Significance
Dysregulation of cell death by necroptosis has been implicated in human inflammatory diseases. Phosphorylation of the MLKL pseudokinase, the terminal effector in the pathway, by RIPK3 is a hallmark of pathway activation, but additional checkpoints downstream in the pathway remain incompletely understood. Here, we generated synthetic protein ligands, monobodies, against MLKL to dissect key checkpoints in the pathway. We identified monobodies that inhibit necroptotic signaling by binding to an indispensable site on the α4 helix of human MLKL’s N-terminal four-helix bundle “killer” domain, which we propose blocks recruitment of essential adaptor(s). These data distinguish assembly of MLKL into higher-order complexes from membrane translocation as separate key checkpoints, and underscore the utility of monobodies as reagents to dissect signal transduction mechanisms.
Keywords: protein interactions, cell death, RIPK3, programmed necrosis, protein engineering
Abstract
The necroptosis cell death pathway has been implicated in host defense and in the pathology of inflammatory diseases. While phosphorylation of the necroptotic effector pseudokinase Mixed Lineage Kinase Domain-Like (MLKL) by the upstream protein kinase RIPK3 is a hallmark of pathway activation, the precise checkpoints in necroptosis signaling are still unclear. Here we have developed monobodies, synthetic binding proteins, that bind the N-terminal four-helix bundle (4HB) “killer” domain and neighboring first brace helix of human MLKL with nanomolar affinity. When expressed as genetically encoded reagents in cells, these monobodies potently block necroptotic cell death. However, they did not prevent MLKL recruitment to the “necrosome” and phosphorylation by RIPK3, nor the assembly of MLKL into oligomers, but did block MLKL translocation to membranes where activated MLKL normally disrupts membranes to kill cells. An X-ray crystal structure revealed a monobody-binding site centered on the α4 helix of the MLKL 4HB domain, which mutational analyses showed was crucial for reconstitution of necroptosis signaling. These data implicate the α4 helix of its 4HB domain as a crucial site for recruitment of adaptor proteins that mediate membrane translocation, distinct from known phospholipid binding sites.
Necroptosis is a caspase-independent cell death pathway that has been implicated in host defense to counter pathogens (1–8), and its dysregulation, in the pathology of inflammatory diseases (9–13). Necroptotic signaling can be initiated by death receptor ligands, including tumor necrosis factor (TNF), engaging their cognate receptors. Typically, the effector kinase, Receptor interacting protein kinase (RIPK)-1, is ubiquitylated by the Inhibitors of apoptosis proteins (IAPs) E3 ligases and engages in proinflammatory signaling downstream of TNF receptor activation. However, in scenarios when RIPK1 ubiquitylation is inhibited, death signaling is initiated. IAP depletion or inhibition (such as by compounds termed Smac mimetics or IAP antagonists) downstream of death receptor activation leads to activation of the proteolytic enzyme caspase-8 and cell death via the extrinsic apoptosis pathway. In cases where caspase-8 activity is compromised, a high molecular weight complex termed the “necrosome” is formed, and serves as a platform for initiation of necroptotic signaling. At the heart of this complex, the RIPK1 and RIPK3 kinases form a heterooligomeric complex in which RIPK3 is activated by autophosphorylation. Although the details of the next steps in necroptosis signaling are incompletely understood, necrosomal RIPK3 is thought to recruit and phosphorylate MLKL to promote its oligomerization, membrane translocation, and lytic permeabilization of the plasma membrane (8, 14–21).
Much of our current understanding of these necroptotic signaling events comes from studies performed with mouse cells (15, 22, 23). However, recent data indicate mechanistic differences between mouse and human necroptotic signaling, where phosphorylation of the MLKL pseudokinase domain by RIPK3 appears to serve different functions in MLKL activation (8, 16, 24). In mouse cells, RIPK3-mediated phosphorylation of the MLKL pseudokinase domain relieves repression of the N-terminal killer four-helix bundle (4HB) domain to induce cell death (15, 22, 25). In the human system, MLKL activation relies on recruitment to RIPK3 via its pseudokinase domain (17, 24), rather than simply phosphorylation by RIPK3. Consequently, the role of RIPK3-mediated phosphorylation in human cell necroptosis and the precise order of signaling events remains to be defined.
Here we report dissection of human necroptotic signaling using monobodies that we developed to selectively bind the killer 4HB domain of human MLKL. Monobodies are synthetic binding proteins constructed using a human fibronectin type III (FN3) domain (26, 27). Monobodies have strong tendency to bind to a functional surface within their targets even when no selection bias for such a functional site is employed during the development processes (27). Because monobodies do not have disulfide bonds, they can be expressed in a fully functional form in the reducing, intracellular environment. Such genetically encoded monobodies have been effective in discovering and validating functional sites in diverse signaling proteins (27, 28). Therefore, we envisioned that new monobodies would enable discovery of functionally important sites in MLKL and the dissection of key signaling events in the necroptosis pathway. Using these ligands, we dissected two previously inseparable necroptosis signaling steps, namely the assembly of MLKL into high molecular pronecroptotic complexes and the translocation of activated MLKL to membranes. The monobody binding site on the MLKL killer 4HB domain is distinct from the formerly described phospholipid binding interface, yet was found to be indispensable for necroptosis, thereby implicating the 4HB domain α4 helix in recruitment of essential adaptors that mediate membrane translocation.
Results
Monobodies that Bind the 4HB Domain of Human MLKL Block Necroptosis in Human Cells.
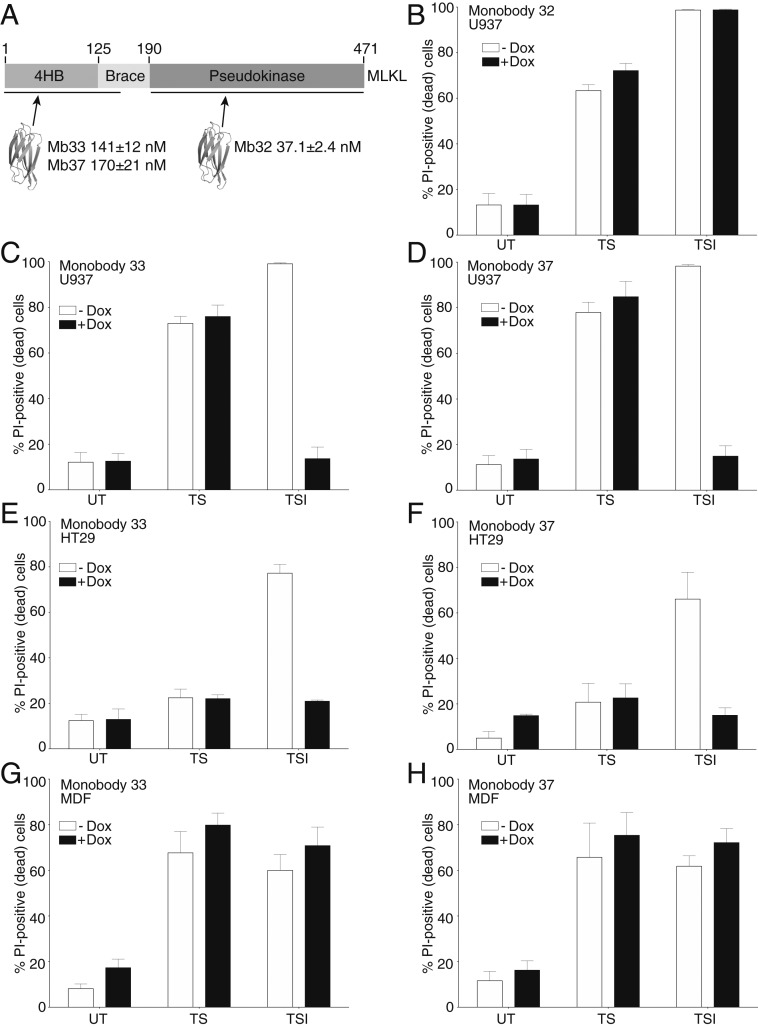
We developed monobodies for the component domains of human MLKL (Fig. 1A). The two monobodies, Mb(MLKL_33) and Mb(MLKL_37) (referred to as Mb33 and Mb37 hereafter for brevity), bound the human MLKL N-terminal 4HB domain and first brace helix (termed the N-terminal region or NTR hereafter) with Kd values of 141 ± 12 nM and 170 ± 21 nM, respectively (SI Appendix, Fig. S1 A and B). Competition binding experiments revealed that the inhibitory monobodies, Mb33 and Mb37, bound to overlapping sites on the human MLKL NTR (SI Appendix, Fig. S1C). These monobodies showed no detectable binding to the mouse MLKL NTR (SI Appendix, Fig. S1D), which was expected because the mouse and human MLKL 4HB domains share only 54% amino acid sequence identity. Another monobody, Mb(MLKL_32) (referred to as Mb32 hereafter), bound the pseudokinase domain with a Kd of 37.1 ± 2.4 nM (SI Appendix, Fig. S1 A and B).
Fig. 1.
Monobodies targeting the human MLKL 4HB killer domain prevent necroptosis. (A) Scheme showing the domain architecture of MLKL with the domains targeted by monobodies developed in this work. The corresponding KD values are shown as mean ± SD of triplicate experiments. The impact of doxycycline-induced expression of Mb32, Mb33, and Mb37 in human U937 cells (B–D) and Mb33 and Mb37 in HT29 (E and F) and MDFs (G and H) was evaluated in untreated (UT), apoptotic (TS), or necroptotic (TSI) conditions. Cell death was measured by PI uptake and flow cytometry; death data represent mean ± SEM of three independent assays.
To dissect necroptosis signaling, we stably introduced monobodies under a doxycycline (dox)-inducible promoter into cells typically used to study necroptosis signaling: the human histiocytic lymphoma line U937, the human colon cancer line HT29, and mouse dermal fibroblast (MDF) cells (Fig. 1 B–H). The monobody constructs bear N-terminal FLAG and C-terminal GFP tags for ease of detection (SI Appendix, Fig. S1F). We then tested the effects of dox-induced monobody expression on necroptosis induced by the “TSI stimulus” [with TNF, a Smac mimetic IAP antagonist (compound A) and pan-Caspase inhibitor (IDN-6556)] and on apoptosis induced by the “TS stimulus” (with TNF and the Smac mimetic, compound A). While other death ligands, interferons, and Toll-like receptor ligands are known to induce extrinsic apoptosis and necroptosis, we used TNF in these experiments because it is the most widely used laboratory stimulus. We observed that the monobodies that bind the human MLKL NTR, Mb33 and Mb37, potently inhibited necroptosis (induced by TSI) in human HT29 and U937 cells, while not impacting apoptosis (TS). This is consistent with these monobodies acting upon MLKL, the terminal effector in the necroptosis pathway, which has no role in apoptotic signaling. As expected from the absence of binding of Mb33 and Mb37 to recombinant mouse MLKL (SI Appendix, Fig. S1D), these monobodies inhibited neither mode of death in mouse cells. Contrary to Mb33 and Mb37, Mb32, which targets the human MLKL pseudokinase domain, did not block necroptosis in either human or mouse cells, and was thus used as a negative control in subsequent studies.
Inhibitory Monobodies Block MLKL Membrane Translocation, but Not RIPK3-Mediated MLKL Phosphorylation or Oligomerization.
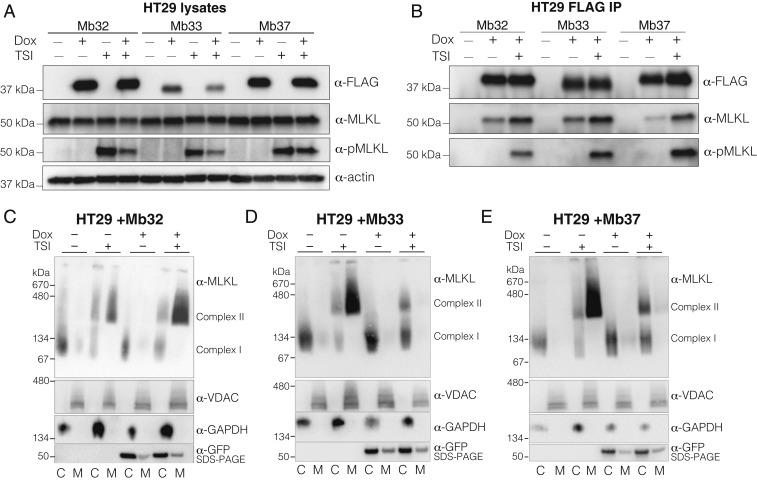
We next sought to define which step(s) of the necroptosis signaling pathway were inhibited by Mb33 and Mb37. First, we examined whether the binding of Mb33 and Mb37 to MLKL blocks interaction with the upstream regulator, RIPK3. By Western blot, the MLKL expression level or phosphorylation was not markedly impacted by expression of the inhibitory monobodies, Mb33 and Mb37, or the negative control, Mb32, in HT29 cells following treatment with the necroptotic stimulus TSI (Fig. 2A). We further examined whether Mb33 and Mb37 exerted their inhibitory effects on necroptosis via interaction with MLKL. Immunoprecipitation of Mb32, Mb33, and Mb37 from HT29 lysates validated their interaction with MLKL in Western blots (Fig. 2B). This interaction was enhanced upon treatment with the necroptosis stimulus TSI, indicating that phosphorylated and/or oligomeric MLKL is more readily accessed by all three monobodies. In parallel, we examined the repertoire of Mb33 and Mb37 binding partners by mass spectrometry. MLKL was the protein most significantly enriched by Mb33 and Mb37 pulldown (SI Appendix, Fig. S2), and we observed known interactors from the necrosome, RIPK3, RIPK1, Caspase-8, and FADD, to be coenriched with MLKL, albeit to a lesser extent than MLKL itself. Notably, the other proteins enriched by each monobody have not been implicated in necroptosis signaling and these interactomes differ between the two monobodies. These data indicate that, except for interaction with MLKL, Mb33 and Mb37 have distinct binding repertoires. Together with their specificity for human MLKL over mouse MLKL, this supports the notion that it is the direct interaction of Mb33 and Mb37 with human MLKL that negates necroptosis signaling. Crucially, none of the tested monobodies block MLKL recruitment to necrosomal RIPK3 or RIPK3-mediated MLKL phosphorylation, which in turn suggests that Mb33 and Mb37 inhibit a step downstream of these events.
Fig. 2.
Mb33 and Mb37 inhibit necroptosis by blocking MLKL membrane translocation. Immunoblotting (A) and Mb immunoprecipitation (B) of lysates of HT29 cells with and without monobody expression. Mb33 and Mb37 did not prevent MLKL phosphorylation by RIPK3 following TSI stimulation. Expression of Mb32 (C), Mb33 (D), or Mb37 (E) in HT29 cells was induced by doxycycline (dox) treatment. Assembly of MLKL into higher-order species (“complex II”) and membrane translocation were assessed by blue-native PAGE ±7.5 h TSI treatment. Separation into cytoplasmic (“C”) and membrane (“M”) fraction was validated by blotting BN-PAGE for VDAC1 (membrane) and GAPDH (cytoplasmic). Expression of monobodies was verified by reducing SDS-PAGE blots for GFP. All images and blots are representative of two independent experiments.
To examine whether the monobodies prevent pMLKL oligomerization and influence MLKL cellular localization, we fractionated untreated or TSI-stimulated HT29 cells into crude membrane and cytoplasmic fractions and examined MLKL assembly into high molecular weight complexes [previously designated “complex II” (22)] by blue-native PAGE. In the absence of the expression of monobodies, TSI stimulation resulted in high molecular weight MLKL complexes, which were present in the membrane fractions (Fig. 2 B–D). Expression of the noninhibitory, pseudokinase domain-binding monobody, Mb32, did not prevent higher-order MLKL complex formation and membrane translocation (Fig. 2B), as expected. In contrast, expression of either of the inhibitory monobodies, Mb33 (Fig. 2C) or Mb37 (Fig. 2D), prevented MLKL translocation to membranes, but did not prevent assembly of high molecular MLKL complexes, which were retained in the cytoplasmic fraction. These data illustrate that inhibitory monobodies do not prevent the transition of the dormant, basal cytoplasmic form into higher-order, pronecroptotic oligomers. Instead, these inhibitory monobodies function by blocking translocation of higher-order MLKL assemblies to the plasma membrane, thus preventing membrane disruption that causes cell death. Consequently, using these monobodies, we have disentangled MLKL oligomerization and membrane translocation as distinct checkpoints downstream of MLKL phosphorylation in the necroptosis pathway.
Mb33 Binds the Human MLKL via an Epitope Centered on the α4 Helix of the 4HB Domain.
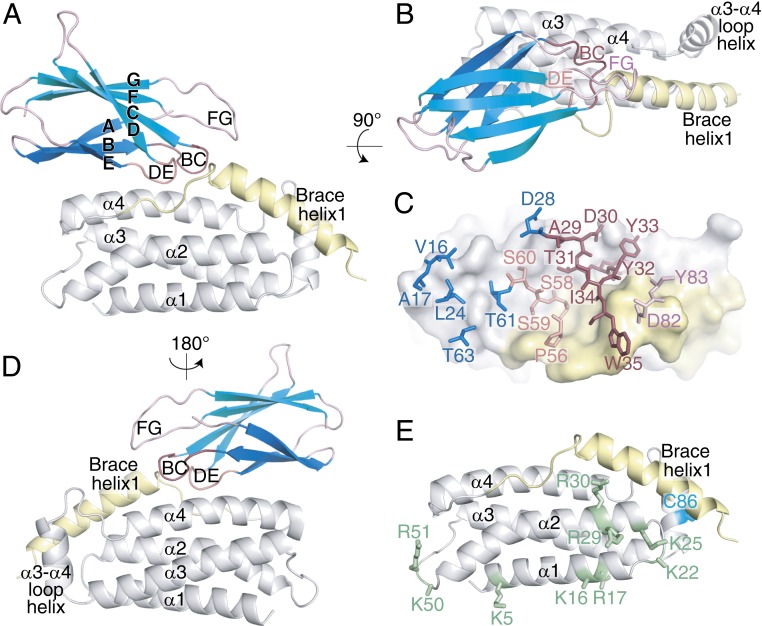
To define the mechanism of action of the inhibitory monobodies to block translocation of activated MLKL to the membrane, we crystallized the complex of Mb33 and the N-terminal region (NTR) of human MLKL encompassing the 4HB domain and the first of the two brace helices (residues 2 to 154) and determined its crystal structure at 2.5-Å resolution (Fig. 3A and Table 1) (29). As expected, the 4HB domain and first brace helix structure closely resembles the structures previously determined using NMR spectroscopy (rmsd 1.68 Å across Cα atoms to 2MSV; rmsd 2.00 Å across Cα atoms to 6D74) (30, 31) (SI Appendix, Fig. S3 A and B). The greatest deviations between the structure herein and the NMR structures were observed for the loops that connect helices and the disposition of the brace helix, which forms part of the Mb33 binding interface. The monobody binds atop the α4 helix, the N-terminal portion of the adjacent first brace helix, and the intervening loop (Fig. 3 A–C). The monobody and MLKL contribute ∼880 and 850 Å2 of solvent-accessible surface areas, respectively, to the interface. Many regions of the monobody contact MLKL, including the BC, DE, and FG loops that were diversified in the library, as well as residues in the ABE sheet (Fig. 3 A–C and SI Appendix, Fig. S1A). Residues in each of these loops bind at the interface of the 4HB domain and the first brace helix (Fig. 3C). Interestingly, the interface by which the monobody engages the 4HB domain of MLKL does not occlude the α3–α4 loop residue, C86, the target for covalent binding by the human MLKL inhibitor necrosulfonamide (NSA) (17) (Fig. 3E). Indeed, modification of recombinant human MLKL NTR with NSA only modestly decreased binding to Mb33 and Mb37 (SI Appendix, Fig. S1E). While there is unlikely to be substantial overlap in the footprint of NSA and Mb binding sites within the NTR, we cannot eliminate the possibility that NSA and Mb33/Mb37 prevent human MLKL binding to a common protein partner that facilitates membrane translocation. The MLKL interface bound by Mb33 also resides on the face opposite from sites of phospholipid and inositol phosphate binding previously identified using NMR spectroscopy: the α1–α2 helix junction; and a second site centered on the N terminus of the α1 helix and the adjacent α2–α3 helix loop (30–33) (Fig. 3E). Consequently, the monobodies inhibited membrane localization of MLKL via a mechanism that is not attributable to occlusion of essential membrane binding residues, and thus they have revealed important functions mediated by the α4 helix.
Fig. 3.
The inhibitory monobody Mb33 blocks necroptosis by binding the human MLKL 4HB domain α4 helix and first brace helix. (A, B, and D) Transverse views of the Mb33:human MLKL 4HB domain-first brace helix cocrystal structure. Mb33 (β-sheets shown in teal) binds atop the 4HB domain (gray) α4 helix and the N-terminal portion of brace helix 1 (yellow). (C) Mb33 residues in FG (pink), BC (raspberry), and DE (salmon) loop and β-sheet (blue) located within 4.5 Å of the MLKL 4HB-brace helix (gray and yellow) are shown as sticks. (E) Residues previously implicated in phospholipid and inositol phosphate binding (green sticks) and the target of necrosulfonamide modification (C86; cyan sticks) are located on the opposing face to the monobody interaction interface (compare with A).
Table 1.
X-ray crystallography data collection and refinement statistics
| Structural parameters | Mb33:human MLKL (2-154) |
| Wavelength | 0.9537 |
| Resolution range | 44.76–2.5 (2.59–2.5) |
| Space group | P 21 21 21 |
| Unit cell dimensions | 58.29 59.42 68.04 90 90 90 |
| Total reflections | 46172 (4177) |
| Unique reflections | 8552 (820) |
| Multiplicity | 5.4 (5.1) |
| Completeness, % | 99.53 (99.51) |
| Mean I/sigma(I) | 7.02 (1.21) |
| Wilson B-factor | 47.58 |
| R-meas | 0.1891 (1.744) |
| R-pim | 0.0804 (0.7699) |
| CC1/2 | 0.994 (0.47) |
| Reflections used in refinement | 8545 (820) |
| Reflections used for R-free | 468 (36) |
| R-work | 0.246 (0.367) |
| R-free | 0.296 (0.321) |
| RMS(bonds) | 0.001 |
| RMS(angles) | 0.37 |
| Ramachandran favored, % | 97.41 |
| Ramachandran allowed, % | 2.59 |
| Ramachandran outliers, % | 0 |
| Rotamer outliers, % | 1.03 |
| Clashscore | 2.22 |
| Average B-factor | 65.1 |
| Number of TLS groups | 10 |
Statistics for the highest-resolution shell are shown in parentheses.
Identification of Residues in the α4 Helix of the MLKL 4HB Domain that Are Essential for Membrane Translocation.
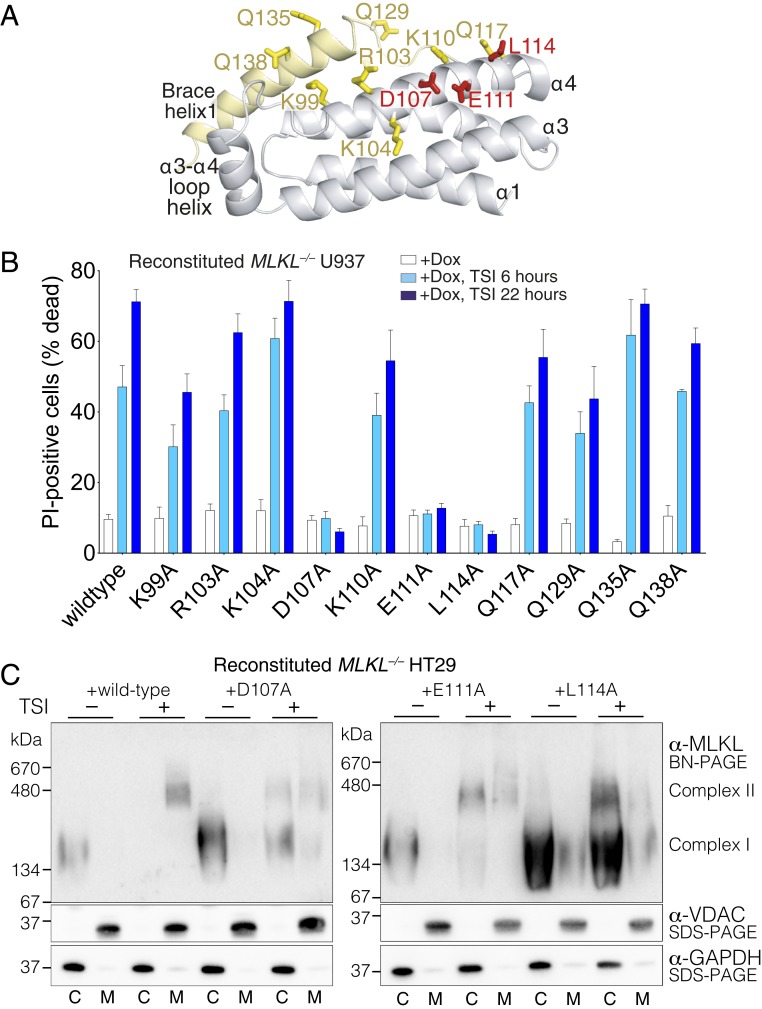
Based on our Mb33:human MLKL NTR structure, we tested the involvement of a total of 11 MLKL residues that are located within and adjacent to the Mb33 epitope (Fig. 4A). We mutated each residue to Ala in the context of full-length human MLKL and stably transduced each of these dox-inducible constructs (or the wild-type counterpart) into MLKL−/− U937 cells to examine their capacity to reconstitute necroptotic signaling. As previously reported (24), expression of wild-type human MLKL restored the cell death response following 6 or 22 h of treatment with a necroptotic stimulus (TSI; Fig. 4B). In contrast, alanine substitution of the MLKL α4 helix residues D107, E111, and L114, located at the core of the structural epitope bound by Mb33, completely abrogated necroptotic signaling (Fig. 4B), even though expression levels of these mutants were comparable to those of wild-type MLKL and the other mutants that did not affect signaling (SI Appendix, Fig. S1G). All other mutants were capable of reconstituting the pathway, with modest attenuation evident for the α4 helix mutant K99A and the α4 helix-brace loop mutant Q129A. D107, E111, and L114 reside at the heart of the structural epitope of Mb33 binding, suggesting that the inhibitory monobodies preclude yet-to-be-identified protein–protein interactions mediated by these residues to prevent MLKL membrane translocation and necroptotic cell death. Consistent with this idea, blue-native PAGE analyses of D107A, E111A, and L114A MLKL mutants revealed deficits in membrane translocation relative to their wild-type counterparts (Fig. 4C). Upon fractionation of cell lysates into cytosolic (C) and crude membrane (M) fractions, we observed that D107A, E111A, and L114A MLKL mutants could assemble into higher-order complexes, like their wild-type counterparts. However, unlike wild-type MLKL, these high molecular weight complexes were retained in the cytoplasmic fraction (Fig. 4C), thus accounting for their signaling deficiencies. The precise mechanism by which this functional epitope mediates membrane translocation, and which chaperones/coeffectors direct this process, remains of outstanding interest and the subject of future studies.
Fig. 4.
A functional site on the human MLKL 4HB domain α4 helix is crucial for MLKL to induce necroptotic death. (A) Cartoon of the 4HB domain-first brace helix structure rotated 30° about the x-axis relative to the depiction in Fig. 3D. MLKL residues proximal to Mb33 in the complex were selected for alanine substitution and are shown as sticks; essential residues for necroptotic signaling are shown as red sticks. (B) Effects of Ala substitutions of the indicated residues within full-length human MLKL on the capacity to reconstitute necroptotic signaling in MLKL−/− human U937 cells. Wild-type or mutant MLKL expression was induced with doxycycline, and death was measured by PI uptake and flow cytometry in the absence of stimulus or the presence of TSI stimulation for 6 or 22 h. Exogenes were expressed in 2 to 3 independent MLKL−/− U937 clones (one clone for Q135A) and assayed independently to a combined n = 3 to 10 for each MLKL variant. Data are plotted as mean ± SEM. (C) Expression of wild-type, D107A, E111A, and L114A human MLKL in MLKL−/− HT29 cells was induced by doxycycline (dox) treatment. Assembly of MLKL into higher-order species (“complex II”) and membrane translocation were assessed by blue-native PAGE ±7.5 h TSI treatment. Separation into cytoplasmic (“C”) and membrane (“M”) fractions was validated by blotting SDS-PAGE for VDAC1 (membrane) and GAPDH (cytoplasmic). All blots are representative of two independent experiments.
Discussion
Several steps in the pathway that culminate in MLKL-mediated membrane permeabilization have been posited (16, 21). However, in the absence of reagents to dissect the pathway, many of these checkpoints have remained hypothetical. Recent studies validated the recruitment of human MLKL to phosphorylated/activated necrosomal RIPK3 as a crucial checkpoint in MLKL activation (8, 24), an event that precedes RIPK3-mediated phosphorylation of MLKL. MLKL phosphorylation is widely considered a signature of necroptosis pathway activation (15–17, 23, 34), although the existence and identities of downstream checkpoints remain to be established.
Here, using monobody inhibitors of the human MLKL 4HB killer domain, we have divorced the assembly of high molecular weight MLKL complexes [termed complex II (22)] from the process of membrane translocation and permeabilization. Monobody inhibition of membrane translocation led to MLKL oligomer accumulation in the cytoplasm, and thus prevented MLKL from permeabilizing the plasma membrane to induce necroptotic cell death. Notably, because monobody binding centered on the 4HB and the first brace helix of human MLKL, we can exclude these regions of MLKL as important for oligomer formation. This view is consistent with earlier studies attributing the role of promoting oligomerization to the second helix in the brace region that connects 4HB and pseudokinase domains (32, 35). The crystal structure enabled us to identify three key MLKL residues, D107, E111, and L114, that are essential for necroptosis (Fig. 4). These residues reside on the α4 helix of the MLKL 4HB domain, on the face opposite from the two clusters of basic residues previously implicated in phospholipid and inositol phosphate binding (14, 30–33), indicating that they perform distinct functions from mediating membrane association and activation by inositol phosphates. Previous alanine scanning mutagenesis of the mouse MLKL NTR to introduce R105A/D106A, E109A/E110A, and LLLL112-115AAAA (SI Appendix, Fig. S3C), where underlined residues are the counterparts of human MLKL D107, E111, and L114, respectively, lost the capacity to constitutively kill MDFs (22). Introduction of R105A/R106A and E109A/E110A substitutions into full-length mouse MLKL prevented reconstitution of necroptosis signaling in Mlkl−/− MDFs, thus implicating this region in cell killing by mouse MLKL (18). However, whether this loss of function was attributable to loss of membrane localization was not examined. In light of the present study, these and our data suggest an evolutionarily conserved role for α4 helix residues in directing MLKL transport downstream of assembly into high molecular weight complexes.
Our previous structural mass spectrometry study of the conformational transition of human MLKL from inactive monomer to pronecroptotic tetramer identified E111 as proximal to the αC helix in the pseudokinase domain in the basal, monomeric state (24). Introduction of the D107A/E111A substitutions into human MLKL were found to ablate cell death (24), contrary to the hypothesis that these mutations would disrupt a charged pair between E111 in the 4HB domain α4 helix and the αC helix residues K255/K256 in the pseudokinase domain to promote transition to an oligomeric, killer conformer. The mechanism underlying this paradox was unclear. Here, our finding that inhibitory monobodies prevent MLKL translocation to membranes suggests that, while E111 of the 4HB domain mediates interactions with K255/K256 in the pseudokinase domain in the dormant monomer form of MLKL, its dominant role is to support downstream membrane translocation. Previous biophysical studies of the human MLKL NTR led to a model in which the first brace helix functions as a “plug” that binds the 4HB domain α4 helix to inhibit its interaction with phospholipids and liposome permeabilization in vitro (31, 32). However, this does not appear to be the case in the context of full-length human MLKL. Double alanine substitution of two key 4HB α4 helix residues, D107 and E111, in the context of recombinant full-length human MLKL did not impact phospholipid binding and liposome permeabilization activities in vitro (24), indicating that these residues are not directly involved in lipid interaction. These data negate the possibility that inhibitory monobodies prevent MLKL membrane translocation simply by locking the human MLKL NTR in an inhibited conformation or directly blocking MLKL–membrane interaction. Instead, they implicate inhibitory monobodies in blocking intermolecular interaction(s) critical for a downstream signaling event.
The observation that our inhibitory monobodies did not prevent MLKL assembly into oligomers further validates a model of the MLKL tetramer derived from solution SAXS and cross-linking/mass spectrometry data in which each 4HB domain is solvent-exposed in the oligomers (24). Such an assembly would avail the 4HB domain α4 helix to interaction with the yet-to-be-identified downstream coeffectors. To date, aside from the upstream regulator kinase RIPK3, few MLKL interactors have been identified. While the HSP90-Cdc37 cochaperones have been implicated in MLKL activation (25, 36, 37), the underlying mechanism is unclear, and such an interaction would be expected to be mediated via the MLKL pseudokinase domain, and not the 4HB domain, because HSP90 and Cdc37 are best characterized as cochaperones for protein kinases and pseudokinases (38). Similarly, the recent implication of TAM kinases in promotion of MLKL-mediated cell death is likely to be a distinct process because the reported substrate residue, Y376, is located in the pseudokinase domain, rather than the NTR. This study proposed that TAM kinase phosphorylation promotes MLKL oligomerization postmembrane translocation (39); however, our data support a reverse chronology, where MLKL oligomerization precedes membrane localization. Other coeffectors have been proposed to negate necroptosis. Recent studies have implicated the ESCRT-III complex in regulating MLKL levels at membranes, including in enabling activated MLKL to be jettisoned from the plasma membrane as “necroptotic bubbles” or vesicles, to negate necroptotic death (40–42). A complementary pathway involving flotillin-mediated endocytosis has also been described, which was proposed to similarly diminish membrane levels of phospho-MLKL to limit membrane damage via lysosomal degradation (43). Because both processes rely on lipid raft formation, and MLKL accumulation therein, it seems unlikely that these proteins would function as the MLKL membrane translocation chaperone. Future interactome analyses utilizing the monobodies and the MLKL mutants developed here may help identify MLKL interactors important for specific steps in necroptosis signaling.
The human MLKL α3–α4 loop helix harbors C86, which is the target for the covalent inhibitor of human MLKL, necrosulfonamide (NSA) (17). NSA modification did not prevent human MLKL phosphorylation or oligomerization, but did prevent membrane translocation (19, 44), which is reminiscent of the effects mediated by the inhibitory monobodies reported herein. However, the underlying mechanisms are likely to differ. C86 resides on the α3–α4 loop helix distal to the monobody-targeting α4 helix (Fig. 3E), and NSA modification did not substantively impact human MLKL NTR binding to Mb33 and Mb37, consistent with our monobodies and NSA occupying different sites on human MLKL, each of which is likely to engage distinct interaction partners. Thioredoxin-1 was recently proposed as one such regulator of human MLKL activation via C86 modification (45). Owing to the lack of conservation of this cysteine, this is unlikely to be a universal mechanism. In contrast, the importance of the 4HB domain α4 helix residues to both mouse and human MLKL necroptotic signaling suggests that interactions mediated by the MLKL 4HB domain α4 helix are likely to be broadly conserved mechanisms across species. Currently, the identity of the protein(s) involved in MLKL 4HB domain binding and delivery of MLKL to the plasma membrane to enable cell death to proceed remains to be discovered and is the subject of ongoing interest. The identification of membrane translocation as a regulated and essential checkpoint in necroptosis signaling distinct from assembly of MLKL into high molecular weight complexes opens new avenues to target this process therapeutically. The crystal structure of the inhibitory monobody may serve as a guide for designing such compounds.
Materials and Methods
Complete descriptions of experimental procedures are included in the SI Appendix. Brief summaries of procedures are included as follows.
Recombinant Protein Expression and Purification.
Recombinant human MLKL 4HB domain-first brace helix (residues 2 to 154) and monobodies were expressed and purified from Escherichia coli BL21-Codon Plus (DE3)-RIL using established methods (18, 24, 46). For monobody screening, bait proteins were expressed with a C-terminally fused flexible penta-Ser linker and AviTag (ASSSSSGLNDIFEAQKIEWHE) and enzymatically biotinylated using recombinant BirA. The human MLKL (2-154) AviTag fusion (synthesized by Bioneer) was expressed and purified from E. coli BL21-Codon Plus (DE3)-RIL, and pseudokinase domain (residues 190 to 471) and full length human MLKL were expressed and purified from Sf21 insect cells via the Bac-to-Bac system (Invitrogen) using established procedures (24, 48).
Monobody Development.
Phage-display library designs, library sorting using phage display and yeast display, and affinity measurement using yeast display flow cytometry assay were performed as described previously (48, 49). The amino acid sequences of the monobodies are shown in SI Appendix, Fig. S1.
Protein Crystallization and Structure Determination.
Recombinant human MLKL (2-154) and Mb33 were coeluted by Superdex-200 size exclusion chromatography and concentrated to 4.7 mg/mL. Crystals grown in 25% PEG-MME 550, 0.1 M MES, pH 6.5, 0.01 M zinc acetate at 20 °C were flash-cooled in liquid nitrogen, and X-ray diffraction data were collected at the Australian Synchrotron MX2 beamline (50). Data were indexed, integrated in XDS, and then merged and scaled in aimless (51, 52). Phases were solved by molecular replacement using a monobody structure (PDB ID code 6D0J) and the 4HB domain of mouse MLKL (PDB ID code 4BTF) as search models in phaser (53). Manual model building and phase refinement were performed using iterative real-space and reciprocal-space refinement in Coot and phenix.refine, respectively (54, 55). The protein interaction interfaces were analyzed using the PISA server (56).
Expression Constructs.
The monobody-encoding genes were cloned into a derivative of the doxycycline-inducible, puromycin-selectable vector pF TRE3G PGK puro (15, 18, 22) encoding an N-terminal FLAG and C-terminal GFP sequence (synthesized by ATUM). Mutations were introduced into a human MLKL DNA template (from DNA2.0) using oligonucleotide-directed overlap PCR or were synthesized by ATUM and subcloned into pF TRE3G PGK puro. Vector DNA was cotransfected into HEK293T cells with pVSVg and pCMV ΔR8.2 helper plasmids to generate lentiviral particles as described previously (15, 22).
Cell Death Assays.
The human histiocytic lymphoma U937 (and their MLKL−/− counterparts), human colorectal adenocarcinoma HT29, and MDF cell lines were cultured in human tonicity RPMI medium (in-house), DMEM, and DMEM (Gibco), respectively, supplemented with 8% vol/vol fetal calf serum (FCS; Sigma), with puromycin (5 μg/mL; StemCell Technologies) added for lines stably transduced with inducible MLKL or monobody constructs as described before (8, 24). Following 3 h induction of exogene expression by addition of doxycycline (20 ng/mL), cells were treated with TNF (100 ng/mL) and the Smac-mimetic compound A (500 nM; TS) to induce apoptosis or with TS in the presence of the pan-caspase inhibitor IDN-6556 (10 μM) to induce necroptosis. Cell death was quantified by propidium iodide (PI; 1 μg/mL) uptake using flow cytometry 24 h poststimulation as described previously (8, 24). Combined data from replicate experiments using 1 to 3 clonal cell lines are presented as mean ± SEM.
Western Blot and Blue Native PAGE.
Two times SDS Laemmli lysis buffer was added to cells, sonicated, boiled at 100 °C for 5 min, and then resolved by 4 to 15% Tris-Glycine gel (Bio-Rad). After transfer to PVDF, membranes were blocked with 5% skim milk and then probed with antibodies as indicated. For blue native PAGE, monobody expression was induced in wild-type HT29 cells with 10 ng/mL doxycycline for 3 h, then treated with TSI (7 h) or left untreated as indicated. Cells were fractionated into cytoplasmic and membrane fractions as previously described (18, 22). Fractions were resolved by 4 to 16% Bis-Tris Native PAGE gel (Thermo Fisher), then transferred to PVDF for Western blot analyses.
Reagents and Antibodies.
Antibodies and the dilutions used in this study are detailed in the SI Appendix. Recombinant hTNF-Fc, produced in-house, and the Smac mimetic, compound A, have been previously described (57, 58). The pan-caspase inhibitor IDN-6556/emricasan was provided by Tetralogic Pharmaceuticals.
Data Availability.
All reagents are available under material transfer agreement. All data, sequences, and protocols are available on request. The Mb33:human MLKL NTR structure atomic coordinates have been deposited in the Protein Data Bank (accession 6UX8).
Supplementary Material
Acknowledgments
We thank the Australian Synchrotron MX beamline staff for assistance with data collection and the CSIRO C3 Facility for assistance with crystallization. We are grateful to the National Health and Medical Research Council for fellowship (541951 to J.M.H.; 1079700 to P.E.C.; 1105754 and 1172929 to J.M.M.), project grant (1124735, 1124737), and infrastructure (9000587) support. We thank the Victorian State Government for Operational Infrastructure Support and acknowledge Australian Government Research Training Program Stipend PhD scholarship support for S.E.G. and K.A.D., and an Australian Institute of Nuclear Science and Engineering Postgraduate Research Award for K.A.D. This research was undertaken in part using the MX2 beamline at the Australian Synchrotron, part of the Australian Nuclear Science and Technology Organization, and made use of the Australian Cancer Research Foundation (ACRF) detector.
Footnotes
Competing interest statement: E.J.P., J.M.H., S.E.G., A.L.S., C.F., S.N.Y., P.E.C., and J.M.M. contribute to a project developing necroptosis inhibitors with Anaxis Pharma Pty Ltd. A.K. and S.K. are listed as inventors on issued and pending patents on the monobody technology filed by The University of Chicago (US Patent 9512199 B2 and related pending applications).
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates were deposited in the Protein Data Bank, https://www.rcsb.org (accession code 6UX8).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919960117/-/DCSupplemental.
References
- 1.Cho Y. S., et al. , Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137, 1112–1123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Upton J. W., Kaiser W. J., Mocarski E. S., Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7, 302–313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo H., et al. , Herpes simplex virus suppresses necroptosis in human cells. Cell Host Microbe 17, 243–251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitur K., et al. , Necroptosis promotes Staphylococcus aureus clearance by inhibiting excessive inflammatory signaling. Cell Rep. 16, 2219–2230 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearson J. S., et al. , EspL is a bacterial cysteine protease effector that cleaves RHIM proteins to block necroptosis and inflammation. Nat. Microbiol. 2, 16258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson J. S., Murphy J. M., Down the rabbit hole: Is necroptosis truly an innate response to infection? Cell. Microbiol. 19, e12750 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Nailwal H., Chan F. K., Necroptosis in anti-viral inflammation. Cell Death Differ. 26, 4–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrie E. J., et al. , Viral MLKL homologs subvert necroptotic cell death by sequestering cellular RIPK3. Cell rep. 28, 3309–3319.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Müller T., et al. , Necroptosis and ferroptosis are alternative cell death pathways that operate in acute kidney failure. Cell. Mol. Life Sci. 74, 3631–3645 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton K., et al. , RIPK3 deficiency or catalytically inactive RIPK1 provides greater benefit than MLKL deficiency in mouse models of inflammation and tissue injury. Cell Death Differ. 23, 1565–1576 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rickard J. A., et al. , TNFR1-dependent cell death drives inflammation in Sharpin-deficient mice. eLife 3, e03464 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rickard J. A., et al. , RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell 157, 1175–1188 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Hildebrand J. M., et al. , Missense mutations in the MLKL ‘brace’ region lead to lethal neonatal inflammation in mice and are present in high frequency in humans. bioRxiv:10.1101/628370 (21 May 2019).
- 14.Dondelinger Y., et al. , MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 7, 971–981 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Murphy J. M., et al. , The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 39, 443–453 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Petrie E. J., Czabotar P. E., Murphy J. M., The structural basis of necroptotic cell death signaling. Trends Biochem. Sci. 44, 53–63 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Sun L., et al. , Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Tanzer M. C., et al. , Evolutionary divergence of the necroptosis effector MLKL. Cell Death Differ. 23, 1185–1197 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H., et al. , Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell 54, 133–146 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Zhao J., et al. , Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc. Natl. Acad. Sci. U.S.A. 109, 5322–5327 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy J. M., The killer pseudokinase mixed lineage kinase domain-like protein (MLKL). Cold Spring Harb. Perspect. Biol., 10.1101/cshperspect.a036376 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hildebrand J. M., et al. , Activation of the pseudokinase MLKL unleashes the four-helix bundle domain to induce membrane localization and necroptotic cell death. Proc. Natl. Acad. Sci. U.S.A. 111, 15072–15077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanzer M. C., et al. , Necroptosis signalling is tuned by phosphorylation of MLKL residues outside the pseudokinase domain activation loop. Biochem. J. 471, 255–265 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Petrie E. J., et al. , Conformational switching of the pseudokinase domain promotes human MLKL tetramerization and cell death by necroptosis. Nat. Commun. 9, 2422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen A. V., et al. , HSP90 activity is required for MLKL oligomerisation and membrane translocation and the induction of necroptotic cell death. Cell Death Dis. 7, e2051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koide A., Bailey C. W., Huang X., Koide S., The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 284, 1141–1151 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Sha F., Salzman G., Gupta A., Koide S., Monobodies and other synthetic binding proteins for expanding protein science. Protein Sci. 26, 910–924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spencer-Smith R., et al. , Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol. 13, 62–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birkinshaw R. W., Petrie E. J., Czabotar P. E., Murphy J. M., Structure of monobody 33 MLKL N-terminal domain complex. RCSB Protein Data Bank. https://www.rcsb.org/structure/6ux8. Deposited 7 November 2019.
- 30.McNamara D. E., et al. , Direct activation of human MLKL by a select repertoire of inositol phosphate metabolites. Cell Chem. Biol. 26, 863–877.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su L., et al. , A plug release mechanism for membrane permeation by MLKL. Structure 22, 1489–1500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quarato G., et al. , Sequential engagement of distinct MLKL phosphatidylinositol-binding sites executes necroptosis. Mol. Cell 61, 589–601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dovey C. M., et al. , MLKL requires the inositol phosphate code to execute necroptosis. Mol. Cell 70, 936–948 e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez D. A., et al. , Characterization of RIPK3-mediated phosphorylation of the activation loop of MLKL during necroptosis. Cell Death Differ. 23, 76–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies K. A., et al. , The brace helices of MLKL mediate interdomain communication and oligomerisation to regulate cell death by necroptosis. Cell Death Differ. 25, 1567–1580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bigenzahn J. W., et al. , An inducible retroviral expression system for tandem affinity purification mass-spectrometry-based proteomics identifies mixed lineage kinase domain-like protein (MLKL) as an heat shock protein 90 (HSP90) client. Mol. Cell. Proteomics 15, 1139–1150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao X. M., et al. , Hsp90 modulates the stability of MLKL and is required for TNF-induced necroptosis. Cell Death Dis. 7, e2089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taipale M., et al. , Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150, 987–1001 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Najafov A., et al. , TAM kinases promote necroptosis by regulating oligomerization of MLKL. Mol. Cell 75, 457–468.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Gong Y. N., et al. , ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell 169, 286–300.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon S., Kovalenko A., Bogdanov K., Wallach D., MLKL, the protein that mediates necroptosis, also regulates endosomal trafficking and extracellular vesicle generation. Immunity 47, 51–65 e7 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Zargarian S., et al. , Phosphatidylserine externalization, “necroptotic bodies” release, and phagocytosis during necroptosis. PLoS Biol. 15, e2002711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan W., et al. , Flotillin-mediated endocytosis and ALIX-syntenin-1-mediated exocytosis protect the cell membrane from damage caused by necroptosis. Sci. Signal. 12, eaaw3423 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Murai S., et al. , A FRET biosensor for necroptosis uncovers two different modes of the release of DAMPs. Nat. Commun. 9, 4457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynoso E., et al. , Thioredoxin-1 actively maintains the pseudokinase MLKL in a reduced state to suppress disulfide bond-dependent MLKL polymer formation and necroptosis. J. Biol. Chem. 292, 17514–17524 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy J. M., et al. , Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL. Biochem. J. 457, 369–377 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Murphy J. M., et al. , A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem. J. 457, 323–334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koide A., Wojcik J., Gilbreth R. N., Hoey R. J., Koide S., Teaching an old scaffold new tricks: Monobodies constructed using alternative surfaces of the FN3 scaffold. J. Mol. Biol. 415, 393–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sha F., et al. , Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains. Proc. Natl. Acad. Sci. U.S.A. 110, 14924–14929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aragão D., et al. , MX2: A high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian Synchrotron. J. Synchrotron Radiat. 25, 885–891 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabsch W., Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afonine P. V., et al. , Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 66, 1153–1163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Vince J. E., et al. , IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 131, 682–693 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Bossen C., et al. , Interactions of tumor necrosis factor (TNF) and TNF receptor family members in the mouse and human. J. Biol. Chem. 281, 13964–13971 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All reagents are available under material transfer agreement. All data, sequences, and protocols are available on request. The Mb33:human MLKL NTR structure atomic coordinates have been deposited in the Protein Data Bank (accession 6UX8).