Significance
For ∼225 million y, all seed plants were woody trees, shrubs, or vines. Shortly after the origin of flowering plants ∼140 million y ago, Nymphaeales (water lilies) became one of the first seed plant lineages to become herbaceous through loss of the meristematic cell population known as the vascular cambium. We sequenced and assembled the draft genome of the water lily Nymphaea thermarum and compared it to genomes of other plants that have retained or lost the vascular cambium. By using both genome-wide and candidate-gene analysis, we found lineage-specific patterns of gene loss and divergence associated with cambium loss. Our results reveal divergent genomic signatures of convergent trait loss in a system characterized by complex gene-trait relationships.
Keywords: Nymphaea, vascular cambium, angiosperm evolution, genome, trait loss
Abstract
For more than 225 million y, all seed plants were woody trees, shrubs, or vines. Shortly after the origin of angiosperms ∼140 million y ago (MYA), the Nymphaeales (water lilies) became one of the first lineages to deviate from their ancestral, woody habit by losing the vascular cambium, the meristematic population of cells that produces secondary xylem (wood) and phloem. Many of the genes and gene families that regulate differentiation of secondary tissues also regulate the differentiation of primary xylem and phloem, which are produced by apical meristems and retained in nearly all seed plants. Here, we sequenced and assembled a draft genome of the water lily Nymphaea thermarum, an emerging system for the study of early flowering plant evolution, and compared it to genomes from other cambium-bearing and cambium-less lineages (e.g., monocots and Nelumbo). This revealed lineage-specific patterns of gene loss and divergence. Nymphaea is characterized by a significant contraction of the HD-ZIP III transcription factors, specifically loss of REVOLUTA, which influences cambial activity in other angiosperms. We also found the Nymphaea and monocot copies of cambium-associated CLE signaling peptides display unique substitutions at otherwise highly conserved amino acids. Nelumbo displays no obvious divergence in cambium-associated genes. The divergent genomic signatures of convergent loss of vascular cambium reveals that even pleiotropic genes can exhibit unique divergence patterns in association with independent events of trait loss. Our results shed light on the evolution of herbaceousness—one of the key biological innovations associated with the earliest phases of angiosperm evolution.
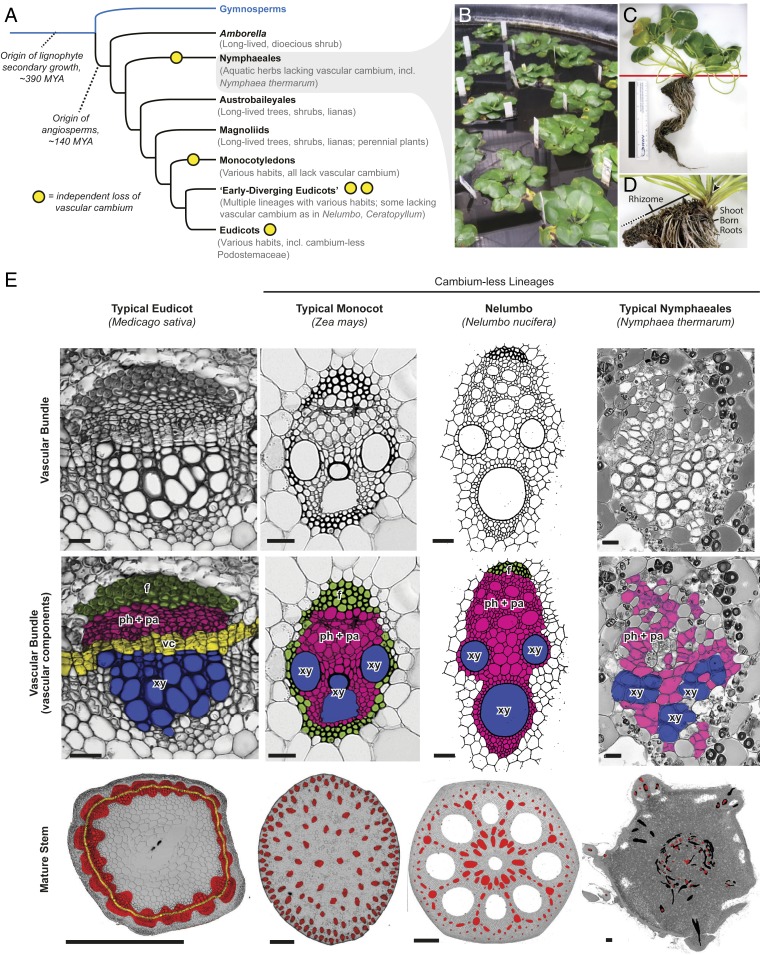
In all seed plants, vascular tissue (xylem and phloem) initially develops in distinct bundles near apical meristems as part of primary growth and differentiation. In stems of most seed plants, the subsequent activation of a population of meristematic cells within and between the bundles forms a continuous layer of vascular cambium, which then produces rings of secondary xylem and phloem responsible for increases in stem or root girth (Fig. 1) (1). Secondary growth provides the additional fluid transport capacity and mechanical support necessary for large, photosynthetic canopies to operate. A vascular cambium and production of secondary xylem and phloem are plesiomorphic for angiosperms. Loss of a vascular cambium is rare (2–5)—even most herbaceous taxa (including Arabidopsis) form a vascular cambium at some point in their ontogenies. Although additional losses may have occurred, only five losses of vascular cambium have been well-documented across angiosperms (6) (Nymphaeales, Ceratophyllum, monocots, Nelumbo, and Podostemaceae), which span ∼140 million y (7) and a diverse array of morphological adaptations and growth habits (Fig. 1A).
Fig. 1.
Vascular cambium evolution and stem vascular anatomy in flowering plants. (A) Abbreviated phylogenetic relationships of major clades of seed plants, with notes on diversity of growth habits found in each group. MYA, million years ago. (B) Growth habit of Nymphaea thermarum. (C) One individual of N. thermarum. Red line indicates in situ soil level. (Scale bar, 15 cm.) (D) Close-up of rhizome from C. Arrowhead, apical meristem; black line, living rhizome section; dashed line, dead rhizome section. (E) Stem vascular anatomy in clades of flowering plants. Cross-sections were taken from maturing (Medicago) or mature (Zea, Nelumbo, Nymphaea) stems. (Top Row) Micrographs (Medicago, Zea, Nymphaea) or drawings based on prepared slides (Nelumbo) of vascular bundles. (Scale bars, 20 μm.) (Middle Row) Vascular components labeled and color-coded. f (green), fibers; ph + pa (pink), phloem and vascular parenchyma; vc (yellow), vascular cambium; xy (blue), xylem. (Bottom Row) Whole stem sections. red, vascular bundles and tissue in cross-section; yellow, vascular cambium; black, vascular bundles that are part of a leaf or flower trace; gray, nonvascular tissue. (Scale bars, 0.5 mm.) Nelumbo panels adapted with permission from ref. 27.
Although primary and secondary vascular tissues in seed plants are derived from different meristematic cell populations, both are produced by precise patterning of the same suite of cell types. Accordingly, some of the same gene families or genetic modules function in both developmental contexts (1, 6, 8–11): the CLAVATA (CLE)–WUSCHEL (WOX) regulatory loop promotes meristem identity at the cost of xylem formation (10–13) while multiple HD-ZIP III family members promote xylem differentiation and bipolar patterning of daughter cell types (14–18) (SI Appendix, Fig. S1). The functions of HD-ZIP III members and CLE–WOX modules have been documented during primary and secondary vascular differentiation in distantly related eudicots, including Arabidopsis thaliana, Populus trichocarpa, and Zinnia elegans, using loss-of-function and/or overexpression studies (1, 8, 19, 20). Whereas at least partial expression or functional redundancy between gene family members is common (15, 18), subfamilies of HD-ZIP III and CLE genes display ontogenic specialization.
The complexity of vascular differentiation and its regulation therefore raises the question of how relevant regulatory genes might evolve (21, 22) in association with the loss of the vascular cambium and secondary tissues. Convergent loss or divergence of relevant genes has been documented in cases of convergent trait loss (23–25), suggesting a consistent, predictable association between trait loss and evolution of relevant genetic components. We use the repeated loss of the vascular cambium in angiosperms to test this association in a system characterized by pleiotropy and genetic redundancy.
Results
The loss of vascular cambium during the early evolution of the water lily lineage (Nymphaeales) likely represents one of the oldest vascular cambium losses among seed plants. Stem vascular structure is sparsely documented within the Nymphaeales (26) and undescribed in Nymphaea thermarum. We investigated rhizome (underground stem) anatomy in N. thermarum and found no evidence of a vascular cambium or secondary vascular tissues (Fig. 1D). Additionally, N. thermarum vascular bundles contained no fibers, which differs from other cambium-less lineages (27) (Fig. 1E). Thus, all vascular tissues in N. thermarum are primary tissues derived from apical meristems.
Genomes for representatives of two of the five seed plant lineages that have independently lost the vascular cambium (monocots and Nelumbo) are already available. To create a genomic resource for N. thermarum, we combined data generated from short insert libraries (∼40× coverage) and mate-pair libraries (∼20× coverage) to create a draft genome assembly (Dataset S1). The total length of the assembled genome size was 368,014,730 base pairs (bp) after discarding scaffolds <1 kilobase (kb), and a k-mer analysis of short insert reads estimated the genome size at 497,339,103 bp. These values are ∼74% and ∼100%, respectively, of the 1C size estimate of 0.51 pg as evaluated by flow cytometry (28) (roughly 498,780,000 bp). For comparison, the 1C (the total amount of DNA contained within a single [haploid] set of chromosomes) genome sizes of some popular systems for plant genetics are 0.16 pg (A. thaliana), 0.50 pg (Oryza sativa), and 2.70 pg (Zea mays); known 1C values of other members of the ANA-grade lineages (Amborellales, Nymphaeales, and Austrobaileyales), which diverge from the rest of angiosperms at basal nodes, range from 0.46 pg to 14.92 pg (mean = 3.29, median = 1.98, n= 47) (29). We also generated and assembled transcriptomes from leaves, roots, and reproductive material. When combined with available plant protein data, we annotated 25,760 protein-coding genes that overlapped input transcript or protein evidence, a Pfam domain, and/or a hit to the UniProt database (SI Appendix, Fig. S2). By comparison, the genome of Amborella (30) is annotated with 26,846 protein-coding genes (SI Appendix, Fig. S2). The N. thermarum assembly contained 865 of 956 (90%) of conserved plant proteins (31), suggesting that its genic regions were well-represented. Whereas changes in noncoding regions likely are important for morphological evolution (25), we limited our analyses to coding regions due to higher confidence in their recovery and annotation across multiple genomes.
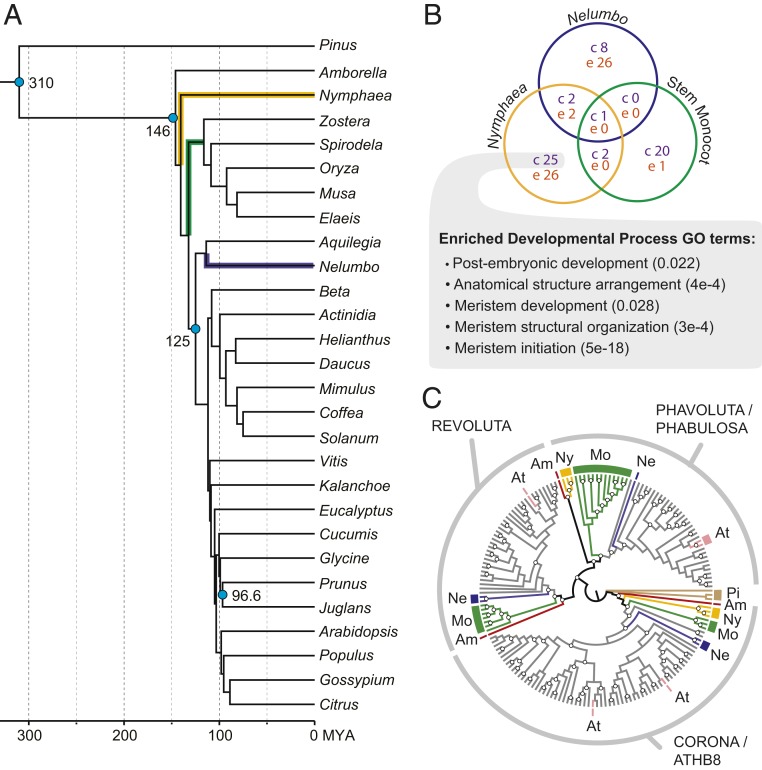
To detect significant contraction of gene families in cambium-less lineages, we first identified ortholog clusters from 28 genomes of seed plants, including three of five lineages that have independently lost the vascular cambium (i.e., Nymphaea, stem monocot, and Nelumbo) (Dataset S2). We estimated gene trees for 1,439 clusters that passed stringent filtering criteria (32) and then used them to estimate species trees with both concatenation and coalescent methods, including different subsets of the data (i.e., fast-evolving sites, slow-evolving sites, or all sites). All method and dataset combinations produced species trees with similar, strongly supported topologies for relationships of interest, including support for Amborella as sister to all other extant angiosperm lineages (SI Appendix, Fig. S3). We used CAFE (33) to detect significant ortholog cluster contraction and expansion across all branches of our species phylogeny on a fossil-calibrated species tree (ASTRAL-II, all rate classes dataset) and the set of gene cluster sizes (Fig. 2A and SI Appendix, Fig. S4). Among a total of 8,147 ortholog clusters included in the analysis, we found that 28 clusters underwent significant expansion within the Nymphaeales while 30 significantly contracted (Dataset S3 and SI Appendix, Fig. S4). Few clusters were contracted in more than one cambium-less lineage (Fig. 2B). Clusters expanded in N. thermarum were enriched for Gene Ontology (GO) terms related to secondary metabolism (including carbohydrate and polysaccharide metabolism), biosynthesis, and response to stimuli. Contracted clusters in Nymphaea were most strongly enriched for nucleosome assembly, fatty acid biosynthetic process, response to auxin stimulus, and meristem initiation (Fig. 2B and SI Appendix, Fig. S5 and Dataset S4).
Fig. 2.
Gene family contraction in cambium-less lineages. (A) Fossil-calibrated phylogenomic analysis, including representatives that have independently lost the vascular cambium: Nymphaea (yellow), monocots (stem lineage, green), and Nelumbo (indigo). Blue dots indicate age and placement of fossils or other estimates used for calibration; fossils and molecular-based estimates are detailed in SI Appendix, Supplementary Materials and Methods. (B) Venn diagram showing number of significantly contracted gene families in each cambium-less lineage (c, number of significantly contracted gene families; e, number of significantly expanded gene families). The set of gene families uniquely contracted in Nymphaea is enriched for “developmental process” GO terms related to meristem development and function (enrichment test P value in parentheses). (C) Phylogeny of HD-ZIP III genes, with branches colored to reflect identity of genes from major lineages of seed plants (complete tree in SI Appendix, Fig. S6). White dots denote branches with >70% bootstrap support. Am, Amborella trichopoda; At, Arabidopsis thaliana; Mo, monocots; Ne, Nelumbo nucifera; Ny, Nymphaeaceae; Pi, Pinus taeda.
The contracted cluster associated with the meristem initiation GO term comprises class III HD-ZIP genes, a gene family known to regulate differentiation of both primary and secondary vascular tissues (34, 35). Specifically, N. thermarum lacks a copy that belongs to the REVOLUTA (REV) subgroup (Fig. 2C and SI Appendix, Fig. S6). We further determined that no REV homolog was present in the transcriptomes of several N. thermarum tissues or in an expressed sequence tag (EST) database of Nuphar advena (36), another member of the Nymphaeales (Fig. 2C and SI Appendix, Fig. S6). The presence of a REV gene in Amborella and many other angiosperms (Fig. 2C and SI Appendix, Fig. S6) suggests that the lack of REV in Nymphaea and Nuphar represents a loss within the Nymphaeales. While some REV functions overlap with those of other HD-ZIP III members in embryogenesis and primary growth, REV misexpression is known to impact both cambial initiation and xylary fiber differentiation during secondary growth in stems of A. thaliana and P. trichocarpa (14, 15, 18, 37). The combined absence of xylary fibers, vascular cambium initiation, and REV in N. thermarum (Fig. 1D and 2C and SI Appendix, Fig. S6) is consistent with the hypothesis that REV is a key regulator of vascular cambium activity and differentiation of secondary tissues throughout angiosperms.
We further found that significant contraction of the HD-ZIP III family occurred along only two other branches (Fig. 2A and Dataset S3): the terminal branches leading to Zostera (sea grass) and Beta (beet). These species are characterized by either the absence of secondary tissues (Zostera) or by a highly anomalous pattern of vascular cambium activity that does not produce continuous rings of secondary xylem and phloem (Beta) (38–40). In contrast, some cambium-less lineages (some monocots and Nelumbo) retain homologs from all HD-ZIP III subfamilies (Fig. 2C and SI Appendix, Fig. S6). Therefore, while all lineages with a significant contraction of the HD-ZIP III gene family display a loss of normal vascular cambium development, complete loss of HD-ZIP III genes is not consistently associated with loss of vascular cambium. This result demonstrates variability in the patterns of gene loss associated with independent, homoplasious evolutionary events.
In addition to corroborating the importance of HD-ZIP III genes in vascular development, the ortholog clusters that are contracted in cambium-less lineages can be used to identify candidate genes for vascular cambium regulation. The set of clusters contracted in N. thermarum include genes that code for the auxin-responsive SAUR proteins (41) and the BIM family of BES1-interacting proteins. Intriguingly, BIM and BES1 proteins regulate SAUR gene expression in other developmental contexts (42), and BES1 is known to regulate secondary growth (1) (SI Appendix, Fig. S1). Together with the contraction of the BIM and SAUR gene families in a species that lacks a vascular cambium, this suggests that BIM and SAUR genes may constitute a novel component of vascular cambium regulation. This remains to be experimentally tested.
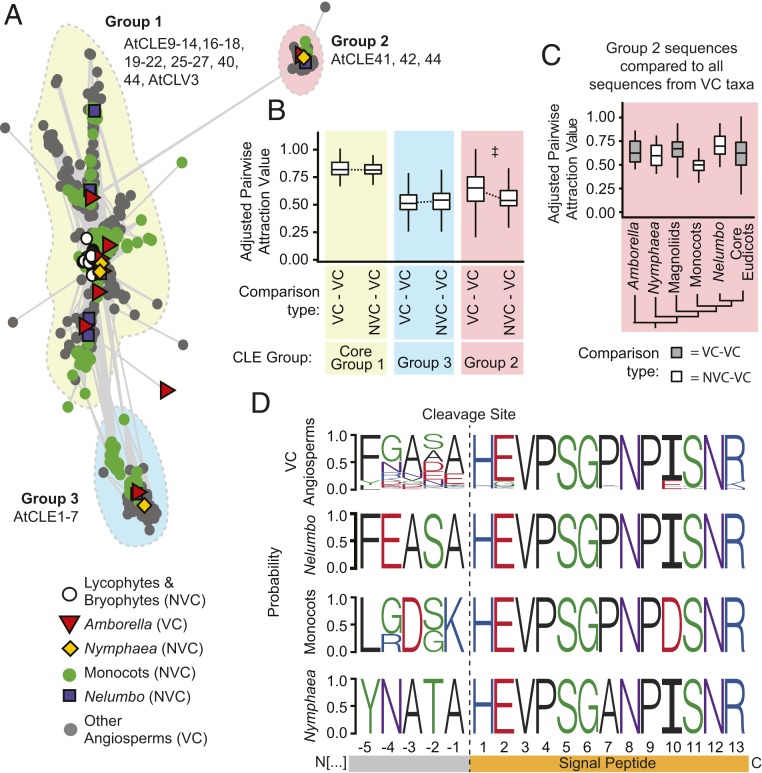
The analysis of ortholog cluster expansion and contraction, however, did not include all gene families of interest. The CLE (CLAVATA3/EMBRYO SURROUNDING REGION-RELATED) family of short (∼21 amino acids) signaling peptides are known to regulate both primary and secondary tissue differentiation in flowering plants (43), but low sequence conservation among members of this gene family precludes many types of analysis. We therefore used CLANS clustering to assess patterns of relatedness between all putative CLE homologs (10) from Phytozome v12 (44) genomes, genomes of N. thermarum and Nelumbo nucifera (45), available angiosperm transcriptomes from the OneKP initiative (46), and N. thermarum transcriptomes (Fig. 3A and SI Appendix, Fig. S7). We recovered major groups similar to results from previous studies, including a distinct CLE 41/42/44 group, called group 2 (10, 47). Group 2 is associated with vascular differentiation and patterning (48) while the other groups are associated with root nodulation (group 3) or a wide range of developmental processes, including the activity of shoot and root apical meristems (group 1) (47). After adjusting for evolutionary distance of comparisons (SI Appendix, Fig. S8) to calculate APAV (adjusted pairwise attraction values) (scaled from 0 to 1, with 1 indicating the highest sequence similarity), group 2 amino acid sequences from species without a vascular cambium (NVC) and species with a vascular cambium (VC) were overall less similar than sequences from any two species that retain the vascular cambium (VC–VC) (Fig. 3B). This relationship was only present and significant (t test, P value < 0.05 and a >1% difference) for comparisons within group 2 (SI Appendix, Table S1). Therefore, divergence of CLE sequences from cambium-less taxa has occurred within group 2, but not within the other CLE groups that are associated with primary root and shoot development.
Fig. 3.
Evolution of vascular development-associated CLE peptides. (A) Two-dimensional CLANS clustering of conserved amino acid sequences of CLE peptides in land plants. Dots/shapes represent individual sequences, coded to indicate whether they are from species within clades that have vascular cambium (VC) or do not have vascular cambium (NVC). Lines indicate PAV < 1e−5. (B) APAV for all pairwise comparisons of CLE sequences between two VC taxa (VC–VC) or between one NVC and one VC taxa (NVC–VC), for each group of CLE peptides. Higher APAV indicate higher sequence similarity. ‡ indicates t test adjusted P value < 0.05 and median APAV difference >1% (Dataset S1). (C) APAV values for all sequences within group 2 CLE peptides, for comparisons between all VC taxa and taxa within particular clades of flowering plants. (D) Conserved peptide region of group 2, including the 13-amino acid signal peptide at the C terminus and five amino acids upstream of the peptide cleavage site. For box plots: center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; outliers removed from plots for clarity, but not from analysis.
The low similarity of group 2 NVC–VC comparisons was largely driven by the divergence of monocot sequences (Fig. 3C). However, comparison of the C-terminal residues (Fig. 3D) revealed that both monocot and N. thermarum sequences show lineage-specific divergences of the five amino acids that precede the signaling peptide. These residues are immediately adjacent to a cleavage site. Changes to residues surrounding the cleavage site may impact cleavage, and therefore mobility, of the peptide (47). Furthermore, in the N. thermarum sequence, a proline (P7) that is conserved across almost all angiosperm CLE peptides (47) is substituted with an alanine. Hydroxylation of this proline (as well as P4) is required for maturation of active group 2 CLE peptides in Arabidopsis (49, 50). Additionally, prolines are necessary for certain types of kink formation (51); a substitution with alanine could negatively impact the ability of the peptide to conform to a receptor binding pocket (50, 52). In monocot sequences, a nonpolar C-terminal binding anchor site (I9) is replaced with a negatively charged residue (D9). Intriguingly, cooption of group 1 CLE peptides, but not group 2, has been associated with secondarily derived vascular cambia within monocots (53). This supports our conclusion that the observed divergence of group 2 CLE peptides within some cambium-less taxa might render them nonfunctional during vascular cambium development and activity.
Discussion
Studies of convergent trait loss often document convergent patterns of sequence loss/divergence (23–25, 54). However, many clear examples of gene loss in association with trait loss involve genes with one or few functions (55–57), and highly interconnected genes are less likely to be lost or modified (58). We find, however, that N. thermarum is characterized by both divergence of a highly conserved CLE amino acid position and a significant contraction of the HD-ZIP III gene family of transcription factors—specifically loss of REVOLUTA, the HD-ZIP III member most closely associated with cambial activity and xylary fiber production in other angiosperms. In contrast, other lineages display unique signatures of cambium loss: All monocot taxa displayed a shared divergence of cambium-associated CLE signaling peptides, supporting a single loss of the cambium early in monocot evolution. Meanwhile, Nelumbo displays little obvious evidence of gene loss or divergence of key vascular cambium regulators. Our results confirm that not only can modification of pleiotropic genes occur in systems characterized by complex gene-trait relationships involving pleiotropy and genetic redundancy, but that genomic signatures of trait loss vary between lineages that represent homoplasious loss events.
Loss of the vascular cambium is highly correlated to transitioning from a terrestrial to an aquatic habit: Nymphaeales, Nelumbo, Ceratophyllum, and Podostemaceae are all aquatic lineages (59). Even in the case of monocots, the last common ancestor of the lineage has been proposed to have been semiaquatic (60). Although there are many types of aquatic habits, submersion in or proximity to water means that aquatic plants typically do not require extensive mechanical reinforcement or high fluid transport capacity. Thus, the loss of selection for the functions performed by the vascular cambium appears to lead readily to the loss of vascular cambium formation. While the conditions that lead to cambium loss may be consistent across independent evolutionary events, our results suggest that the associated changes to genetic components of cambium regulation are not.
The exploration of new growth habits, such as nonwoody aquatic herbaceousness in Nymphaeales, is a hallmark of early angiosperm evolution (3). Yet, the genetic basis for the evolutionary diversification of early angiosperms has remained largely inaccessible due to the lack of a tractable system for genetic analyses outside of monocots or eudicots. Publication of the Amborella genome (30) represented a significant advance for understanding the genetic basis of flowering plant origins. However, extant members of the ANA-grade lineages, which diverge from the rest of angiosperms at basal nodes, commonly exhibit habits ill-suited to maintaining large populations in controlled laboratory environments, long generation times (Fig. 1A), and large genomes (>1 gigabase [Gb]) (29, 61). N. thermarum, the smallest member of the Nymphaeaceae, has a short generation time of 4 to 5 mo, has one of the smallest genomes described for any member of the Nymphaeales, and can both self-fertilize and outcross (28, 62, 63). A draft genome of N. thermarum represents a critical step in developing a system for functional genetics from within the ANA-grade lineages—a tool that holds great promise for addressing Darwin’s “abominable mystery” regarding the early radiation of the most species-rich clade of plants on earth (64, 65).
Methods
Extended methods are available in SI Appendix, Supplementary Materials and Methods.
Genome Sample and Sequence Collection.
Whole genome sequence data were collected from a live specimen of N. thermarum at the Arnold Arboretum at Harvard University. Genomic DNA was extracted using a modified cetyltrimethylammonium bromide extraction protocol (66). Short insert and mate-pair libraries were prepared for sequencing using 1 μg of DNA each with the Illumina TruSeq Library Kit and the Illumina Nextera Mate Pair Library Kit. Size selection was conducted using a Sage Science Pippin Prep machine with a target insert size of 3 kb. Both libraries were sequenced on an Illumina HiSeq 2500 sequencer with v4 chemistry (2× 250-bp reads for the short insert library and 2× 125-bp reads for the mate-pair library).
Transcriptome Sequence Collection, Assembly, and Annotation.
RNA was extracted from various tissues (leaves, roots, floral buds, and ovules) of multiple N. thermarum individuals grown at the Arnold Arboretum of Harvard University using a hot acid–phenol protocol (67). Libraries for RNA-Seq analysis were prepared by the Whitehead Institute Genome Technology Core, from total RNA (200 to 500 ng) with the Apollo 324 system from WaferGen Biosystems using the WaferGen Prep-X Directional RNA-Seq kit according to the manufacturer’s protocols to produce strand-specific complementary DNA (cDNA) libraries. Adapter-ligated cDNA fragments were enriched and amplified with 15 cycles of PCR using the HiFi NGS Library Amplification kit from KAPA Biosystems. Libraries were multiplexed at equimolar concentration and sequenced on the Illumina HiSeq 2500 for 1× 40 bases. Ribosomal RNA (rRNA) reads were filtered out with Bowtie (68) by alignment to rRNA sequences from multiple Nymphaeales. Remaining reads were assembled using the Trinity pipeline (69, 70). TransDecoder was applied to contigs with the unstranded option to identify open reading frames (ORFs) (70). BLASTN, querying against the TAIR10 Arabidopsis cDNA database at an E-value of 0.05, was used to assign identities to the ORF encoded by the mRNA.
Genome Assembly and Annotation.
Reads from the short insert library were assembled into contigs using DISCOVAR de novo v52488 (71). We next used the mate-pair data and SSPACE v3.0 (72) to join contigs into scaffolds, which resulted in a draft de novo assembly of 368,014,730 bp in 6,225 scaffolds with an N50 (the minimum contig length needed to cover 50% of the genome) of 275,242 bp (scaffolds <1 kb were discarded). Repeats were masked with RepeatMasker v4.0.5 (73). The quality of the draft genome was assessed with BUSCO v1.1 (31) for conserved plant genes.
This assembly was then annotated using MAKER v2.31.8 (74) using protein evidence (protein sequences from select angiosperm reference genomes) and transcript evidence (transcripts of N. thermarum). Four iterations of MAKER were completed, each with annotations limited to >20 amino acids on scaffolds >5 kb in length. In the first run, we initialized gene models for the ab initio software SNAP (75), and, in subsequent runs, the gene models were refined using the best ∼2,500 genes, and the ab initio program AUGUSTUS (76) was also added. MAKER generated 60,427 annotations, of which 25,760 had an annotation edit distance <1, a Pfam domain (searched with InterProScan), or a blastp hit to the UniProt database. We used Proteinortho v5.11 (77), with default settings, to identify shared and unique orthogroups between N. thermarum, A. thaliana, and Amborella trichopoda.
Phylogenomic Analysis.
We clustered homologs via an all-vs.-all pairwise search with BLASTP v2.2.25 (78) with an E-value of 1e−20, followed by grouping with MCL v09-308 with an inflation value of 5.0 (79). We clustered in-paralogs at 98.5% identity using CD-HIT v4.6 (80) and retained the longest amino acid, or chose one randomly in case of ties. We required clusters to 1) include at least four species with 2) at least one sequence from Pinus (for outgroup rooting), Amborella, and Nymphaea each, 3) include at least 100 amino acids for each sequence (81), 4) have a mean of less than five homologous sequences per species, and 5) have a median of less than two sequences per species (32). We aligned retained genes with MUSCLE v3.8.31 (82). We removed high-entropy regions of the alignment with TrimAl v1.2rev59 (83) and back-translated the amino acid alignment to codons using PAL2NAL v14 (84). We calculated homolog trees on the back-translated codons using the GTRGAMMA model in RAxML v8.2.8 (85) with Pinus designated as the outgroup, running 100 rapid bootstraps, and selecting the best-scoring maximum likelihood tree. We inferred orthologs following the “Maximal Occupancy” methods of Yang and Smith (86). Using only the inferred ortholog sequences, we then made multiple alignments, filtered high-entropy regions, back-translated to codons, and calculated gene trees.
We concatenated the alignments into a supermatrix and identified parsimony informative sites using FASconCAT v1.02 (87). We calculated observed variability for every alignment position in the supermatrix as described by Goremykin et al. (88): For a given alignment position, the sum of all pairwise differences is divided by the total number of characters. We created two synthetic alignments for each gene cluster by partitioning parsimony-informative alignment positions into those evolving faster and slower than the median. We concatenated the fast and slow synthetic alignments into respective supermatrices. We calculated species trees on all three supermatrices and gene trees for the fast and slow evolving synthetic alignments using RAxML as described above. We built species trees from gene trees for all three rate categories using ASTRAL-II v4.10.6 (89) and MP-EST v1.4 (90).
Phylogenomic Dating, Estimation of Gene Family Expansion/Contraction, and GO Enrichment Analysis.
We used the ASTRAL II all-rate classes tree for phlyogenomic dating with r8s v1.5 (91). Four nodes were fixed or constrained, based on fossil evidence (7). Clusters used for phylogenomic analysis were filtered for a mean cluster size variance per taxa of less than or equal to 140, with a median size greater than or equal to one, leaving a set of 8,147 clusters included for further analysis with CAFE v3.1 (33).
For nodes of interest within the species tree, clusters for which cluster size changed significantly were identified (P value of change from previous node <0.05). Within a cluster of interest, the gene IDs of all A. thaliana sequences were then used as the input for AgriGO Singular Enrichment Analysis (92) (against TAIR9 reference, Fischer test with Yekutieli multiple testing correction).
Evolution of HD-ZIP III Gene Family.
The cluster that contained HD-ZIP III members was defined by the presence of A. thaliana copies of REVOLUTA, PHAVOLUTA, PHABULOSA, CORONA, and ATHB8. This cluster was comprised of two clearly delineated subclades. One of these distinct subclades contained the HD-ZIP III genes, and only this subclade was used in further gene family analysis. Nuphar putative homologs were identified using HMMR v3.1b1 (93) with aligned nucleic acid sequences of all Nymphaea and Arabidopsis homologs [aligned with MUSCLE v3.8.31 (82) and then manually trimmed to exclude poorly aligned regions] against the Nuphar EST database (36) from the Ancestral Angiosperm Genome Project. ORFs were identified within the Nuphar sequences using TransDecoder v2.0.1 (70), and tBLASTx was used to identify Nuphar ORFs with a sequence similarity to A. thaliana HD-ZIP III genes. Nuphar sequences were added to the HD-ZIP III nucleotide alignment, which was then used to estimate the gene family phylogeny with RAxML v8.2.8 (85) (GTRGAMMA model, 100 bootstrap replicates). Trees were viewed in FigTree v1.3.1 (94).
Evolution of CLE Peptides.
Putative CLE peptide sequences were collected using a HMMR v3.1b1 (93) search of aligned nucleic acid sequences of all Arabidopsis homologs (aligned with MUSCLE and then manually trimmed to exclude the most poorly aligned regions) queried against a broad selection of plant genomes and transcriptomes. Amino acid sequences were aligned using MUSCLE and then trimmed to exclude the most poorly aligned regions. The resulting alignment of ∼100 amino acids was used for CLANS (95) cluster analysis.
In order to normalize for different evolutionary distances represented by comparisons, the effect of evolutionary distance on cross-genera pairwise attraction values (PAV) was modeled in R v3.5.1, using the lm function and divergence times collected from TimeTree (96). The model was then used to remove the effect of evolutionary distance on PAV, to give APAV (SI Appendix, Fig. S8). Taxa were classified by whether (VC) or not (NVC) they have a vascular cambium. For sequences for each CLE groups of interest, APAV were sorted into VC–VC and NVC–VC categories depending on the taxa they originate from. Two-sample t tests in R v3.5.1 were used to assess differences between APAV of the VC–VC and NVC–VC categories within each CLE subgroup. APAV were also compared between the sets of all sequences from taxa or clades of interest and the set of all sequences from VC taxa.
Microscopy.
N. thermarum rhizomes older than 1 y were collected from specimens at the Arnold Arboretum of Harvard University, embedded in JB4 resin (Electron Microscopy Sciences), and processed for microscopy (97). Sections were stained with periodic acid–Schiff reagent and toluidine blue (98). Prepared slides of Medicago truncatula and Z. mays stems were purchased from Triarch Incorporated (Ripon, WI). Bright-field and differential interference contrast images were recorded with a Zeiss Axio Imager Z2 microscope equipped with a Zeiss HR Axiocam digital camera (Zeiss, Oberkochen, Germany).
Data Availability.
Raw sequence data, whole-genome assembly, and transcriptomes of N. thermarum are available in the National Center for Biotechnology Information (NCBI) database under BioProject PRJNA508901. Biological material and all other data are available in Datasets S1–S4 or SI Appendix, or from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We acknowledge support from National Science Foundation Grants IOS-0919986 (to W.E.F.), DEB-1120243 (to C.C.D.), MCB-1453459 (to M.G.), IOS-1416825 (to S.M.), and IOS-1812116 (to R.A.P.). Botanische Gärten der Universität Bonn provided original plant material for propagation. We thank S. Conway for assistance with archival research. We thank Schweizerbart Scientific Publishing (http://www.schweizerbart.de/144001100) for granting permission to reproduce and adapt original drawings from ref. 27.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Raw sequence data, whole-genome assembly, and transcriptomes of Nymphaea thermarum are available in to the National Center for Biotechnology Information (NCBI) database, https://www.ncbi.nlm.nih.gov/, under BioProject PRJNA508901.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922873117/-/DCSupplemental.
References
- 1.Nieminen K., Blomster T., Helariutta Y., Mähönen A. P., Vascular cambium development. Arabidopsis Book 13, e0177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donoghue M. J., Key innovations, convergence, and success: Macroevolutionary lessons from plant phylogeny. Paleobiology 31, 77–93 (2005). [Google Scholar]
- 3.Feild T. S., Arens N. C., Form, function and environments of the early angiosperms: Merging extant phylogeny and ecophysiology with fossils. New Phytol. 166, 383–408 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Rowe N., Speck T., Plant growth forms: An ecological and evolutionary perspective. New Phytol. 166, 61–72 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Rothwell G. W., Sanders H., Wyatt S. E., Lev-Yadun S., A fossil record for growth regulation: The role of auxin in wood evolution. Ann. Mo. Bot. Gard. 95, 121–134 (2008). [Google Scholar]
- 6.Spicer R., Groover A., Evolution of development of vascular cambia and secondary growth. New Phytol. 186, 577–592 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Magallón S., Gómez-Acevedo S., Sánchez-Reyes L. L., Hernández-Hernández T., A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 207, 437–453 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Zhang J., Nieminen K., Serra J. A. A., Helariutta Y., The formation of wood and its control. Curr. Opin. Plant Biol. 17, 56–63 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Campbell L., Turner S., Regulation of vascular cell division. J. Exp. Bot. 68, 27–43 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Oelkers K., et al. , Bioinformatic analysis of the CLE signaling peptide family. BMC Plant Biol. 8, 1–15 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etchells J. P., Turner S. R., The PXY-CLE41 receptor ligand pair defines a multifunctional pathway that controls the rate and orientation of vascular cell division. Development 137, 767–774 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Etchells J. P., Mishra L. S., Kumar M., Campbell L., Turner S. R., Wood formation in trees is increased by manipulating PXY-regulated cell division. Curr. Biol. 25, 1050–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kucukoglu M., Nilsson J., Zheng B., Chaabouni S., Nilsson O., WUSCHEL-RELATED HOMEOBOX4 (WOX4)-like genes regulate cambial cell division activity and secondary growth in Populus trees. New Phytol. 215, 642–657 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Emery J. F., et al. , Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13, 1768–1774 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Robischon M., Du J., Miura E., Groover A., The Populus class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol. 155, 1214–1225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y., Song D., Sun J., Wang X., Li L., PtrHB7, a class III HD-Zip gene, plays a critical role in regulation of vascular cambium differentiation in Populus. Mol. Plant 6, 1331–1343 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Ramachandran P., Carlsbecker A., Etchells J. P., Class III HD-ZIPs govern vascular cell fate: An HD view on patterning and differentiation. J. Exp. Bot. 68, 55–69 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Zhu Y., Song D., Xu P., Sun J., Li L., A HD-ZIP III gene, PtrHB4, is required for interfascicular cambium development in Populus. Plant Biotechnol. J. 16, 808–817 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baucher M., El Jaziri M., Vandeputte O., From primary to secondary growth: Origin and development of the vascular system. J. Exp. Bot. 58, 3485–3501 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Guerriero G., Sergeant K., Hausman J. F., Wood biosynthesis and typologies: A molecular rhapsody. Tree Physiol. 34, 839–855 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Varela-Lasheras I., et al. , Breaking evolutionary and pleiotropic constraints in mammals: On sloths, manatees and homeotic mutations. Evodevo 2, 11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavličev M., Cheverud J. M., Constraints evolve: Context dependency of gene effects allows evolution of pleiotropy. Annu. Rev. Ecol. Evol. Syst. 46, 413–434 (2015). [Google Scholar]
- 23.Chan Y. F., et al. , Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhen Y., Aardema M. L., Medina E. M., Schumer M., Andolfatto P., Parallel molecular evolution in an herbivore community. Science 337, 1634–1637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sackton T. B., et al. , Convergent regulatory evolution and loss of flight in paleognathous birds. Science 364, 74–78 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Weidlich W. H., The organization of the vascular system in the stems of the Nymphaeaceae. I. Nymphaea subgenera Castalia and Hydrocallis. Am. J. Bot. 63, 499–509 (1976). [Google Scholar]
- 27.Wigand A., Dennert E., Nelumbium speciosum W. Eine monographische Studie. Bibl. Bot. 11, 1–68 (1888). [Google Scholar]
- 28.Pellicer J., Kelly L. J., Magdalena C., Leitch I. J., Insights into the dynamics of genome size and chromosome evolution in the early diverging angiosperm lineage Nymphaeales (water lilies). Genome 56, 437–449 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Leitch I. J., Johnston E., Pellicer J., Hidalgo O., Bennett M. D., Plant DNA c-values database, release 7.1. (2019). https://cvalues.science.kew.org/. Accessed 14 February 2020.
- 30.Amborella Genome Project , The Amborella genome and the evolution of flowering plants. Science 342, 1241089 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Xi Z., Liu L., Rest J. S., Davis C. C., Coalescent versus concatenation methods and the placement of Amborella as sister to water lilies. Syst. Biol. 63, 919–932 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Han M. V., Thomas G. W. C., Lugo-Martinez J., Hahn M. W., Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 30, 1987–1997 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Roodbarkelari F., Groot E. P., Regulatory function of homeodomain-leucine zipper (HD-ZIP) family proteins during embryogenesis. New Phytol. 213, 95–104 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Merelo P., Paredes E. B., Heisler M. G., Wenkel S., The shady side of leaf development: The role of the REVOLUTA/KANADI1 module in leaf patterning and auxin-mediated growth promotion. Curr. Opin. Plant Biol. 35, 111–116 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Duarte J. M., et al. , Utility of Amborella trichopoda and Nuphar advena expressed sequence tags for comparative sequence analysis. Taxon 57, 1110–1122 (2008). [Google Scholar]
- 37.Prigge M. J., Clark S. E., Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 8, 350–361 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Artschwager E., Anatomy of the vegetative organs of the sugar beet. J. Agric. Res. 33, 143–176 (1926). [Google Scholar]
- 39.Philipson W. R., Ward J. M., The ontogeny of the vascular cambium in the stem of seed plants. Biol. Rev. 40, 534–579 (1965). [Google Scholar]
- 40.Carlquist S., Successive cambia revisited: Ontogeny, histology, diversity, and functional significance. J. Torrey Bot. Soc. 134, 301–332 (2007). [Google Scholar]
- 41.Kong Y., et al. , Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol. 54, 609–621 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Yin Y., et al. , A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Dao T. Q., Fletcher J. C., CLE peptide-mediated signaling in shoot and vascular meristem development. Front. Biol. 12, 406–420 (2017). [Google Scholar]
- 44.Goodstein D. M., et al. , Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ming R., et al. , Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.). Genome Biol. 14, R41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matasci N., et al. , Data access for the 1,000 Plants (1KP) project. Gigascience 3, 17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goad D. M., Zhu C., Kellogg E. A., Comprehensive identification and clustering of CLV3/ESR-related (CLE) genes in plants finds groups with potentially shared function. New Phytol. 216, 605–616 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Hirakawa Y., Bowman J. L., A role of TDIF peptide signaling in vascular cell differentiation is conserved among Euphyllophytes. Front Plant Sci 6, 1048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito Y., et al. , Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Zhang H., Lin X., Han Z., Qu L.-J., Chai J., Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res. 26, 543–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Betts M. J., Russell R. B., “Amino-acid properties and consequences of substitutions” in Bioinformatics for Geneticists, Barnes M. R., Gray I. C., Eds. (John Wiley & Sons, Ltd; ), pp. 311–342. [Google Scholar]
- 52.Morita J., et al. , Crystal structure of the plant receptor-like kinase TDR in complex with the TDIF peptide. Nat. Commun. 7, 12383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zinkgraf M., Gerttula S., Groover A., Transcript profiling of a novel plant meristem, the monocot cambium. J. Integr. Plant Biol. 59, 436–449 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Zhang W., Kramer E. M., Davis C. C., Similar genetic mechanisms underlie the parallel evolution of floral phenotypes. PLoS One 7, e36033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang W., et al. , The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nat. Commun. 5, 3311 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olsen J. L., et al. , The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530, 331–335 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Zhang G. Q., et al. , The Apostasia genome and the evolution of orchids. Nature 549, 379–383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lammers M., Kraaijeveld K., Mariën J., Ellers J., Gene expression changes associated with the evolutionary loss of a metabolic trait: Lack of lipogenesis in parasitoids. BMC Genomics 20, 309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cook C. D. K., The number and kinds of embryo-bearing plants which have become aquatic: A survey. Perspect. Plant Ecol. Evol. Syst. 2, 79–102 (1999). [Google Scholar]
- 60.Givnish T. J., et al. , Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. Am. J. Bot. 105, 1888–1910 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Pellicer J., Hidalgo O., Dodsworth S., Leitch I. J., Genome size diversity and its impact on the evolution of land plants. Genes (Basel) 9, 88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischer E., Rodriguez C. M., 690. NYMPHAEA THERMARUM. Curtis’s Bot. Mag. 27, 318–327 (2010). [Google Scholar]
- 63.Povilus R. A., Diggle P. K., Friedman W. E., Evidence for parent-of-origin effects and interparental conflict in seeds of an ancient flowering plant lineage. Proc. Biol. Sci. 285 20172491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Friedman W. E., The meaning of Darwin’s ‘abominable mystery’. Am. J. Bot. 96, 5–21 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Chen F., et al. , Water lilies as emerging models for Darwin’s abominable mystery. Hortic. Res. 4, 17051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doyle J. J., Doyle J. L., A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytolog. Bull. 19, 11–15 (1987). [Google Scholar]
- 67.van Tunen A. J., et al. , Cloning of the two chalcone flavanone isomerase genes from Petunia hybrida: Coordinate, light-regulated and differential expression of flavonoid genes. EMBO J. 7, 1257–1263 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grabherr M. G., et al. , Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haas B. J., et al. , De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weisenfeld N. I., et al. , Comprehensive variation discovery in single human genomes. Nat. Genet. 46, 1350–1355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boetzer M., Henkel C. V., Jansen H. J., Butler D., Pirovano W., Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Smit A. F. A., Hubley R., Green P., RepeatMasker Open-4.0 1996–2010. http://www.repeatmasker.org. Accessed 6 December 2015.
- 74.Holt C., Yandell M., MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Korf I., Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stanke M., et al. , AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lechner M., et al. , Proteinortho: Detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics 12, 124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Enright A. J., Van Dongen S., Ouzounis C. A., An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li W., Godzik A., Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Liu Q., Xue Q., Comparative studies on codon usage pattern of chloroplasts and their host nuclear genes in four plant species. J. Genet. 84, 55–62 (2005). [DOI] [PubMed] [Google Scholar]
- 82.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Suyama M., Torrents D., Bork P., PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang Y., Smith S. A., Orthology inference in nonmodel organisms using transcriptomes and low-coverage genomes: Improving accuracy and matrix occupancy for phylogenomics. Mol. Biol. Evol. 31, 3081–3092 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kück P., Meusemann K., FASconCAT: Convenient handling of data matrices. Mol. Phylogenet. Evol. 56, 1115–1118 (2010). [DOI] [PubMed] [Google Scholar]
- 88.Goremykin V. V., Nikiforova S. V., Bininda-Emonds O. R. P., Automated removal of noisy data in phylogenomic analyses. J. Mol. Evol. 71, 319–331 (2010). [DOI] [PubMed] [Google Scholar]
- 89.Mirarab S., Warnow T., ASTRAL-II: Coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31, i44–i52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhong B., Liu L., Yan Z., Penny D., Origin of land plants using the multispecies coalescent model. Trends Plant Sci. 18, 492–495 (2013). [DOI] [PubMed] [Google Scholar]
- 91.Sanderson M. J., r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 19, 301–302 (2003). [DOI] [PubMed] [Google Scholar]
- 92.Du Z., Zhou X., Ling Y., Zhang Z., Su Z., agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, W64–W70 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnson L. S., Eddy S. R., Portugaly E., Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11, 431 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rambaut A., FigTree v1.3. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 4 December 2012.
- 95.Frickey T., Lupas A., CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704 (2004). [DOI] [PubMed] [Google Scholar]
- 96.Kumar S., Stecher G., Suleski M., Hedges S. B., TimeTree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819 (2017). [DOI] [PubMed] [Google Scholar]
- 97.Povilus R. A., Losada J. M., Friedman W. E., Floral biology and ovule and seed ontogeny of Nymphaea thermarum, a water lily at the brink of extinction with potential as a model system for basal angiosperms. Ann. Bot. 115, 211–226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Feder N., O’Brien T. P., Plant microtechnique: Some principles and new methods. Bot. Soc. Am. 55, 123–139 (1968). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data, whole-genome assembly, and transcriptomes of N. thermarum are available in the National Center for Biotechnology Information (NCBI) database under BioProject PRJNA508901. Biological material and all other data are available in Datasets S1–S4 or SI Appendix, or from the corresponding authors upon request.