caused by dsAcABCG4. A “+” indicates with the treatments, and a “−” indicates without the treatments. All of the mean (±SE) is based on four biological replicates. normal, wingless adult under normal N4-WD, fourth instar winged nymph. (E) Proposed model of miR-9b/ABCG4 in the regulation of wing dimorphism and wing development in aphids. Red color indicates increase and blue color indicates decrease. All of the mean (±SE) is based on four biological replicates. The significant differences among different treatments are indicated by lowercase letters above each bar (one-way ANOVA followed by the LSD test, P < 0.05) in A and B (data from normal and N4-WD were compared separately) and in D. The significant differences between treatment and control are indicated by asterisks (Student’s t test, **P < 0.01; ***P < 0.001) in C.
Discussion
Population density is a key environmental factor inducing winged morphs in aphid species (3, 17, 18). We found a key regulator, miR-9b, that controls crowding-induced wing dimorphism in aphids. miRNAs jointly regulate wing development in nonwing dimorphic insects (19–21). Based on a target-miRNA reverse prediction strategy, the insulin receptor gene of the IIS pathway was targeted by miR-34 in a planthopper (22). Here, based on an RNA-seq approach, we found that only the expression of aci-miR-9b was altered in both the wing dimorphism and the wing development period but not in wingless development from fourth instar nymphs to adults. The differential expression of miR-9b between winged and wingless morphs might reflect a maternal effect of the crowding. Indeed, via experimental manipulation of miR-9b, the wing dimorphism and development were altered. Inhibition of miR-9b increased the winged offspring proportion while overexpression of miR-9b could decline the high proportion of winged offspring induced by crowding. During wing development, the overexpression of miR-9b also led to adults with malformed wings. Upon miR-9b inhibitor exposure, the treated aphids could not produce more winged offspring than other treatments under crowding for two possible reasons: 1) The expression of miR-9b could not be completely depleted through feeding on the miR-9b inhibitor (the expression of aci-miR-9b declined 44 to 87%, SI Appendix, Fig. S8). A genome-editing approach may clarify this in future studies (23). 2) Alternatively, different genetic backgrounds of aphids may exhibit variation of winged offspring proportion in a range from “weak-inducible” to “high-inducible” genotypes after aphids perceiving environmental cues (e.g., crowding) (9). In the current study, the additional miR-9b inhibitor in crowding did not increase the proportion of winged offspring in A. citricidus. Since the proportions of winged offspring under the miR-9b inhibitor treatment alone or under crowding conditions were both ∼50%, this suggested that an ∼50% winged proportion might be the threshold for this genetic background, which confirms that miR-9b is the key regulator in crowding-induced wing dimorphism in A. citricidus.
We found that miR-9b down-regulates ABCG4 and also regulates the activity of the carbohydrate-stimulated IIS pathway. Phosphorylation by the corresponding ligands is an important indicator to reflect the activity of the IIS pathway. In addition to phosphorylation, the expression levels of downstream intracellular signaling can also reflect the diminished or enhanced IIS activities (24). The link between miR-9b and ABCG4 to IIS through ILP3 was based on the following: 1) The expression patterns of ILP3 were consistent with ABCG4 upon miR-9b inhibitor/dsABCG4/miR-9b mimic treatments. 2) Silencing of ILP3 resulted in malformed wings, which was the same phenotype caused by the overexpression of miR-9b or silencing of ABCG4. 3) Feeding bovine insulin to A. citricidus rescued the malformed wing phenotypes caused by dsABCG4 treatment. 4) The expression patterns of downstream components of IIS exhibited similar dynamics as ABCG4 upon miR-9b inhibitor/dsABCG4/miR-9b mimic. It is known that there is a transgenerational effect of wing dimorphism in several aphid species, which implies that the parent senses the environmental cues (e.g., crowding) and the proportion of winged offspring is altered (7, 18, 25), which was confirmed in A. citricidus and A. pisum in this study. In addition, we also observed that the expression of aci-miR-9b was reduced in nymphs (N3-WD and N3-WL) upon crowding (SI Appendix, Fig. S18), which indicated that the wing dimorphism could also occur throughout nymphal stages. A previous study showed that the interactions among the insulin signaling pathway, ecdysone signaling, and juvenile hormone signaling are involved in wing dimorphism (22). In this study, we found that this physiological process was regulated by the cascade “miR-9b-ABCG4-insulin signaling.” However, how this signal transduces to the offspring (embryo) in aphids, and the sensitive period of wing dimorphism in aphids still requires further study.
As primary-active proteins that bind and hydrolyze ATP, ABC transporters facilitate the transportation of many substrates such as amino acids, sugars, lipids, and toxic metabolites across membranes (26). However, beyond miR-9b in regulating ABCG4, it is not yet clear how ABCG4 participates in the carbohydrate-stimulated IIS pathway in aphids. In mouse pancreatic β cells (MIN6 cells), miR-463 targets ABCG4 and controls insulin secretion, which is based on the concentration of glucose (27). High carbohydrate content increased wing growth (4, 28) while the triglyceride content showed an opposite trend with trehalose level during wing dimorphism and development (SI Appendix, Fig. S19). This is also supported by transcriptome and proteome-based analysis, which revealed a high level of glycolysis/gluconeogenesis as well as pyruvate metabolism in winged morphs of A. citricidus and A. pisum (29, 30).
The insect fat body is necessary for energy storage and metabolism, and it may function as a nutrient sensor through regulation of ILP secretion (31). In the fruit fly Drosophila melanogaster and the yellow fever mosquito Aedes aegypti, only ILP6 was secreted by the fat body (32, 33). In the present study, we found that two ILPs (ILP3 and ILP5) were highly expressed in the fat body of A. citricidus, and ILP3 appeared to be specifically related to wing dimorphism.
In conclusion, we found a miRNA, aci-miR-9b, that is a critical regulator of wing dimorphism and wing development in aphids. For wing dimorphism, aphids perceive crowding which decreases the expression of miR-9b, thereby increasing the expression of its target gene ABCG4 and carbohydrate content, which subsequently activate the IIS pathway leading to an increase in the proportion of winged morphs. This cascade was termed as miR-9b-ABCG4-insulin signaling. For wing development, the same cascade, miR-9b-ABCG4-insulin signaling, was again involved in the transition from fourth instar winged nymph to winged adult. (Fig. 4E). The results increase our knowledge of miRNA as a sensory factor that can help insects adapt to a changing environment. The information may stimulate the development of methods for controlling the dispersal of aphid pests and virus transmission.
Materials and Methods
Insects, sample preparation for RNA-seq, transcriptome and small RNA sequencing, luciferase activity assay, RNA immunoprecipitation assay, dsRNA synthesis and RNAi assay, RT-qPCR, determination of trehalose content, Western blot, and statistics used in this study are included in SI Appendix.
See SI Appendix, Materials and Methods for details.
Data Availability.
Sequence data have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive (accession nos. PRJNA578247 and PRJNA576278).
Supplementary Material
Acknowledgments
We thank Dr. Guy Smagghe (Ghent University) for his critical comments on experimental design. This research was supported by the National Key R&D Program of China (Grant 2017YFD0200900); the China Postdoctoral Science Foundation (Grant 2018M640894); the 111 Project (Grant B18044); and the earmarked fund for the Modern Agro-industry (Citrus) Technology Research System of China (Grant CARS-26).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence data have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA578247 and PRJNA576278).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919204117/-/DCSupplemental.
References
- 1.Hayes A. M., Lavine M. D., Gotoh H., Lin X., Lavine L. C., Mechanisms regulating phenotypic plasticity in wing polyphenic insects. Adv. Insect Physiol. 56, 43–72 (2019). [Google Scholar]
- 2.Zhang C. X., Brisson J. A., Xu H. J., Molecular mechanisms of wing polymorphism in insects. Annu. Rev. Entomol. 64, 297–314 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Braendle C., Davis G. K., Brisson J. A., Stern D. L., Wing dimorphism in aphids. Heredity 97, 192–199 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Xu H. J., et al. , Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519, 464–467 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Lin X., et al. , JNK signaling mediates wing form polymorphism in brown planthoppers (Nilaparvata lugens). Insect Biochem. Mol. Biol. 73, 55–61 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Lin X., Yao Y., Wang B., Lavine M. D., Lavine L. C., FOXO links wing form polyphenism and wound healing in the brown planthopper, Nilaparvata lugens. Insect Biochem. Mol. Biol. 70, 24–31 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Vellichirammal N. N., Gupta P., Hall T. A., Brisson J. A., Ecdysone signaling underlies the pea aphid transgenerational wing polyphenism. Proc. Natl. Acad. Sci. U.S.A. 114, 1419–1423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J., Zhou Y., Li X., Cai W., Hua H., Silencing of juvenile hormone epoxide hydrolase gene (Nljheh) enhances short wing formation in a macropterous strain of the brown planthopper, Nilaparvata lugens. J. Insect Physiol. 102, 18–26 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Parker B. J., Brisson J. A., A laterally transferred viral gene modifies aphid wing plasticity. Curr. Biol. 29, 2098–2103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asgari S., MicroRNA functions in insects. Insect Biochem. Mol. Biol. 43, 388–397 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Lucas K., Raikhel A. S., Insect microRNAs: Biogenesis, expression profiling and biological functions. Insect Biochem. Mol. Biol. 43, 24–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brisson J. A., Ishikawa A., Miura T., Wing development genes of the pea aphid and differential gene expression between winged and unwinged morphs. Insect Mol. Biol. 19 (suppl. 2), 63–73 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Linz D. M., Tomoyasu Y., Dual evolutionary origin of insect wings supported by an investigation of the abdominal wing serial homologs in Tribolium. Proc. Natl. Acad. Sci. U.S.A. 115, E658–E667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X., et al. , Decapentaplegic function in wing vein development and wing morph transformation in brown planthopper, Nilaparvata lugens. Dev. Biol. 449, 143–150 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Koyama T., Mendes C. C., Mirth C. K., Mechanisms regulating nutrition-dependent developmental plasticity through organ-specific effects in insects. Front. Physiol. 4, 263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Q., Brown M. R., Signaling and function of insulin-like peptides in insects. Annu. Rev. Entomol. 51, 1–24 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Brisson J. A., Aphid wing dimorphisms: Linking environmental and genetic control of trait variation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 605–616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa A., Miura T., Transduction of high‐density signals across generations in aphid wing polyphenism. Physiol. Entomol. 38, 150–156 (2013). [Google Scholar]
- 19.Rubio M., Belles X., Subtle roles of microRNAs let-7, miR-100 and miR-125 on wing morphogenesis in hemimetabolan metamorphosis. J. Insect Physiol. 59, 1089–1094 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Aparicio R., Simoes Da Silva C. J., Busturia A., MicroRNA miR-7 contributes to the control of Drosophila wing growth. Dev. Dyn. 244, 21–30 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Ling L., et al. , MiR-2 family targets awd and fng to regulate wing morphogenesis in Bombyx mori. RNA Biol. 12, 742–748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye X., et al. , miR-34 modulates wing polyphenism in planthopper. PLoS Genet. 15, e1008235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Trionnaire G., et al. , An integrated protocol for targeted mutagenesis with CRISPR-Cas9 system in the pea aphid. Insect Biochem. Mol. Biol. 110, 34–44 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y., Huang G., Zhang W., Mutations in NlInR1 affect normal growth and lifespan in the brown planthopper Nilaparvata lugens. Insect Biochem. Mol. Biol. 115, 103246 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Ogawa K., Miura T., Aphid polyphenisms: Trans-generational developmental regulation through viviparity. Front. Physiol. 5, 1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dermauw W., Van Leeuwen T., The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance. Insect Biochem. Mol. Biol. 45, 89–110 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Hou X., Wu W., Yin B., Liu X., Ren F., MicroRNA-463-3p/ABCG4: A new axis in glucose-stimulated insulin secretion. Obesity 24, 2368–2376 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Lin X., Xu Y., Jiang J., Lavine M., Lavine L. C., Host quality induces phenotypic plasticity in a wing polyphenic insect. Proc. Natl. Acad. Sci. U.S.A. 115, 7563–7568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shang F., et al. , Differential expression of genes in the alate and apterous morphs of the brown citrus aphid, Toxoptera citricida. Sci. Rep. 6, 32099 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song L., Gao Y., Li J., Ban L., iTRAQ-based comparative proteomic analysis reveals molecular mechanisms underlying wing dimorphism of the pea aphid, Acyrthosiphon pisum. Front. Physiol. 9, 1016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arrese E. L., Soulages J. L., Insect fat body: Energy, metabolism, and regulation. Annu. Rev. Entomol. 55, 207–225 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamoto N., et al. , A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev. Cell 17, 885–891 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling L., Raikhel A. S., Serotonin signaling regulates insulin-like peptides for growth, reproduction, and metabolism in the disease vector Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 115, E9822–E9831 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive (accession nos. PRJNA578247 and PRJNA576278).