Abstract
Deubiquitinases (DUBs) are emerging as important regulators of many pathways germane to cancer. They may regulate the stability of key oncogenes, exemplified by USP28 stabilisation of c‐Myc. Alternatively they can negatively regulate ubiquitin‐dependent signalling cascades such as the NF‐κB activation pathway. We review the current literature that associates DUBs with cancer and discuss their suitability as drug targets of the future. © 2010 IUBMB IUBMB Life, 62(2): 140–157, 2010
Keywords: signal transduction, signalling, drug discovery
INTRODUCTION
Over the past few decades, ubiquitination has emerged as one of the most versatile of post‐translational modifications, with roles in the regulation of a diverse array of cellular processes such as cell‐cycle control, DNA damage repair and membrane trafficking (1). Ubiquitin is conjugated through the sequential action of three components: E1 ubiquitin activating enzymes, E2 ubiquitin conjugating enzymes and finally E3 ubiquitin ligases, which provide for specificity. Polyubiquitin chains are formed through the serial addition of ubiquitin molecules linked to lysine residues in the preceding ubiquitin. A total of seven internal lysine residues (K6, K11, K27, K29, K33, K48, and K63) allows for diversity of chain configurations.
Ubiquitination plays a central role in degradation of proteins both through proteasomal targeting and by direct sorting to the lysosome. However, it is now becoming clear that reversible ubiquitination is also a crucial mediator within intracellular signalling cascades as exemplified by Nuclear Factor‐κB (NF‐kB) signalling. Protein networks can be formed by interaction with specific ubiquitin binding domains of which there are at least 20 classes within the human genome (2).
Reversibility of ubiquitination is accomplished through deubiquitinases (DUBs). Approximately one hundred human DUBs fall into five classes; ubiquitin specific proteases (USP), ubiquitin C‐terminal hydrolases (UCH), ovarian tumour proteases (OTU), Josephins and the Jab1/MPN/MOV34 metalloenzymes (JAMM, also known as MPN+). The structure and function of these enzymes has recently been reviewed elsewhere (3, 4). In this review, we will focus on their emergence as attractive anti‐cancer targets and discuss what is known about their function within pathways germane to cancer. In general terms, DUBs may influence the stability of key oncogenes or they may negatively regulate ubiquitin mediated signalling (Fig. 1).
Figure 1.
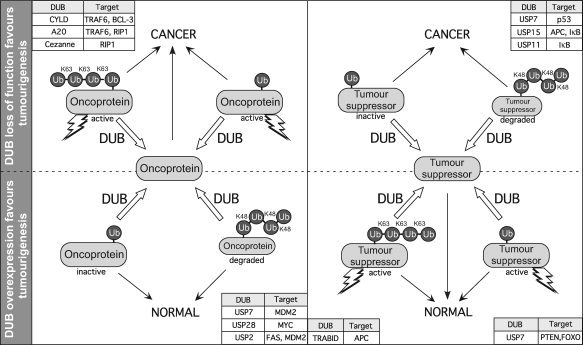
Deubiquitinases are important regulators of oncogenes and tumour suppressors. Both overexpression and loss of function of DUBs can promote cancer. Ubiquitination of oncoproteins and tumour suppressors can promote their destabilization by targeting them for degradation (e.g., K48‐linked poly‐ubiquitination specifies proteasomal degradation), or regulate their activity (activation or inactivation). Activation here may refer to a variety of processes like translocation to the nucleus (e.g., PTEN and FOXO), or engagement in signalling protein interaction networks (TRAF6, RIP1). Specific DUBs implicated in tumourigenesis are shown with their cognate targets. Note that not all targets are shown here.
Both oncogenic and tumour suppressive functions have been ascribed to individual DUBs. However, DUBs may have multiple substrates, thereby making it difficult to determine if a DUB has a net oncogenic or tumour suppressive function in vivo (Fig. 1). Although knockout and overexpression models are helpful, there is evidence to suggest that function may vary between tissue types and stage of malignancy. For a subset of DUBs, mutations and/or altered expression in cancer specimens and cell lines have been described (Table 1). Initiatives such as the Catalogue Of Somatic Mutations in Cancer (COSMIC) (87) and collation of gene expression data through Oncomine (88) provide important resources for the association of DUBs with cancer.
Table 1.
Deubiquitinases and cancer
| DUB name | Pathway/mechanism | Mutations and translocations | Published abnormalities in expression/protein level | Oncomine | |
|---|---|---|---|---|---|
| Upregulation | Downregulation | ||||
| USP1 | Deubiquitinates and negatively regulates FANCD2 and DNA repair (5). Also shown to deubiquitinate and inactivate PCNA, an important component of the DNA repair translesion synthesis pathway (6). | None reported | Upregulated in hydatidiform mole (7). | Brain, liver, cervical, gastric, sarcoma. (13 others) | Leukaemia, testicular, ovarian, prostate |
| USP2 | Upregulated in response to androgen, leading to stabilization of fatty acid synthase (8). Additionally deubiquitinates and stabilizes MDM2 (9). | 1/2 bladder cancer cell linesa | Overexpressed in ovarian carcinoma (10). Overexpressed and associated with poor prognosis in prostate cancer (8). | Brain | Brain, renal (3), HNSCC (2), Colon (2), lung. (3 others) |
| USP3 | Deubiquitinates histone H2A/H2B. Knockdown leads to defects in cell cycle progression and increased DNA damage (11). | None reported | None reported | Brain, bladder, prostate, HNSCC, testicular. (2 others) | Brain, leukaemia |
| USP4 | Transforms NIH 3T3 cells (12). Although known to bind to Rb (13), its mechanism of action remains unknown. A recent study has indicated it negatively regulates the WNT signaling pathway (14). | 1/2 bladder cancer cell linesa | Upregulated in adrenocortical carcinoma (15), small cell and adenoncarcinoma of the lung (16). Downregulation in small cell lung cancer cell lines (17). | Myeloma (2), liver, melanoma, brain, bladder. (1 other) | Testicular, lung (3), HNSCC (5), renal, brain. |
| USP5 | Knockdown leads to an increase in level and activity of p53 (18). Mechanism thought to be indirect and involve accumulation of free ubiquitin chains. | None reported | None reported. | Lung, colon(2), leukaemia, ovarian(2), liver. (2 others) | Brain (3), lymphoma, cervical |
| USP6 | Identified as oncogene through transformation of NIH 3T3 cells (19), mechanism unclear. Interacts with CDC2 and RAC1 to promote actin remodeling (20). TBC domain may regulate endosomal trafficking through activation of ARF6 (21). | Chromosomal translocations associated with aneurysmal bone cysts (22), usually but not exclusively, benign (23) | None reported | Myeloma, salivary gland, sarcoma | Brain (3), lymphoma, testis, myeloma, HNSCC. (1 other) |
| USP7 | Deubiquitinates and affects stability of both p53 and MDM2. Additionally involved in the regulation of localisation and activity of PTEN and FOXO (see text). | None reported | Increased expression in prostate cancer (24), However reduced expression reported in a study of NSCLC (25). | Colon, lung, testicular, myeloma, bladder (2). (5 others) | Brain, HNSCC, testicular, breast, melanoma. (3 others) |
| USP8 | Required for RTK downregulation following internalization, mechanism involving deubiquitination and stabilization of ESCRT components (26, 27, 28). | 1/11 lung cancer cell linesa | None reported | Brain (2), cervical, myeloma, lymphoma, brain. (1 other) | Leukaemia, lymphoma (2), testicular, lung (2) |
| USP9X | Interacts with and stabilizes beta‐catenin (29, 30). Also promotes TGF‐beta signaling through deubiquitination of SMAD4 (31). Additionally implicated in AMPK family kinase regulation. | None reported. | Overexpression reported in breast cancer (32). | Brain (2), gastric, cervical, colon, leukaemia. (3 others) | Brain, bladder, testicular, leukaemia (2), lymphoma (2) |
| USP11 | Deubiquitinates and stabilizes E7 (human papilloma virus (HPV) protein that targets Rb1‐ involved in pathogenesis of cervical cancer) (33). Interacts with BRCA2 (34) and stabilizes IkB (35). | None reported | None reported | Lung, myeloma, melanoma, HNSCC, skin. (1 other) | Brain (3), renal (2), testis, pancreas, HNSCC. (3 others) |
| USP15 | Deubiquitinates E6 (HPV protein which leads to ubiquitination and degradation of p53‐ involved in pathogenesis of cervical cancer) (36). Implicated in stabilization of APC (37). Associates with COP9 signalosome and has been shown to deubiquitinate IκB (38). | None reported | Downregulated in paclitaxel resistant ovarian cancer (39). | Vulva, brain, breast, lymphoma. | Brain, bladder, testicular, liver, melanoma. (3 others) |
| USP17 | Negatively regulates the activity of ras‐converting enzyme 1, thereby blocking ras membrane localisation and activation (40). | None reported | None reported | No available data | No available data |
| USP18 | Upregulates EGFR, mechanism uncertain but likely to occur at level of translation(41). Reported specificity for the ubiquitin‐like modifier ISG15 (42). | None reported | None reported | Bladder, ovarian, lung, brain, prostate. (1 other) | None |
| USP20 | Deubiquitinates and stabilizes HIF1‐alpha (43). | None reported | None reported | None | Lymphoma, leukaemia, testis, lung. |
| USP21 | Deubiquitinates histone H2A, activating transcription (44). | None reported | None reported | Leukaemia, bladder (2), liver, colon. | Testis, HNSCC. |
| USP22 | Deubiquitinates both Histone H2A and H2B, and is required for progression through cell cycle (45, 46). | None reported | Overexpressed and part of an 11‐gene signature that predicts poor prognosis in a wide range of malignancies (47). | Lung (2), bladder, leukaemia, melanoma, salivary. (4 others) | Brain (3), leukaemia (2), colon, liver, lymphoma. |
| USP28 | Required for MYC stability(48). Additionally stabilizes Chk2 and 53BP1 in response to DNA damage (49). | Somatic mutation reported in case of lobular breast cancer (50). 1/101 renal cell linesa. | Overexpression in colon and breast cancer (48). | Testicular, cervical, HNSCC, myeloma. | None |
| USP33 | Interacts with Robo1 and is required for Slit signaling (51, 52). | None reported | Overexpression in paediatric ALL (53). | Lung (2), lymphoma, Prostate (2), Myeloma, bladder. (1 other) | Brain (3), leukaemia (2), lung (3), breast. |
| USP39 | Involved in the mitotic spindle checkpoint, influences Aurora B expression, possibly through mRNA splicing (54). USP39 is catalytically inactive. | None reported | None reported | Leukaemia, ovarian, brain, lung, cervical. (6 others) | Brain, pancreas. |
| USP44 | Critical regulator of spindle checkpoint. Depletion of USP44 leads to defects in chromatin segregation (55) | 1/11 lung cancer cell linesa | None reported | Leukaemia. | Renal, brain, gastric, testis (2). |
| CYLD | Negative regulation of the NF‐κB pathway (see text). | Mutations (catalytic domain) in Cylindromatosis and trichoepithelioma. 1/128 lymphomaa | Several reports of down regulation in cancers including lung (56), liver (57), colon (57), multiple myeloma (58). Downregulation associated with progression in melanoma (59). | Leukaemia, renal (2), lung, testis, myeloma. (2 others) | Brain, ovarian, lung (3), HNSCC (2), bladder. (8 others) |
| AMSH | Involved in endosomal sorting/trafficking and promotes recycling of RTK from multivesicular body (60). Also reported as a positive regulator of BMP signaling (61). | None reported | None reported | Lung, liver, bladder, leukaemia (2), colon. (6 others) | Leukaemia (2) |
| AMSH‐LP | Potentiates TGFß signaling through interaction with inihibitory I‐SMADs (62) | None reported | None reported | Kidney (2), liver, brain, HNSCC. | Brain, testicular, leukaemia, brain. |
| BRCC36 | Component of the BRCA1 and BRCA2 containing complex (BRCC) involved in maintaining the G2 checkpoint and in the response to ionizing radiation (63). | Gene locus is at a chromosomal break point associated with translocations in T‐cell PLL (64). 3/36 lung cancer cell linesa | Overexpressed in breast cancer (63). | Lung, Testicular, colon, ovary, prostate. (6 others) | Brain (2). |
| MYSM1 | Deubiquitinates histone H2A and participates in androgen receptor dependant transcription (65). | None reported | None reported | Brain (2), testicular, HNSCC (2) | Brain. |
| POH1 | Component of the 19s proteasomal lid complex (66). Regulates ErbB2 ubiquitination, albeit without affecting its turnover (67). Deubiquitinates and stabilizes c‐jun (68). | None reported | None reported | Lung (3), colon, gastric, HNSCC (2), brain. (8 others) | Leukaemia (2), breast, lymphoma, testicular. |
| BAP1 | Discovered through its interaction with BRCA1 (69), BAP1 has poorly understood tumour suppressive functions. A recent study has shown interaction with the cell cycle regulator HCF‐1 to be critical (70). | Mutations and deletions in breast and lung cancer (69, 71, 72) | None reported | Testicular, lung (2), brain (2), kidney (2), cervical. (2 others) | Ovary, oesophageal, BCC, breast, lymphoma. (2 others) |
| UCHL1 | Function and mechanism of action uncertain (see text). | None reported (in cancer) | Upregulated in several malignancies (e.g. lung, colon) (73, 74, 75, 76, 77). However there have also been recent reports of methylation and reduced expression (see text). | Ovary, HNSCC (2), lung (3), gastric, oesophagus. (1 other) | Brain (2), ovarian, kidney, bladder, colon. (4 others) |
| UCHL5 | Reported to interact with SMADs and regulate TGFβ signaling (78). | None reported | None reported | Lung, breast, ovarian, vulva, parathyroid. | Brain, pancreas, breast. |
| OTUB1 | OTUB1 interacts with estrogen receptor alpha and negatively regulates ER‐alpha‐mediated transcription (79) | None reported | None reported | Bladder (3), lung, prostate, HNSCC, breast. (3 others) | Brain (2), HNSCC (2), Testis, Cervical, renal, sarcoma. (4 others) |
| A20 | Negative regulator of NF‐κB signaling (see text). | Chromosomal deletions and inactivating mutations found in several lymphoma subtypes (80, 81, 82, 83). 2 out of 11 lung cancera | Study showing overexpression in Hodgkins and anaplastic B‐cell lymphomas, with downregulation in other lymphoma types (84). | HNSCC (4), leukaemia (2), lung, brain (2), cervical. (6 others) | Bladder, ovary (2), lung, lymphoma (2), sarcoma. |
| Cezanne | Downregulates NF‐κB signalling through the deubiquitination and inactivation of RIP1 (85). | None reported | None reported | Liver, myeloma. | Ovarian. |
| TRABID | Positive regulator of WNT signaling required for TCF‐ mediated transcription (86). | 1/ 202 kidney cancer | None reported | Brain, testicular, leukaemia, oesophagus, liver. | Brain (2), leukaemia, liver, testicular, bladder. (2 others) |
Published data on DUB mutations and aberrations in expression remain sparse. A review of the COSMIC (Catalogue Of Somatic Mutations In Cancer http://www.sanger.ac.uk/genetics/CGP/cosmic (87)) and Oncomine (http://www.oncomine.org) (88) databases was therefore performed. Oncomine (Compedia bioscience, Ann Arbor, MI) was used for analysis and visualization of microarray data. Expression in cancer and normal tissue was compared, and studies in which statistically significant differences in expression (P < 10−4) are collated above. Up to five cancer types are named; further studies meeting the aforementioned criteria are numerated but not named. Tumour subtypes were not analysed. Together with differences in stage etc, this may explain the observation of both over and under‐expression within the same tumour group. Testicular refers to germ cell tumours of the testis.
Mutations identified by COSMIC, expressed as a fraction of tumours of that type that were assessed. Arf6, ADP ribosylation factor 6; BAP1, BRCA1 associated protein 1; BRCC36, BRCA1/BRCA2 containing complex subunit 3; HNSCC, Head and Neck Squamous Cell Carcinoma; Rb1, retinoblastoma 1; Robo1, Roundabout 1.
DEUBIQUITINASES AND RECEPTOR TYROSINE KINASE SIGNALLING CASCADES
Targeting receptor tyrosine kinases (RTKs) and their downstream effectors, has recently proven to be one of the most successful avenues in drug development, with several such targeted therapies already in clinical practice. Examples include trastuzumab (Herceptin) and imatinib (Gleevec), which have contributed significantly to the treatment of ErbB2 overexpressing breast cancer (89) and gastrointestinal stromal tumours (90) respectively. However, for the majority of inhibitors, only a small proportion of patients respond to treatment, and responses are often short lived. Innate or acquired resistance frequently occurs through up‐regulation of other signalling pathways (such as the Met pathway in patients treated with EGFR inhibitors (91, 92)) or downstream activating mutations. A deeper understanding of the intricate regulation of RTK cascades is therefore essential for future rational drug development and in particular, combination therapy.
The significance of endocytic growth factor receptor trafficking in cancer is reflected in the multitude of aberrations in the endocytic machinery that have now been described (93). It is well established that ubiquitination of RTKs directs their lysosomal degradation and that mutants which escape ubiquitination are frequently transforming (94). Following receptor internalisation, ubiquitin‐dependent receptor sorting occurs in the sorting endosome where receptors can be incorporated into lumenal vesicles destined for degradation in the lysosome or otherwise recycled to the plasma membrane (95).
Two endosomal DUBs exert opposite effects upon the fate of internalised epidermal growth factor receptor (EGFR). AMSH (associated molecule with the SH3 domain of STAM) is a member of the JAMM family of metalloproteases with specific activity against K63 ubiquitin linkages (60, 96, 97). Knockdown of AMSH leads to an increased degradation rate of activated EGFR (60, 98). A working model to explain this observation suggests that AMSH deubiquitination of receptors at the sorting endosome leads to their recycling to the plasma membrane (60, 99). Thus, a balance between ubiquitinating and deubiquitinating activity will control receptor fate. Interestingly, in a review of the Oncomine database, AMSH expression was found to be elevated in lung, liver, bladder and colon cancer, consistent with a tumour promoting potential that may correlate with its endocytic function.
On the other hand USP8, also called UBPY, is required for the downregulation of EGFR and Met, the receptor for Hepatocyte growth factor (HGF) (26, 98, 100, 101). Its functions on the endocytic pathway are likely to be pleiotropic, but include the deubiquitination, and stabilisation of components of the endosomal sorting machinery, HRS and STAM (26, 99).
We recently reported an siRNA screen aimed at identifying DUBs that regulate the stability of ErbB2, a member of the EGFR family, which does not undergo activity dependent endocytosis (67). We identified POH1 (26S proteasome‐associated pad1 homologue, also known as PSMD14; Rpn11 in yeast), a component of the proteasome lid, as a critical DUB controlling the ubiquitination status of ErbB2, albeit without necessarily affecting its turnover. Nevertheless, this highlights that one unanticipated effect of treatment with the proteasome inhibitor Bortezimib, may be an altered activity of ErbB2 receptors that could have clinical implications.
To date, the major established role of ubiquitin following RTK stimulation has been to promote lysosomal degradation of receptors. We recently screened for DUBs that may be involved in hepatocyte growth factor (HGF)‐dependent cell scattering of A549 lung carcinoma cells (102). The implication of up to 12 DUBs in this process, independent of effects on Met receptor levels and down‐regulation, suggests hitherto unappreciated roles of ubiquitin‐linked events in regulating the outcomes of HGF‐signaling. Although, the requirement for catalytic activity was not assessed, it is reasonable to suggest that this set of DUBs will contain some, which could be considered as anti‐metastatic targets. The relevant substrates are, however, yet to be identified.
USP18 has recently been shown to influence EGFR protein expression levels, with a 50–80% reduction observed following USP18 knockdown (41). Conversely, overexpression of catalytically active USP18 resulted in an increase in EGFR expression. Amongst several RTKs tested (Met and ErbB2), only EGFR protein levels were affected and, although the exact mechanism remains uncertain, regulation appears to occur at the level of translation.
CYLD, CYLINDROMATOSIS, AND NF‐κB SIGNALLING
Cylindromatosis is a rare familial condition characterised by the development of multiple skin tumours, known as cylindromas, mostly on the scalp. Early work into its aetiology led to the discovery of CYLD, a member of the USP class of DUBs (103). Inherited in an autosomal dominant fashion, cylindromatosis has been shown to be due to mutations in the cyld gene. Individual tumours are characterised by loss of heterozygosity whereby the second copy of cyld is mutated and non‐functional. The majority of mutations that have been described affect the catalytic domain (104, 105). A second familial condition, trichoepithelioma, is also caused by CYLD mutations, identical to those causing cylindromatosis but resulting in tumours of the hair follicle rather than the sebaceous gland as occurs in cylindromatosis. Malignant transformation in both conditions is very rare but has been described (106, 107).
In a series of groundbreaking experiments, Brummelkamp et al identified CYLD as a negative regulator of NF‐κB signalling. Using a shRNA library targeting 50 DUBs to identify candidates involved in the regulation of cancer related pathways, they demonstrated that knockdown of CYLD enhanced NF‐κB signalling following Tumour Necrosis Factor (TNF) stimulation (108). Two other groups identified CYLD in yeast two‐hybrid screens for proteins interacting with NEMO (nuclear factor kappa B essential modifier), a key adaptor protein in NF‐κB signaling (109, 110). This association of CYLD with the NF‐κB pathway suggests the application of topical aspirin derivatives, which inhibit NF‐κB activity, may be a valid therapeutic strategy in familial cylindrimatosis (108).
The role of CYLD in NF‐κB signalling has been extensively reviewed elsewhere and is described only briefly here (111, 112, 113). Receptor stimulation leads to activation of the IκB kinase (IKK) complex, (comprising two catalytic subunits and the regulatory protein NEMO), which in turn phosphorylates IκB. Phosphorylated IκB dissociates from NF‐κB, undergoes K48 polyubiquitination and is degraded by the proteasome leaving NF‐κB to activate downstream proliferative signals (114). Several proteins involved in signal transduction (NEMO, TNF receptor‐associated factor 2 and 6 (TRAF2 and TRAF6)), are modified by the addition of K63 linked polyubiquitin chains, which results in their activation. CYLD shows preferential activity for K63‐linked polyubiquitin chains and its expression is upregulated by NF‐κB, providing negative feedback regulation of this pathway through deubiquitination of NEMO, TRAF2, and TRAF6 (109, 110, 115, 116). CYLD has also been shown to deubiquitinate (and inactivate) the coactivator BCL‐3 (117), which switches the transcriptional properties of NF‐κB from repressive to activating, and is known to strongly promote cell proliferation and oncogenesis (118, 119). Recently, IκB kinase epsilon (IKKϵ), an oncoprotein overexpressed in 30% of breast cancers (120), which plays a critical role in interferon signalling pathways, has been shown to phosphorylate and inactivate CYLD (121). Significantly inactivation of CYLD was necessary for IKKϵ‐driven transformation (121).
Emerging evidence indicates CYLD has yet other, NF‐κB independent roles, which may be associated with malignancy, including (1) regulating mitotic entry and potentially cytokinesis (122, 123), (2) deubiquitination of TrkA, the receptor for neuronal growth factor (124), and (3) facilitating cell migration (125). Whilst in the latter cases, CYLD may function as a positive regulator of mitogenic and motogenic processes, it is loss of function that is more commonly associated with tumourigenesis and correlates with its negative role in the NF‐κB pathway. There have been several reports of down‐regulation of CYLD expression in cancer including lung (56), liver (57) and colon (57) cancer and multiple myeloma (58). Reduced expression has also been correlated with poor prognosis in melanoma (59). Furthermore, cyld−/− mice exhibit increased susceptibility to colon cancers on a background of induced colitis (126) as well as skin tumours (117).
A second DUB, A20 (also known as tumour necrosis factor‐α‐induced protein 3, TNFAIP3) negatively regulates NF‐κB signalling. A member of the OTU class of DUBs, A20 exerts its regulatory activity through the removal of activating K63‐linked ubiquitin chains from TRAF6 and Ral interacting protein 1 (RIP1) (127, 128). CYLD and A20 target an overlapping set of NF‐κB pathway effectors (e.g. TRAF6, RIP1, NEMO) and it has been proposed that CYLD may largely be responsible for the constitutive inactivation of the cascade, whilst A20 protein levels and activity are induced in response to a negative feedback mechanism (112, 113). In vitro, A20 is only poorly active towards free K63‐ compared with K48‐linked ubiquitin chains, but instead has been shown to remove these chains en bloc from TRAF6 (129, 130). A20 has also been reported to possess E3‐ligase activity, which adds K48‐linked chains to RIP1 and TRAF6, although this function of A20 may also involve the E3‐ligase ITCH (131). Thus A20 can edit ubiquitin chain modifications by co‐ordinating removal of one type of ubiquitin chain (K63) promoting NF‐κB activation from RIP‐1 and its replacement with K48‐linked ubiquitin chains specifying degradation. A20 is frequently inactivated in B‐cell lymphomas (132, 133), and its restoration in A20 null cell lines leads to inhibition of cell growth and induction of apoptosis (132). These studies establish A20 as an important tumour suppressor in lymphoid malignancies.
Three further DUBs have been implicated in negative regulation of NF‐κB signalling: the OTU DUB Cezanne (also known as OTUD7B), USP11 and USP15 (112). Cezanne is induced by TNF‐alpha in cultured cells leading to down‐regulation of NF‐κB signalling through the deubiquitination and inactivation of RIP1, thereby providing a negative feedback loop in pro‐inflammatory signalling (85). USP11, as well as its COP9‐signalosome associated paralogue USP15, have both been shown to deubiquitinate and stabilise IκB, which binds and sequesters NF‐κB in the cytoplasm (35, 38). However, in the case of USP11, a catalytically inactive mutant is still able to partially inhibit NF‐κB activation, indicating a further, as yet unknown, mechanism of action.
The NF‐κB pathway is constitutively active in a wide variety of cancers, and is intimately linked to cancer growth and metastasis (134). The involvement of five DUBs in NF‐κB regulation underlines the importance of its tight regulation in normal cells, and in particular the role of reversible deubiquitination (135).
USP9X, AMSH‐LP, UCHL‐5, AND THE TGFß PATHWAY
The Transforming Growth Factor (TGF)‐ß pathway plays a dual role in tumour development: at early stages, this signaling cascade has a growth inhibitory effect whilst at later stages, the same growth factor promotes epithelial to mesenchymal transition (EMT) with pro‐metastatic outcomes (136). Signal transduction in this pathway is primarily mediated by the SMAD effectors, and both SMADs as well as the TGFß receptors are regulated by reversible ubiquitination (137). USP9X (also called FAM) was identified as a positive regulator of TGFß signaling by means of a siRNA screen. It acts by removing monoubiquitin from the Co‐SMAD, SMAD4, thereby permitting its association with phospho‐SMAD2 and subsequent activation of TGFß/SMAD‐responsive gene targets (31). USP9X has also been shown to control AMP‐activated protein kinase (AMPK)‐related kinase activity through direct removal of non‐canonical K29/K33‐linked ubiquitin chains (138). UCHL‐5 (also called UCH37) has been reported to interact with the inhibitory SMAD7 and regulate TGFß‐receptor ubiquitination, stability and TGFß‐dependent transcription (78). Likewise, the closely related JAMM DUBs AMSH and AMSH‐LP (AMSH‐like protein) have been implicated as positive regulators in the bone morphogenetic protein (BMP) and TGFß responses respectively through their interaction with inhibitory I‐SMADs (61, 62).
USP4, USP15, TRABID, AND THE WNT SIGNALING PATHWAY
Various DUBs have been shown to influence the WNT signaling pathway, a key mediator of cell polarity, proliferation and cellular homeostasis (139, 140), which is widely implicated in malignancy, particularly colon cancer (141). WNT signaling increases the stability of ß‐catenin by promoting its dissociation from the Axin/GSK‐3/APC complex. This allows ß‐catenin to enter the nucleus and activate TCF/Lef family transcription factors to promote specific gene expression. A siRNA screen of human DUBs identified USP4 as a suppressor of ß‐catenin dependent transcription, possibly through direct deubiquitination of the transcription factor TCF4 (14). USP15, a COP9 signalosome associated paralogue of USP4, has also been implicated as a negative regulator of this pathway by stabilizing the tumour suppressor adenomatous polyposis coli (APC), thereby promoting the degradation of ß‐catenin by the proteasome (37). In contrast to USP4 and USP15, the Otubain DUB, TRABID (TRAF‐binding protein domain, also called ZRANB1) is a positive regulator of WNT signaling. It interacts with and deubiquitinates APC without affecting its stability, in line with its demonstrated specificity for K63‐linked polyubiquitin chains (86, 97). TRABID is required for TCF‐mediated transcription in cells with high WNT activity, including colorectal cancer cell lines in which it may affect assembly or activity of the TCF/ß‐catenin transcription complex (86).
USP7 AND THE TUMOUR SUPPRESSORS p53, PTEN AND FOXO
Initially described as a herpes simplex encoded protein required for lytic infection and termed Herpes associated USP (HAUSP), USP7 has subsequently been linked with the regulation of three vital tumour suppressors; p53 (142), PTEN (phosphatase and tensin homolog) (24), and FOXO (Forkhead box O) (143). The role of USP7 in p53 regulation is complex, as USP7 deubiquitinates both p53 and the E3 ligase MDM2 (murine double minute 2 homolog), which itself ubiquitinates and downregulates p53. Thus USP7 would be posited to have opposing effects depending on whether it predominantly deubiquitinates and rescues p53 or MDM2. Current evidence is that USP7 preferentially binds and deubiquitinates MDM2, thereby leading to increased degradation of p53, and consequent anti‐apoptotic functions. In this scenario USP7 can be considered to be an oncogene (144). However, there may be situations in which the balance is reversed and USP7 stabilises p53 and acts as a tumour suppressor. For example, USP7 has been proposed to switch from MDM2 to p53 stabilisation in response to DNA damage and subsequent activation of the kinase ATM (ataxia telangiectasia mutated) (145). Other DUBs also influence p53 stability including USP2, which is described further below (9).
The second prominent substrate for USP7 is PTEN, a lipid phosphatase and tumour suppressor which acts to terminate phosphatidylinositol 3‐kinase (PtdIns 3‐kinase) signalling through the hydrolysis of PtdIns(3,4,5)P3 to PtdIns(4,5)P2 (146). Possibly the second most important tumour suppressor after p53, PTEN is mutated in a wide range of malignancies. Interestingly, nuclear exclusion of PTEN has been shown to result in a more aggressive phenotype in malignancies (147, 148, 149), although the tumour suppressive functions of nuclear PTEN remain unclear (150). Monoubiquitination of PTEN promotes its shuttling into the nucleus where it is sequestered in PML bodies. USP7 in turn deubiquitinates PTEN and promotes its exclusion from the nucleus (24). The same study also showed overexpression of USP7 in prostate cancer with associated nuclear exclusion of PTEN.
The FOXO proteins (FOXO1, 3, 4, and 6) are transcription factors, which have tumour suppressive functions in mouse models. Phosphorylation of FOXOs, in response to PtdIns(3,4,5)P3 mediated PKB/AKT activation, promotes their nuclear export, poly‐ubiquitination and consequent proteasomal degradation. Conversely, mono‐ubiquitination of FOXO3 and FOXO4 has been shown to result in nuclear retention and promote their transcriptional activity. As described above for PTEN, this ubiquitination is reversed by USP7 (143).
Ongoing work will hopefully decipher the various contributions of the different functions in vivo, clarifying the net result of inhibition of USP7 in individual cells and whole organisms. However, at least at face value, inhibition of USP7 would appear a promising anticancer strategy, by promoting stabilisation of p53. Early preclinical work on USP7 inhibitors has recently been reported (151).
USP2, FATTY ACID SYNTHASE AND PROSTATE CANCER
In 2004, Graner et al identified the rat homologue of USP2 as an androgen regulated DUB (8) in a prostate cancer cell line. Expression of USP2 was upregulated in LNCaP cells exposed to dihydrotestosterone, and inhibited by the anti‐androgen bicalutamide. USP2 was shown to associate with and stabilise fatty acid synthase, a protein, which is often overexpressed in biologically aggressive prostate cancer cells. Significantly, siRNA knockdown of USP2 resulted in apoptosis, which could be reversed by overexpression of fatty acid synthase (FAS) (8). However, a subsequent study, using a yeast two‐hybrid screen, identified MDM2 as a further USP2 substrate. Unlike USP7, USP2 does not deubiquitinate p53 (9). Overexpression of USP2 was shown to reduce p53 stability, conversely suppression led to destabilisation of MDM2 and increased p53 level and activation (9). A recent study also reports a similar role for USP2 in the stabilisation of MDMX, another negative regulator of p53 and close structural homologue of MDM2 (152).
In a further study, increased expression of USP2 was found in 44% of prostate tumours studied (153). Overexpression of USP2 in prostate cancer cell lines protected cells from apoptosis and conferred resistance to chemotherapeutic agents, while siRNA knockdown led to apoptosis. Interestingly, gene profiling of prostate cancers overexpressing USP2 showed up‐regulation of pathways associated with both known substrates, p53 and fatty acid synthase (153).
USP2 expression has also recently been shown to correlate with tumour progression and worse prognosis in oral squamous cell carcinomas (154). Whether this reflects a more widespread role for USP2 in carcinogenesis remains to be seen.
USP28 AND c‐MYC: REGULATING CELL PROLIFERATION
Aberrations in the expression and stability of the proto‐oncogene c‐myc, are widespread in malignancy (155, 156). MYC is a transcription factor that binds to ∼10–15% of the genome and acts as a core regulator of cell growth, proliferation and apoptosis (157). MYC is ubiquitinated and rapidly degraded leading toa short half‐life and thereby allowing exquisite control of its cellular level (158). Although mutations affecting phosphorylation and subsequent ubiquitination of MYC have been described, stabilisation of MYC may also occur in the absence of mutations. Identified through a large‐scale shRNA screen, USP28 has recently been shown to control the stability of MYC by antagonising the activity of the SCFFBW7 ubiquitin ligase (48). Significantly, shRNA knockdown of USP28 led to a decrease in MYC in all members of a panel of cancer cell lines, and an associated inhibition in growth. The authors also demonstrated increased expression of USP28 in a significant proportion of colon and breast carcinomas. This study identifies a novel mechanism by which cancer cells may upregulate MYC. USP28 may well prove amenable to inhibition and provide a means of targeting malignancies in which MYC is up‐regulated. In a different study, however, USP28 was shown to stabilise Chk2 and p53 binding protein1 (53BP1) in response to DNA damage and was required for DNA damage induced apoptosis (49). USP28 inhibition may therefore be expected to result in significantly different outcomes, depending on which pathways are activated.
UCHL1
Ubiquitin C‐terminal hydrolase –L1 (UCHL1), a member of the UCH class of DUBs, is one of the most well studied DUBs in view of its association with neurodegenerative conditions, including Parkinsons Disease, and a wide range of malignancies. Despite this, its in vivo functions remain enigmatic.
Normally almost exclusively expressed in neurones, the neuroendocrine system and the gonads, aberrant expression has been described in non‐small cell lung cancer (73, 74), invasive colorectal cancer (75), pancreatic cancer (159), osteosarcoma (76), and oesophageal cancer (77). Overexpression has also been associated with tumour progression, size and invasiveness (75, 159) There have been several reports indicating hypomethylation of the uchl1 gene leads to increased expression in Gall bladder cancer (160), as well as association with progression and metastasis in colorectal cancer (161). On the other hand there have also been reports of methylation mediated silencing of uchl1 in progression of oesophageal squamous cell carcinoma and gastric cancer (162, 163) and in pancreatic cancer cell lines (164). Furthermore, overexpression of UCHL1 has been associated with apoptosis in a breast cancer cell line (165). Although mutations have been described, these have so far been associated with Parkinsons disease rather than cancer, for which altered expression appears to be more common.
Kim et al. (166) recently demonstrated a role for UCHL1 in migration and invasiveness of the non‐small cell lung cancer cell line H157, through siRNA knockdown studies. Additionally they suggested that UCHL1 knockdown resulted in decreased activation of AKT and downstream effectors. Bheda et al have also recently described upregulation of β‐catenin signalling by UCHL1 (167). Moreover, they also showed reciprocal upregulation of UCHL1 expression by β‐catenin, suggesting a positive feedback loop in transformed cells. In a second study, the same authors used a microarray approach to profile genes affected by siRNA knockdown of UCHL1 and have identified several genes involved in apoptosis, cell cycle, proliferation and migration whose expression was altered (168). Although the data described earlier indicate important roles for UCHL1 in malignancy, the mechanism by which this occurs remains uncertain. Structural analysis indicates a prominent loop covering the active site (169), thereby preventing the binding of large protein substrates and limiting activity to small peptide chains (170). It has been hypothesised that UCHL1 may, however, act at protein termini or on unfolded substrates (169).
USP1 AND FANCONI ANAEMIA
Fanconi Anaemia (FA) is a rare congenital syndrome characterised by skeletal abnormalities, microcephaly, progressive bone marrow failure and a predisposition to malignancy later in life (171). Inherited as an autosomal recessive or X‐linked disease, the clinical phenotype of patients with FA varies widely. However, all variants share a high frequency of chromosomal abnormalities and sensitivity to DNA intrastrand crosslinking agents (172). Thirteen complementation groups have been described (designated FA‐A, ‐B etc), each characterized by mutation of a gene involved in a common pathway that regulates the sensing, signaling and repair of DNA intrastrand crosslinks (173).
Central to the FA pathway, FANCD2 (fanconi anaemia, complementation group D2) is monoubiquitinated by a nuclear multiprotein complex incorporating at least eight other FA proteins (173). This event is dependent on phosphorylation of serine 331, which has been shown to be mediated by CHK1 (174). Monoubiquitination of FANCD2 and PCNA (proliferating cell nuclear antigen) promotes DNA repair through chromatin association and enhanced recruitment of enzymes involved in translesion synthesis respectively. In a siRNA screen, Nijman et al identified USP1 as a novel regulator of FANCD2, with USP1 inhibition leading to accumulation of monoubiquitinated FANCD2 (5). Whilst both FANCD2 and PCNA (6) are substrates of USP1, only deubiquitination of FANCD2 is necessary for DNA repair (175), reflecting the need for a dynamic population of FANCD2. Significantly, USP1 knockout mice exhibit a similar, albeit more severe, phenotype to most FA mouse models, with stunted growth, chromosomal instability and sensitivity to DNA cross‐linking agents (176). The severity of the phenotype and the high perinatal lethality likely reflect involvement of other substrates, including PCNA, the ubiquitinated form of which was found to be elevated in these mice.
DUB INHIBITORS
The stage is set to begin thinking about the development of small molecule inhibitors of DUBs. This will be facilitated by the development of suitable high throughput screening (HTS) platforms. Most current screens use fluorescent substrates such as Ub‐AMC (177) or a sandwich of green fluorescing protein (GFP)‐ubiquitin and terbium, which undergoes fluorescence resonance energy transfer (178). One limitation is that these assays will not be suitable for those DUBs, which do not process linear chains (e.g. AMSH) (97). Isopeptide linked substrates are required.
Proof of principle has been achieved through the identification of first generation inhibitor molecules from chemical libraries, which show some specificity. Two unrelated small molecule inhibitors targeted against the SARS coronavirus DUB papain like protease (PLpro) were identified from High‐throughput screening (HTS), and one of these was subsequently optimized to achieve nanomolar inhibition (179, 180). HTS of small molecule libraries against UCHL1 identified several hits with selectivity for UCHL1 over the related UCHL3 (181). Small molecules showing some selectivity for USP7 and USP8 respectively have also been reported (182). HB41,108, is a USP7 inhibitor developed by Hybrigenics, recently shown to inhibit USP7 in the sub‐micromolar range, leading to p53 stabilisation and activation with subsequent apoptosis and inhibition of cancer cell growth (151). Selectivity has so far been demonstrated with relatively small panels of enzymes. More effort needs to be placed upon the development of panels of DUB enzymes for characterisation of selectivity.
CONCLUDING REMARKS
Cancer, although defined by shared clinical characteristics, is driven by a remarkable diversity of genetic and epigenetic aberrations in fundamental cellular processes. Future therapeutic strategies will depend on the ability to define the genetic makeup of individual tumours and to inhibit specific activated pathways. This is in turn reliant on a thorough understanding of the regulation of these pathways. As discussed in this review, members of the DUB superfamily have been implicated in many of these processes. They provide novel targets for drug development, and may also prove useful biomarkers for activation of specific pathways. The early involvement of pharmaceutical companies, combined with a wealth of techniques built up through drug development in analogous areas, has given research in this field a head start. However, the clinical utility of targeting DUBs remains to be proven and will be the subject of extensive future research.
Acknowledgements
JJS is funded by a grant from Clatterbridge Cancer Research. SU is a Cancer Research UK Senior Research Fellow.
References
- 1. Welchman, R. L. , Gordon, C. , and Mayer, R. J. ( 2005) Ubiquitin and ubiquitin‐like proteins as multifunctional signals. Nat. Rev. Mol. Cell. Biol. 6, 599–609. [DOI] [PubMed] [Google Scholar]
- 2. Dikic, I. , Wakatsuki, S. , and Walters, K. J. ( 2009) Ubiquitin‐binding domains—from structures to functions. Nat. Rev. Mol. Cell. Biol. 10, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Komander, D. , Clague, M. J. , and Urbe, S. ( 2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell. Biol. 10, 550–563. [DOI] [PubMed] [Google Scholar]
- 4. Reyes‐Turcu, F. E. , Ventii, K. H. , and Wilkinson, K. D. ( 2009) Regulation and cellular roles of ubiquitin‐specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nijman, S. M. , Huang, T. T. , Dirac, A. M. , Brummelkamp, T. R. , Kerkhoven, R. M. , D'Andrea, A. D. , and Bernards, R. ( 2005) The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 17, 331–339. [DOI] [PubMed] [Google Scholar]
- 6. Huang, T. T. , Nijman, S. M. , Mirchandani, K. D. , Galardy, P. J. , Cohn, M. A. , Haas, W. , Gygi, S. P. , Ploegh, H. L. , Bernards, R. , and D'Andrea, A. D. ( 2006) Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell. Biol. 8, 339–347. [DOI] [PubMed] [Google Scholar]
- 7. Kim, S. J. , Lee, S. Y. , Lee, C. , Kim, I. , An, H. J. , Kim, J. Y. , Baek, K. H. , Kim, E. J. , Kim, J. M. , Lee, J. B. , Lee, J. W. , Jung, W. W. , Chun, T. , and Oh, Y. K. ( 2006) Differential expression profiling of genes in a complete hydatidiform mole using cDNA microarray analysis. Gynecol. Oncol. 103, 654–660. [DOI] [PubMed] [Google Scholar]
- 8. Graner, E. , Tang, D. , Rossi, S. , Baron, A. , Migita, T. , Weinstein, L. J. , Lechpammer, M. , Huesken, D. , Zimmermann, J. , Signoretti, S. , and Loda, M. ( 2004) The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 5, 253–261. [DOI] [PubMed] [Google Scholar]
- 9. Stevenson, L. F. , Sparks, A. , Allende‐Vega, N. , Xirodimas, D. P. , Lane, D. P. , and Saville, M. K. ( 2007) The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 26, 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang, Y. , Hou, J. Q. , Qu, L. Y. , Wang, G. Q. , Ju, H. W. , Zhao, Z. W. , Yu, Z. H. , and Yang, H. J. ( 2007) [Differential expression of USP2, USP14 and UBE4A between ovarian serous cystadenocarcinoma and adjacent normal tissues]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 23, 504–506. [PubMed] [Google Scholar]
- 11. Nicassio, F. , Corrado, N. , Vissers, J. H. , Areces, L. B. , Bergink, S. , Marteijn, J. A. , Geverts, B. , Houtsmuller, A. B. , Vermeulen, W. , Di Fiore, P. P. , and Citterio, E. ( 2007) Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 17, 1972–1977. [DOI] [PubMed] [Google Scholar]
- 12. Gupta, K. , Chevrette, M. , and Gray, D. A. ( 1994) The Unp proto‐oncogene encodes a nuclear protein. Oncogene 9, 1729–1731. [PubMed] [Google Scholar]
- 13. DeSalle, L. M. , Latres, E. , Lin, D. , Graner, E. , Montagnoli, A. , Baker, R. T. , Pagano, M. , and Loda, M. ( 2001) The de‐ubiquitinating enzyme Unp interacts with the retinoblastoma protein. Oncogene 20, 5538–5542. [DOI] [PubMed] [Google Scholar]
- 14. Zhao, B. , Schlesiger, C. , Masucci, M. G. , and Lindsten, K. ( 2009) The ubiquitin specific protease 4 (Usp4) is a new player in the Wnt signalling pathway. J. Cell. Mol. Med. (E‐publication ahead of print, Jan 28 2009.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Velazquez‐Fernandez, D. , Laurell, C. , Geli, J. , Hoog, A. , Odeberg, J. , Kjellman, M. , Lundeberg, J. , Hamberger, B. , Nilsson, P. , and Backdahl, M. ( 2005) Expression profiling of adrenocortical neoplasms suggests a molecular signature of malignancy. Surgery 138, 1087–1094. [DOI] [PubMed] [Google Scholar]
- 16. Gray, D. A. , Inazawa, J. , Gupta, K. , Wong, A. , Ueda, R. , and Takahashi, T. ( 1995) Elevated expression of Unph, a proto‐oncogene at 3p21.3, in human lung tumors. Oncogene 10, 2179–2183. [PubMed] [Google Scholar]
- 17. Frederick, A. , Rolfe, M. , and Chiu, M. I. ( 1998) The human UNP locus at 3p21.31 encodes two tissue‐selective, cytoplasmic isoforms with deubiquitinating activity that have reduced expression in small cell lung carcinoma cell lines. Oncogene 16, 153–165. [DOI] [PubMed] [Google Scholar]
- 18. Dayal, S. , Sparks, A. , Jacob, J. , Allende‐Vega, N. , Lane, D. P. , and Saville, M. K. ( 2009) Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J. Biol. Chem. 284, 5030–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakamura, T. , Hillova, J. , Mariage‐Samson, R. , and Hill, M. ( 1988) Molecular cloning of a novel oncogene generated by DNA recombination during transfection. Oncogene Res. 2, 357–370. [PubMed] [Google Scholar]
- 20. Masuda‐Robens, J. M. , Kutney, S. N. , Qi, H. , and Chou, M. M. ( 2003) The TRE17 oncogene encodes a component of a novel effector pathway for Rho GTPases Cdc42 and Rac1 and stimulates actin remodeling. Mol. Cell. Biol. 23, 2151–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinu, L. , Masuda‐Robens, J. M. , Robertson, S. E. , Santy, L. C. , Casanova, J. E. , and Chou, M. M. ( 2004) The TBC (Tre‐2/Bub2/Cdc16) domain protein TRE17 regulates plasma membrane‐endosomal trafficking through activation of Arf6. Mol. Cell. Biol. 24, 9752–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oliveira, A. M. , Hsi, B. L. , Weremowicz, S. , Rosenberg, A. E. , Dal Cin, P. , Joseph, N. , Bridge, J. A. , Perez‐Atayde, A. R. , and Fletcher, J. A. ( 2004) USP6 (Tre2) fusion oncogenes in aneurysmal bone cyst. Cancer Res. 64, 1920–1923. [DOI] [PubMed] [Google Scholar]
- 23. van de Luijtgaarden, A. C. , Veth, R. P. , Slootweg, P. J. , Wijers‐Koster, P. M. , Schultze Kool, L. J. , Bovee, J. V. , and van der Graaf, W. T. ( 2009) Metastatic potential of an aneurysmal bone cyst. Virchows Arch. (E‐publication ahead of print, Oct 17 2009.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Song, M. S. , Salmena, L. , Carracedo, A. , Egia, A. , Lo‐Coco, F. , Teruya‐Feldstein, J. , and Pandolfi, P. P. ( 2008) The deubiquitinylation and localization of PTEN are regulated by a HAUSP‐PML network. Nature 455, 813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Masuya, D. , Huang, C. , Liu, D. , Nakashima, T. , Yokomise, H. , Ueno, M. , Nakashima, N. , and Sumitomo, S. ( 2006) The HAUSP gene plays an important role in non‐small cell lung carcinogenesis through p53‐dependent pathways. J. Pathol. 208, 724–732. [DOI] [PubMed] [Google Scholar]
- 26. Row, P. E. , Prior, I. A. , McCullough, J. , Clague, M. J. , and Urbe, S. ( 2006) The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down‐regulation. J. Biol. Chem. 281, 12618–12624. [DOI] [PubMed] [Google Scholar]
- 27. Alwan, H. A. and van Leeuwen, J. E. ( 2007) UBPY‐mediated epidermal growth factor receptor (EGFR) de‐ubiquitination promotes EGFR degradation. J. Biol. Chem. 282, 1658–1669. [DOI] [PubMed] [Google Scholar]
- 28. Niendorf, S. , Oksche, A. , Kisser, A. , Lohler, J. , Prinz, M. , Schorle, H. , Feller, S. , Lewitzky, M. , Horak, I. , and Knobeloch, K. P. ( 2007) Essential role of ubiquitin specific protease 8 (UBPy) for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol. Cell. Biol. 27, 5029–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murray, R. Z. , Jolly, L. A. , and Wood, S. A. ( 2004) The FAM deubiquitylating enzyme localizes to multiple points of protein trafficking in epithelia, where it associates with E‐cadherin and beta‐catenin. Mol. Biol. Cell. 15, 1591–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taya, S. , Yamamoto, T. , Kanai‐Azuma, M. , Wood, S. A. , and Kaibuchi, K. ( 1999) The deubiquitinating enzyme Fam interacts with and stabilizes beta‐catenin. Genes Cells 4, 757–767. [DOI] [PubMed] [Google Scholar]
- 31. Dupont, S. , Mamidi, A. , Cordenonsi, M. , Montagner, M. , Zacchigna, L. , Adorno, M. , Martello, G. , Stinchfield, M. J. , Soligo, S. , Morsut, L. , Inui, M. , Moro, S. , Modena, N. , Argenton, F. , Newfeld, S. J. , and Piccolo, S. ( 2009) FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 136, 123–135. [DOI] [PubMed] [Google Scholar]
- 32. Deng, S. , Zhou, H. , Xiong, R. , Lu, Y. , Yan, D. , Xing, T. , Dong, L. , Tang, E. , and Yang, H. ( 2007) Over‐expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD‐PCR and proteomics. Breast Cancer Res. Treat. 104, 21–30. [DOI] [PubMed] [Google Scholar]
- 33. Lin, C. H. , Chang, H. S. , and Yu, W. C. ( 2008) USP11 stabilizes HPV‐16E7 and further modulates the E7 biological activity. J. Biol. Chem. 283, 15681–15688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schoenfeld, A. R. , Apgar, S. , Dolios, G. , Wang, R. , and Aaronson, S. A. ( 2004) BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol. Cell. Biol. 24, 7444–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sun, W. , Tan, X. , Shi, Y. , Xu, G. , Mao, R. , Gu, X. , Fan, Y. , Yu, Y. , Burlingame, S. , Zhang, H. , Rednam, S. P. , Lu, X. , Zhang, T. , Fu, S. , Cao, G. , Qin, J. , and Yang, J. ( 2009) USP11 negatively regulates TNFalpha‐induced NF‐κB activation by targeting on IκBα. Cell Signal. 22, 386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vos, R. M. , Altreuter, J. , White, E. A. , and Howley, P. M. ( 2009) The ubiquitin‐specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. J. Virol. 83, 8885–8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang, X. , Langelotz, C. , Hetfeld‐Pechoc, B. K. , Schwenk, W. , and Dubiel, W. ( 2009) The COP9 signalosome mediates beta‐catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. J. Mol. Biol. 391, 691–702. [DOI] [PubMed] [Google Scholar]
- 38. Schweitzer, K. , Bozko, P. M. , Dubiel, W. , and Naumann, M. ( 2007) CSN controls NF‐kappaB by deubiquitinylation of IκBα. EMBO J. 26, 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu, M. , Takanashi, M. , Oikawa, K. , Tanaka, M. , Nishi, H. , Isaka, K. , Kudo, M. , and Kuroda, M. ( 2009) USP15 plays an essential role for caspase‐3 activation during Paclitaxel‐induced apoptosis. Biochem. Biophys. Res. Commun. 388, 366–371. [DOI] [PubMed] [Google Scholar]
- 40. Burrows, J. F. , Kelvin, A. A. , McFarlane, C. , Burden, R. E. , McGrattan, M. J. , De la Vega, M. , Govender, U. , Quinn, D. J. , Dib, K. , Gadina, M. , Scott, C. J. , and Johnston, J. A. ( 2009) USP17 regulates Ras activation and cell proliferation by blocking RCE1 activity. J. Biol. Chem. 284, 9587–9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Duex, J. E. and Sorkin, A. ( 2009) RNA interference screen identifies Usp18 as a regulator of epidermal growth factor receptor synthesis. Mol. Biol. Cell. 20, 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Malakhov, M. P. , Malakhova, O. A. , Kim, K. I. , Ritchie, K. J. , and Zhang, D. E. ( 2002) UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277, 9976–9981. [DOI] [PubMed] [Google Scholar]
- 43. Li, Z. , Wang, D. , Messing, E. M. , and Wu, G. ( 2005) VHL protein‐interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF‐1alpha. EMBO Rep. 6, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakagawa, T. , Kajitani, T. , Togo, S. , Masuko, N. , Ohdan, H. , Hishikawa, Y. , Koji, T. , Matsuyama, T. , Ikura, T. , Muramatsu, M. , and Ito, T. ( 2008) Deubiquitylation of histone H2A activates transcriptional initiation via trans‐histone cross‐talk with H3K4 di‐ and trimethylation. Genes Dev. 22, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang, X. Y. , Varthi, M. , Sykes, S. M. , Phillips, C. , Warzecha, C. , Zhu, W. , Wyce, A. , Thorne, A. W. , Berger, S. L. , and McMahon, S. B. ( 2008) The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell‐cycle progression. Mol. Cell. 29, 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao, Y. , Lang, G. , Ito, S. , Bonnet, J. , Metzger, E. , Sawatsubashi, S. , Suzuki, E. , Le Guezennec, X. , Stunnenberg, H. G. , Krasnov, A. , Georgieva, S. G. , Schule, R. , Takeyama, K. , Kato, S. , Tora, L. , and Devys, D. ( 2008) A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 29, 92–101. [DOI] [PubMed] [Google Scholar]
- 47. Glinsky, G. V. , Berezovska, O. , and Glinskii, A. B. ( 2005) Microarray analysis identifies a death‐from‐cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Invest. 115, 1503–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Popov, N. , Wanzel, M. , Madiredjo, M. , Zhang, D. , Beijersbergen, R. , Bernards, R. , Moll, R. , Elledge, S. J. , and Eilers, M. ( 2007) The ubiquitin‐specific protease USP28 is required for MYC stability. Nat. Cell. Biol. 9, 765–774. [DOI] [PubMed] [Google Scholar]
- 49. Zhang, D. , Zaugg, K. , Mak, T. W. , and Elledge, S. J. ( 2006) A role for the deubiquitinating enzyme USP28 in control of the DNA‐damage response. Cell 126, 529–542. [DOI] [PubMed] [Google Scholar]
- 50. Shah, S. P. , Morin, R. D. , Khattra, J. , Prentice, L. , Pugh, T. , Burleigh, A. , Delaney, A. , Gelmon, K. , Guliany, R. , Senz, J. , Steidl, C. , Holt, R. A. , Jones, S. , Sun, M. , Leung, G. , Moore, R. , Severson, T. , Taylor, G. A. , Teschendorff, A. E. , Tse, K. , Turashvili, G. , Varhol, R. , Warren, R. L. , Watson, P. , Zhao, Y. , Caldas, C. , Huntsman, D. , Hirst, M. , Marra, M. A. , and Aparicio, S. ( 2009) Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813. [DOI] [PubMed] [Google Scholar]
- 51. Yuasa‐Kawada, J. , Kinoshita‐Kawada, M. , Rao, Y. , and Wu, J. Y. ( 2009) Deubiquitinating enzyme USP33/VDU1 is required for Slit signaling in inhibiting breast cancer cell migration. Proc. Natl. Acad. Sci. USA 106, 14530–14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yuasa‐Kawada, J. , Kinoshita‐Kawada, M. , Wu, G. , Rao, Y. , and Wu, J. Y. ( 2009) Midline crossing and Slit responsiveness of commissural axons require USP33. Nat. Neurosci. 12, 1087–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. De Pitta, C. , Tombolan, L. , Campo Dell'Orto, M. , Accordi, B. , te Kronnie, G. , Romualdi, C. , Vitulo, N. , Basso, G. , and Lanfranchi, G. ( 2005) A leukemia‐enriched cDNA microarray platform identifies new transcripts with relevance to the biology of pediatric acute lymphoblastic leukemia. Haematologica 90, 890–898. [PubMed] [Google Scholar]
- 54. van Leuken, R. J. , Luna‐Vargas, M. P. , Sixma, T. K. , Wolthuis, R. M. , and Medema, R. H. ( 2008) Usp39 is essential for mitotic spindle checkpoint integrity and controls mRNA‐levels of aurora B. Cell Cycle. 7, 2710–2719. [DOI] [PubMed] [Google Scholar]
- 55. Stegmeier, F. , Rape, M. , Draviam, V. M. , Nalepa, G. , Sowa, M. E. , Ang, X. L. , McDonald, E. R.,3rd , Li, M. Z. , Hannon, G. J. , Sorger, P. K. , Kirschner, M. W. , Harper, J. W. , and Elledge, S. J. ( 2007) Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature 446, 876–881. [DOI] [PubMed] [Google Scholar]
- 56. Zhong, S. , Fields, C. R. , Su, N. , Pan, Y. X. , and Robertson, K. D. ( 2007) Pharmacologic inhibition of epigenetic modifications, coupled with gene expression profiling, reveals novel targets of aberrant DNA methylation and histone deacetylation in lung cancer. Oncogene 26, 2621–2634. [DOI] [PubMed] [Google Scholar]
- 57. Hellerbrand, C. , Bumes, E. , Bataille, F. , Dietmaier, W. , Massoumi, R. , and Bosserhoff, A. K. ( 2007) Reduced expression of CYLD in human colon and hepatocellular carcinomas. Carcinogenesis 28, 21–27. [DOI] [PubMed] [Google Scholar]
- 58. Jenner, M. W. , Leone, P. E. , Walker, B. A. , Ross, F. M. , Johnson, D. C. , Gonzalez, D. , Chiecchio, L. , Dachs Cabanas, E. , Dagrada, G. P. , Nightingale, M. , Protheroe, R. K. , Stockley, D. , Else, M. , Dickens, N. J. , Cross, N. C. , Davies, F. E. , and Morgan, G. J. ( 2007) Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood 110, 3291–3300. [DOI] [PubMed] [Google Scholar]
- 59. Massoumi, R. , Kuphal, S. , Hellerbrand, C. , Haas, B. , Wild, P. , Spruss, T. , Pfeifer, A. , Fassler, R. , and Bosserhoff, A. K. ( 2009) Down‐regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J. Exp. Med. 206, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McCullough, J. , Clague, M. J. , and Urbe, S. ( 2004) AMSH is an endosome‐associated ubiquitin isopeptidase. J. Cell. Biol. 166, 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Itoh, F. , Asao, H. , Sugamura, K. , Heldin, C. H. , ten Dijke, P. , and Itoh, S. ( 2001) Promoting bone morphogenetic protein signaling through negative regulation of inhibitory Smads. EMBO J. 20, 4132–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ibarrola, N. , Kratchmarova, I. , Nakajima, D. , Schiemann, W. P. , Moustakas, A. , Pandey, A. , and Mann, M. ( 2004) Cloning of a novel signaling molecule, AMSH‐2, that potentiates transforming growth factor beta signaling. BMC Cell. Biol. 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dong, Y. , Hakimi, M. A. , Chen, X. , Kumaraswamy, E. , Cooch, N. S. , Godwin, A. K. , and Shiekhattar, R. ( 2003) Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome‐like subunit and its role in DNA repair. Mol. Cell. 12, 1087–1099. [DOI] [PubMed] [Google Scholar]
- 64. Fisch, P. , Forster, A. , Sherrington, P. D. , Dyer, M. J. , and Rabbitts, T. H. ( 1993) The chromosomal translocation t(X;14)(q28;q11) in T‐cell pro‐lymphocytic leukaemia breaks within one gene and activates another. Oncogene 8, 3271–3276. [PubMed] [Google Scholar]
- 65. Zhu, P. , Zhou, W. , Wang, J. , Puc, J. , Ohgi, K. A. , Erdjument‐Bromage, H. , Tempst, P. , Glass, C. K. , and Rosenfeld, M. G. ( 2007) A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol. Cell. 27, 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Verma, R. , Aravind, L. , Oania, R. , McDonald, W. H. , Yates, J. R.,3rd , Koonin, E. V. , and Deshaies, R. J. ( 2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615. [DOI] [PubMed] [Google Scholar]
- 67. Liu, H. , Buus, R. , Clague, M. J. , and Urbe, S. ( 2009) Regulation of ErbB2 receptor status by the proteasomal DUB POH1. PLoS One. 4, e5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nabhan, J. F. and Ribeiro, P. ( 2006) The 19 S proteasomal subunit POH1 contributes to the regulation of c‐Jun ubiquitination, stability, and subcellular localization. J. Biol. Chem. 281, 16099–16107. [DOI] [PubMed] [Google Scholar]
- 69. Jensen, D. E. , Proctor, M. , Marquis, S. T. , Gardner, H. P. , Ha, S. I. , Chodosh, L. A. , Ishov, A. M. , Tommerup, N. , Vissing, H. , Sekido, Y. , Minna, J. , Borodovsky, A. , Schultz, D. C. , Wilkinson, K. D. , Maul, G. G. , Barlev, N. , Berger, S. L. , Prendergast, G. C. , and Rauscher, F. J.,III . ( 1998) BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1‐mediated cell growth suppression. Oncogene 16, 1097–1112. [DOI] [PubMed] [Google Scholar]
- 70. Machida, Y. J. , Machida, Y. , Vashisht, A. A. , Wohlschlegel, J. A. , and Dutta, A. ( 2009) The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF‐1. J. Biol. Chem. 284, 34179–34188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wood, L. D. , Parsons, D. W. , Jones, S. , Lin, J. , Sjoblom, T. , Leary, R. J. , Shen, D. , Boca, S. M. , Barber, T. , Ptak, J. , Silliman, N. , Szabo, S. , Dezso, Z. , Ustyanksky, V. , Nikolskaya, T. , Nikolsky, Y. , Karchin, R. , Wilson, P. A. , Kaminker, J. S. , Zhang, Z. , Croshaw, R. , Willis, J. , Dawson, D. , Shipitsin, M. , Willson, J. K. , Sukumar, S. , Polyak, K. , Park, B. H. , Pethiyagoda, C. L. , Pant, P. V. , Ballinger, D. G. , Sparks, A. B. , Hartigan, J. , Smith, D. R. , Suh, E. , Papadopoulos, N. , Buckhaults, P. , Markowitz, S. D. , Parmigiani, G. , Kinzler, K. W. , Velculescu, V. E. , and Vogelstein, B. ( 2007) The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113. [DOI] [PubMed] [Google Scholar]
- 72. Buchhagen, D. L. , Qiu, L. , and Etkind, P. ( 1994) Homozygous deletion, rearrangement and hypermethylation implicate chromosome region 3p14.3–3p21.3 in sporadic breast‐cancer development. Int. J. Cancer 57, 473–479. [DOI] [PubMed] [Google Scholar]
- 73. Hibi, K. , Liu, Q. , Beaudry, G. A. , Madden, S. L. , Westra, W. H. , Wehage, S. L. , Yang, S. C. , Heitmiller, R. F. , Bertelsen, A. H. , Sidransky, D. , and Jen, J. ( 1998) Serial analysis of gene expression in non‐small cell lung cancer. Cancer Res. 58, 5690–5694. [PubMed] [Google Scholar]
- 74. Hibi, K. , Westra, W. H. , Borges, M. , Goodman, S. , Sidransky, D. , and Jen, J. ( 1999) PGP9.5 as a candidate tumor marker for non‐small‐cell lung cancer. Am. J. Pathol. 155, 711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yamazaki, T. , Hibi, K. , Takase, T. , Tezel, E. , Nakayama, H. , Kasai, Y. , Ito, K. , Akiyama, S. , Nagasaka, T. , and Nakao, A. ( 2002) PGP9.5 as a marker for invasive colorectal cancer. Clin. Cancer Res. 8, 192–195. [PubMed] [Google Scholar]
- 76. Liu, X. , Zeng, B. , Ma, J. , and Wan, C. ( 2009) Comparative proteomic analysis of osteosarcoma cell and human primary cultured osteoblastic cell. Cancer Invest. 27, 345–352. [DOI] [PubMed] [Google Scholar]
- 77. Takase, T. , Hibi, K. , Yamazaki, T. , Nakayama, H. , Taguchi, M. , Kasai, Y. , Ito, K. , Akiyama, S. , Nagasaka, T. , and Nakao, A. ( 2003) PGP9.5 overexpression in esophageal squamous cell carcinoma. Hepatogastroenterology. 50, 1278–1280. [PubMed] [Google Scholar]
- 78. Wicks, S. J. , Haros, K. , Maillard, M. , Song, L. , Cohen, R. E. , Dijke, P. T. , and Chantry, A. ( 2005) The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF‐β signalling. Oncogene 24, 8080–8084. [DOI] [PubMed] [Google Scholar]
- 79. Stanisic, V. , Malovannaya, A. , Qin, J. , Lonard, D. M. , and O'Malley, B. W. ( 2009) OTU Domain‐containing ubiquitin aldehyde‐binding protein 1 (OTUB1) deubiquitinates estrogen receptor (ER) alpha and affects ERalpha transcriptional activity. J. Biol. Chem. 284, 16135–16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Honma, K. , Tsuzuki, S. , Nakagawa, M. , Tagawa, H. , Nakamura, S. , Morishima, Y. , and Seto, M. ( 2009) TNFAIP3/A20 functions as a novel tumor suppressor gene in several subtypes of non‐Hodgkin lymphomas. Blood 114, 2467–2475. [DOI] [PubMed] [Google Scholar]
- 81. Novak, U. , Rinaldi, A. , Kwee, I. , Nandula, S. V. , Rancoita, P. M. , Compagno, M. , Cerri, M. , Rossi, D. , Murty, V. V. , Zucca, E. , Gaidano, G. , Dalla‐Favera, R. , Pasqualucci, L. , Bhagat, G. , and Bertoni, F. ( 2009) The NF‐{kappa}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood 113, 4918–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chanudet, E. , Ye, H. , Ferry, J. , Bacon, C. M. , Adam, P. , Muller‐Hermelink, H. K. , Radford, J. , Pileri, S. A. , Ichimura, K. , Collins, V. P. , Hamoudi, R. A. , Nicholson, A. G. , Wotherspoon, A. C. , Isaacson, P. G. , and Du, M. Q. ( 2009) A20 deletion is associated with copy number gain at the TNFA/B/C locus and occurs preferentially in translocation‐negative MALT lymphoma of the ocular adnexa and salivary glands. J. Pathol. 217, 420–430. [DOI] [PubMed] [Google Scholar]
- 83. Honma, K. , Tsuzuki, S. , Nakagawa, M. , Karnan, S. , Aizawa, Y. , Kim, W. S. , Kim, Y. D. , Ko, Y. H. , and Seto, M. ( 2008) TNFAIP3 is the target gene of chromosome band 6q23.3‐q24.1 loss in ocular adnexal marginal zone B cell lymphoma. Genes Chromosomes Cancer. 47, 1–7. [DOI] [PubMed] [Google Scholar]
- 84. Durkop, H. , Hirsch, B. , Hahn, C. , Foss, H. D. , and Stein, H. ( 2003) Differential expression and function of A20 and TRAF1 in Hodgkin lymphoma and anaplastic large cell lymphoma and their induction by CD30 stimulation. J. Pathol. 200, 229–239. [DOI] [PubMed] [Google Scholar]
- 85. Enesa, K. , Zakkar, M. , Chaudhury, H. , Luong le, A. , Rawlinson, L. , Mason, J. C. , Haskard, D. O. , Dean, J. L. , and Evans, P. C. ( 2008) NF‐kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro‐inflammatory signaling. J. Biol. Chem. 283, 7036–7045. [DOI] [PubMed] [Google Scholar]
- 86. Tran, H. , Hamada, F. , Schwarz‐Romond, T. , and Bienz, M. ( 2008) Trabid, a new positive regulator of Wnt‐induced transcription with preference for binding and cleaving K63‐linked ubiquitin chains. Genes Dev. 22, 528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Forbes, S. A. , Bhamra, G. , Bamford, S. , Dawson, E. , Kok, C. , Clements, J. , Menzies, A. , Teague, J. W. , Futreal, P. A. , and Stratton, M. R. ( 2008) The catalogue of somatic mutations in cancer (COSMIC). Curr. Protoc. Hum. Genet. 10, 10.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Rhodes, D. R. , Kalyana‐Sundaram, S. , Mahavisno, V. , Varambally, R. , Yu, J. , Briggs, B. B. , Barrette, T. R. , Anstet, M. J. , Kincead‐Beal, C. , Kulkarni, P. , Varambally, S. , Ghosh, D. , and Chinnaiyan, A. M. ( 2007) Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9, 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hall, P. S. and Cameron, D. A. ( 2009) Current perspective ‐ trastuzumab. Eur J Cancer. 45, 12–18. [DOI] [PubMed] [Google Scholar]
- 90. Sleijfer, S. , Wiemer, E. , and Verweij, J. ( 2008) Drug Insight: gastrointestinal stromal tumors (GIST)—the solid tumor model for cancer‐specific treatment. Nat. Clin. Pract. Oncol. 5, 102–111. [DOI] [PubMed] [Google Scholar]
- 91. Yano, S. , Wang, W. , Li, Q. , Matsumoto, K. , Sakurama, H. , Nakamura, T. , Ogino, H. , Kakiuchi, S. , Hanibuchi, M. , Nishioka, Y. , Uehara, H. , Mitsudomi, T. , Yatabe, Y. , and Sone, S. ( 2008) Hepatocyte growth factor induces gefitinib resistance of lung adenocarcinoma with epidermal growth factor receptor‐activating mutations. Cancer Res. 68, 9479–9487. [DOI] [PubMed] [Google Scholar]
- 92. Engelman, J. A. , Zejnullahu, K. , Mitsudomi, T. , Song, Y. , Hyland, C. , Park, J. O. , Lindeman, N. , Gale, C. M. , Zhao, X. , Christensen, J. , Kosaka, T. , Holmes, A. J. , Rogers, A. M. , Cappuzzo, F. , Mok, T. , Lee, C. , Johnson, B. E. , Cantley, L. C. , and Janne, P. A. ( 2007) MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043. [DOI] [PubMed] [Google Scholar]
- 93. Mosesson, Y. , Mills, G. B. , and Yarden, Y. ( 2008) Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer. 8, 835–850. [DOI] [PubMed] [Google Scholar]
- 94. Peschard, P. and Park, M. ( 2003) Escape from Cbl‐mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell. 3, 519–523. [DOI] [PubMed] [Google Scholar]
- 95. Urbe, S. ( 2005) Ubiquitin and endocytic protein sorting. Essays Biochem. 41, 81–98. [DOI] [PubMed] [Google Scholar]
- 96. McCullough, J. , Row, P. E. , Lorenzo, O. , Doherty, M. , Beynon, R. , Clague, M. J. , and Urbe, S. ( 2006) Activation of the endosome‐associated ubiquitin isopeptidase AMSH by STAM, a component of the multivesicular body‐sorting machinery. Curr. Biol. 16, 160–165. [DOI] [PubMed] [Google Scholar]
- 97. Komander, D. , Reyes‐Turcu, F. , Licchesi, J. D. , Odenwaelder, P. , Wilkinson, K. D. , and Barford, D. ( 2009) Molecular discrimination of structurally equivalent Lys 63‐linked and linear polyubiquitin chains. EMBO Rep. 10, 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Bowers, K. , Piper, S. C. , Edeling, M. A. , Gray, S. R. , Owen, D. J. , Lehner, P. J. , and Luzio, J. P. ( 2006) Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. J. Biol. Chem. 281, 5094–5105. [DOI] [PubMed] [Google Scholar]
- 99. Clague, M. J. and Urbe, S. ( 2006) Endocytosis: the DUB version. Trends. Cell. Biol. 16, 551–559. [DOI] [PubMed] [Google Scholar]
- 100. Row, P. E. , Liu, H. , Hayes, S. , Welchman, R. , Charalabous, P. , Hofmann, K. , Clague, M. J. , Sanderson, C. M. , and Urbe, S. ( 2007) The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J. Biol. Chem. 282, 30929–30937. [DOI] [PubMed] [Google Scholar]
- 101. Mizuno, E. , Kobayashi, K. , Yamamoto, A. , Kitamura, N. , and Komada, M. ( 2006) A deubiquitinating enzyme UBPY regulates the level of protein ubiquitination on endosomes. Traffic 7, 1017–1031. [DOI] [PubMed] [Google Scholar]
- 102. Buus, R. , Faronato, M. , Hammond, D. E. , Urbe, S. , and Clague, M. J. ( 2009) Deubiquitinase activities required for hepatocyte growth factor‐induced scattering of epithelial cells. Curr. Biol. 19, 1463–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Biggs, P. J. , Wooster, R. , Ford, D. , Chapman, P. , Mangion, J. , Quirk, Y. , Easton, D. F. , Burn, J. , and Stratton, M. R. ( 1995) Familial cylindromatosis (turban tumour syndrome) gene localised to chromosome 16q12‐q13: evidence for its role as a tumour suppressor gene. Nat. Genet. 11, 441–443. [DOI] [PubMed] [Google Scholar]
- 104. Blake, P. W. and Toro, J. R. ( 2009) Update of cylindromatosis gene (CYLD) mutations in Brooke‐Spiegler syndrome: novel insights into the role of deubiquitination in cell signaling. Hum. Mutat. 30, 1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Massoumi, R. and Paus, R. ( 2007) Cylindromatosis and the CYLD gene: new lessons on the molecular principles of epithelial growth control. Bioessays 29, 1203–1214. [DOI] [PubMed] [Google Scholar]
- 106. Gerretsen, A. L. , van der Putte, S. C. , Deenstra, W. , and van Vloten, W. A. ( 1993) Cutaneous cylindroma with malignant transformation. Cancer 72, 1618–1623. [DOI] [PubMed] [Google Scholar]
- 107. Fisher, G. H. , Mones, J. , Gill, M. , Celebi, J. T. , and Geronemus, R. G. ( 2005) Mohs surgical extirpation of a basal cell carcinoma in a patient with familial multiple trichoepitheliomas. Dermatol. Surg. 31, 1458–1461. [DOI] [PubMed] [Google Scholar]
- 108. Brummelkamp, T. R. , Nijman, S. M. , Dirac, A. M. , and Bernards, R. ( 2003) Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF‐κB. Nature 424, 797–801. [DOI] [PubMed] [Google Scholar]
- 109. Kovalenko, A. , Chable‐Bessia, C. , Cantarella, G. , Israel, A. , Wallach, D. , and Courtois, G. ( 2003) The tumour suppressor CYLD negatively regulates NF‐κB signalling by deubiquitination. Nature 424, 801–805. [DOI] [PubMed] [Google Scholar]
- 110. Trompouki, E. , Hatzivassiliou, E. , Tsichritzis, T. , Farmer, H. , Ashworth, A. , and Mosialos, G. ( 2003) CYLD is a deubiquitinating enzyme that negatively regulates NF‐κB activation by TNFR family members. Nature 424, 793–796. [DOI] [PubMed] [Google Scholar]
- 111. Courtois, G. ( 2008) Tumor suppressor CYLD: negative regulation of NF‐κB signaling and more. Cell. Mol. Life Sci. 65, 1123–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sun, S. C. ( 2008) Deubiquitylation and regulation of the immune response. Nat. Rev. Immunol. 8, 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Sun, S. C. ( 2010) CYLD: a tumor suppressor deubiquitinase regulating NF‐kappaB activation and diverse biological processes. Cell Death Differ. 17, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Chen, Z. , Hagler, J. , Palombella, V. J. , Melandri, F. , Scherer, D. , Ballard, D. , and Maniatis, T. ( 1995) Signal‐induced site‐specific phosphorylation targets I kappa B alpha to the ubiquitin‐proteasome pathway. Genes Dev. 9, 1586–1597. [DOI] [PubMed] [Google Scholar]
- 115. Jono, H. , Lim, J. H. , Chen, L. F. , Xu, H. , Trompouki, E. , Pan, Z. K. , Mosialos, G. , and Li, J. D. ( 2004) NF‐kappaB is essential for induction of CYLD, the negative regulator of NF‐kappaB: evidence for a novel inducible autoregulatory feedback pathway. J. Biol. Chem. 279, 36171–36174. [DOI] [PubMed] [Google Scholar]
- 116. Komander, D. , Lord, C. J. , Scheel, H. , Swift, S. , Hofmann, K. , Ashworth, A. , and Barford, D. ( 2008) The structure of the CYLD USP domain explains its specificity for Lys63‐linked polyubiquitin and reveals a B box module. Mol. Cell. 29, 451–464. [DOI] [PubMed] [Google Scholar]
- 117. Massoumi, R. , Chmielarska, K. , Hennecke, K. , Pfeifer, A. , and Fassler, R. ( 2006) Cyld inhibits tumor cell proliferation by blocking Bcl‐3‐dependent NF‐kappaB signaling. Cell 125, 665–677. [DOI] [PubMed] [Google Scholar]
- 118. Westerheide, S. D. , Mayo, M. W. , Anest, V. , Hanson, J. L. , and Baldwin, A. S.,Jr. ( 2001) The putative oncoprotein Bcl‐3 induces cyclin D1 to stimulate G(1) transition. Mol. Cell. Biol. 21, 8428–8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Rocha, S. , Martin, A. M. , Meek, D. W. , and Perkins, N. D. ( 2003) p53 represses cyclin D1 transcription through down regulation of Bcl‐3 and inducing increased association of the p52 NF‐kappaB subunit with histone deacetylase 1. Mol. Cell. Biol. 23, 4713–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Eddy, S. F. , Guo, S. , Demicco, E. G. , Romieu‐Mourez, R. , Landesman‐Bollag, E. , Seldin, D. C. , and Sonenshein, G. E. ( 2005) Inducible IkappaB kinase/IkappaB kinase epsilon expression is induced by CK2 and promotes aberrant nuclear factor‐kappaB activation in breast cancer cells. Cancer Res. 65, 11375–11383. [DOI] [PubMed] [Google Scholar]
- 121. Hutti, J. E. , Shen, R. R. , Abbott, D. W. , Zhou, A. Y. , Sprott, K. M. , Asara, J. M. , Hahn, W. C. , and Cantley, L. C. ( 2009) Phosphorylation of the tumor suppressor CYLD by the breast cancer oncogene IKKepsilon promotes cell transformation. Mol. Cell. 34, 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Stegmeier, F. , Sowa, M. E. , Nalepa, G. , Gygi, S. P. , Harper, J. W. , and Elledge, S. J. ( 2007) The tumor suppressor CYLD regulates entry into mitosis. Proc. Natl. Acad. Sci. USA 104, 8869–8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wickstrom, S. A. , Masoumi, K. C. , Khochbin, S. , Fassler, R. , and Massoumi, R. ( 2009) CYLD negatively regulates cell‐cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. (E‐publication ahead of print, Nov 5 2009.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Geetha, T. , Jiang, J. , and Wooten, M. W. ( 2005) Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol. Cell. 20, 301–312. [DOI] [PubMed] [Google Scholar]
- 125. Gao, J. , Huo, L. , Sun, X. , Liu, M. , Li, D. , Dong, J. T. , and Zhou, J. ( 2008) The tumor suppressor CYLD regulates microtubule dynamics and plays a role in cell migration. J. Biol. Chem. 283, 8802–8809. [DOI] [PubMed] [Google Scholar]
- 126. Zhang, J. , Stirling, B. , Temmerman, S. T. , Ma, C. A. , Fuss, I. J. , Derry, J. M. , and Jain, A. ( 2006) Impaired regulation of NF‐kappaB and increased susceptibility to colitis‐associated tumorigenesis in CYLD‐deficient mice. J. Clin. Invest. 116, 3042–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Boone, D. L. , Turer, E. E. , Lee, E. G. , Ahmad, R. C. , Wheeler, M. T. , Tsui, C. , Hurley, P. , Chien, M. , Chai, S. , Hitotsumatsu, O. , McNally, E. , Pickart, C. , and Ma, A. ( 2004) The ubiquitin‐modifying enzyme A20 is required for termination of Toll‐like receptor responses. Nat. Immunol. 5, 1052–1060. [DOI] [PubMed] [Google Scholar]
- 128. Wertz, I. E. , O'Rourke, K. M. , Zhou, H. , Eby, M. , Aravind, L. , Seshagiri, S. , Wu, P. , Wiesmann, C. , Baker, R. , Boone, D. L. , Ma, A. , Koonin, E. V. , and Dixit, V. M. ( 2004) De‐ubiquitination and ubiquitin ligase domains of A20 downregulate NF‐kappaB signalling. Nature 430, 694–699. [DOI] [PubMed] [Google Scholar]
- 129. Lin, S. C. , Chung, J. Y. , Lamothe, B. , Rajashankar, K. , Lu, M. , Lo, Y. C. , Lam, A. Y. , Darnay, B. G. , and Wu, H. ( 2008) Molecular basis for the unique deubiquitinating activity of the NF‐kappaB inhibitor A20. J. Mol. Biol. 376, 526–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Komander, D. and Barford, D. ( 2008) Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem. J. 409, 77–85. [DOI] [PubMed] [Google Scholar]
- 131. Shembade, N. , Harhaj, N. S. , Parvatiyar, K. , Copeland, N. G. , Jenkins, N. A. , Matesic, L. E. , and Harhaj, E. W. ( 2008) The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin—editing enzyme A20. Nat. Immunol. 9, 254–262. [DOI] [PubMed] [Google Scholar]
- 132. Kato, M. , Sanada, M. , Kato, I. , Sato, Y. , Takita, J. , Takeuchi, K. , Niwa, A. , Chen, Y. , Nakazaki, K. , Nomoto, J. , Asakura, Y. , Muto, S. , Tamura, A. , Iio, M. , Akatsuka, Y. , Hayashi, Y. , Mori, H. , Igarashi, T. , Kurokawa, M. , Chiba, S. , Mori, S. , Ishikawa, Y. , Okamoto, K. , Tobinai, K. , Nakagama, H. , Nakahata, T. , Yoshino, T. , Kobayashi, Y. , and Ogawa, S. ( 2009) Frequent inactivation of A20 in B‐cell lymphomas. Nature 459, 712–716. [DOI] [PubMed] [Google Scholar]
- 133. Schmitz, R. , Hansmann, M. L. , Bohle, V. , Martin‐Subero, J. I. , Hartmann, S. , Mechtersheimer, G. , Klapper, W. , Vater, I. , Giefing, M. , Gesk, S. , Stanelle, J. , Siebert, R. , and Kuppers, R. ( 2009) TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J. Exp. Med. 206, 981–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Prasad, S. , Ravindran, J. , and Aggarwal, B. B. ( 2009) NF‐κB and cancer: how intimate is this relationship. Mol. Cell. Biochem. (E‐publication ahead of print, Oct 8 2009.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chiu, Y. H. , Zhao, M. , and Chen, Z. J. ( 2009) Ubiquitin in NF‐κB signaling. Chem. Rev. 109, 1549–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Pardali, K. and Moustakas, A. ( 2007) Actions of TGF‐beta as tumor suppressor and pro‐metastatic factor in human cancer. Biochim. Biophys. Acta. 1775, 21–62. [DOI] [PubMed] [Google Scholar]
- 137. Izzi, L. and Attisano, L. ( 2004) Regulation of the TGFbeta signalling pathway by ubiquitin‐mediated degradation. Oncogene 23, 2071–2078. [DOI] [PubMed] [Google Scholar]
- 138. Al‐Hakim, A. K. , Zagorska, A. , Chapman, L. , Deak, M. , Peggie, M. , and Alessi, D. R. ( 2008) Control of AMPK‐related kinases by USP9X and atypical Lys(29)/Lys(33)‐linked polyubiquitin chains. Biochem. J. 411, 249–260. [DOI] [PubMed] [Google Scholar]
- 139. Logan, C. Y. and Nusse, R. ( 2004) The Wnt signaling pathway in development and disease. Annu. Rev. Cell. Dev. Biol. 20, 781–810. [DOI] [PubMed] [Google Scholar]
- 140. MacDonald, B. T. , Tamai, K. , and He, X. ( 2009) Wnt/beta‐catenin signaling: components, mechanisms, and diseases. Dev Cell. 17, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Polakis, P. ( 2007) The many ways of Wnt in cancer. Curr. Opin. Genet. Dev. 17, 45–51. [DOI] [PubMed] [Google Scholar]
- 142. Li, M. , Chen, D. , Shiloh, A. , Luo, J. , Nikolaev, A. Y. , Qin, J. , and Gu, W. ( 2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416, 648–653. [DOI] [PubMed] [Google Scholar]
- 143. van der Horst, A. , de Vries‐Smits, A. M. , Brenkman, A. B. , van Triest, M. H. , van den Broek, N. , Colland, F. , Maurice, M. M. , and Burgering, B. M. ( 2006) FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 8, 1064–1073. [DOI] [PubMed] [Google Scholar]
- 144. Cummins, J. M. and Vogelstein, B. ( 2004) HAUSP is required for p53 destabilization. Cell Cycle 3, 689–692. [PubMed] [Google Scholar]
- 145. Meulmeester, E. , Pereg, Y. , Shiloh, Y. , and Jochemsen, A. G. ( 2005) ATM‐mediated phosphorylations inhibit Mdmx/Mdm2 stabilization by HAUSP in favor of p53 activation. Cell Cycle 4, 1166–1170. [DOI] [PubMed] [Google Scholar]
- 146. Maehama, T. and Dixon, J. E. ( 1998) The tumour suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5‐triphosphate. J. Biol. Chem. 273, 13375–13378. [DOI] [PubMed] [Google Scholar]
- 147. Fridberg, M. , Servin, A. , Anagnostaki, L. , Linderoth, J. , Berglund, M. , Soderberg, O. , Enblad, G. , Rosen, A. , Mustelin, T. , Jerkeman, M. , Persson, J. L. , and Wingren, A. G. ( 2007) Protein expression and cellular localization in two prognostic subgroups of diffuse large B‐cell lymphoma: higher expression of ZAP70 and PKC‐beta II in the non‐germinal center group and poor survival in patients deficient in nuclear PTEN. Leuk. Lymphoma 48, 2221–2232. [DOI] [PubMed] [Google Scholar]
- 148. Perren, A. , Komminoth, P. , Saremaslani, P. , Matter, C. , Feurer, S. , Lees, J. A. , Heitz, P. U. , and Eng, C. ( 2000) Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. Am. J. Pathol. 157, 1097–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Zhou, X. P. , Loukola, A. , Salovaara, R. , Nystrom‐Lahti, M. , Peltomaki, P. , de la Chapelle, A. , Aaltonen, L. A. , and Eng, C. ( 2002) PTEN mutational spectra, expression levels, and subcellular localization in microsatellite stable and unstable colorectal cancers. Am. J. Pathol. 161, 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Planchon, S. M. , Waite, K. A. , and Eng, C. ( 2008) The nuclear affairs of PTEN. J. Cell. Sci. 121, 249–253. [DOI] [PubMed] [Google Scholar]
- 151. Colland, F. , Formstecher, E. , Jacq, X. , Reverdy, C. , Planquette, C. , Conrath, S. , Trouplin, V. , Bianchi, J. , Aushev, V. N. , Camonis, J. , Calabrese, A. , Borg‐Capra, C. , Sippl, W. , Collura, V. , Boissy, G. , Rain, J. C. , Guedat, P. , Delansorne, R. , and Daviet, L. ( 2009) Small‐molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 8, 2286–2295. [DOI] [PubMed] [Google Scholar]
- 152. Allende‐Vega, N. , Sparks, A. , Lane, D. P. , and Saville, M. K. ( 2009) MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene. (Epublication ahead of print, Oct 19 2009.). [DOI] [PubMed] [Google Scholar]
- 153. Priolo, C. , Tang, D. , Brahamandan, M. , Benassi, B. , Sicinska, E. , Ogino, S. , Farsetti, A. , Porrello, A. , Finn, S. , Zimmermann, J. , Febbo, P. , and Loda, M. ( 2006) The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Res. 66, 8625–8632. [DOI] [PubMed] [Google Scholar]
- 154. da Silva, S. D. , Cunha, I. W. , Nishimoto, I. N. , Soares, F. A. , Carraro, D. M. , Kowalski, L. P. , and Graner, E. ( 2009) Clinicopathological significance of ubiquitin‐specific protease 2a (USP2a), fatty acid synthase (FASN), and ErbB2 expression in oral squamous cell carcinomas. Oral Oncol. 45, e134–139. [DOI] [PubMed] [Google Scholar]
- 155. Oster, S. K. , Ho, C. S. , Soucie, E. L. and Penn, L. Z. ( 2002) The myc oncogene: MarvelouslY Complex. Adv. Cancer Res. 84, 81–154. [DOI] [PubMed] [Google Scholar]
- 156. Meyer, N. and Penn, L. Z. ( 2008) Reflecting on 25 years with MYC. Nat. Rev. Cancer. 8, 976–990. [DOI] [PubMed] [Google Scholar]
- 157. Dang, C. V. , O'Donnell, K. A. , Zeller, K. I. , Nguyen, T. , Osthus, R. C. , and Li, F. ( 2006) The c‐Myc target gene network. Semin. Cancer Biol. 16, 253–264. [DOI] [PubMed] [Google Scholar]
- 158. Salghetti, S. E. , Kim, S. Y. , and Tansey, W. P. ( 1999) Destruction of Myc by ubiquitin‐mediated proteolysis: cancer‐associated and transforming mutations stabilize Myc. EMBO J. 18, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Tezel, E. , Hibi, K. , Nagasaka, T. , and Nakao, A. ( 2000) PGP9.5 as a prognostic factor in pancreatic cancer. Clin. Cancer Res. 6, 4764–4767. [PubMed] [Google Scholar]
- 160. Lee, Y. M. , Lee, J. Y. , Kim, M. J. , Bae, H. I. , Park, J. Y. , Kim, S. G. , and Kim, D. S. ( 2006) Hypomethylation of the protein gene product 9.5 promoter region in gallbladder cancer and its relationship with clinicopathological features. Cancer Sci. 97, 1205–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Mizukami, H. , Shirahata, A. , Goto, T. , Sakata, M. , Saito, M. , Ishibashi, K. , Kigawa, G. , Nemoto, H. , Sanada, Y. , and Hibi, K. ( 2008) PGP9.5 methylation as a marker for metastatic colorectal cancer. Anticancer Res. 28, 2697–2700. [PubMed] [Google Scholar]
- 162. Sato, F. and Meltzer, S. J. ( 2006) CpG island hypermethylation in progression of esophageal and gastric cancer. Cancer 106, 483–493. [DOI] [PubMed] [Google Scholar]
- 163. Mandelker, D. L. , Yamashita, K. , Tokumaru, Y. , Mimori, K. , Howard, D. L. , Tanaka, Y. , Carvalho, A. L. , Jiang, W. W. , Park, H. L. , Kim, M. S. , Osada, M. , Mori, M. , and Sidransky, D. ( 2005) PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Res. 65, 4963–4968. [DOI] [PubMed] [Google Scholar]
- 164. Kumagai, T. , Akagi, T. , Desmond, J. C. , Kawamata, N. , Gery, S. , Imai, Y. , Song, J. H. , Gui, D. , Said, J. and Koeffler, H. P. ( 2009) Epigenetic regulation and molecular characterization of C/EBPalpha in pancreatic cancer cells. Int. J. Cancer. 124, 827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Wang, W. J. , Li, Q. Q. , Xu, J. D. , Cao, X. X. , Li, H. X. , Tang, F. , Chen, Q. , Yang, J. M. , Xu, Z. D. , and Liu, X. P. ( 2008) Over‐expression of ubiquitin carboxy terminal hydrolase‐L1 induces apoptosis in breast cancer cells. Int. J. Oncol. 33, 1037–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Kim, H. J. , Kim, Y. M. , Lim, S. , Nam, Y. K. , Jeong, J. , and Lee, K. J. ( 2009) Ubiquitin C‐terminal hydrolase‐L1 is a key regulator of tumor cell invasion and metastasis. Oncogene 28, 117–127. [DOI] [PubMed] [Google Scholar]
- 167. Bheda, A. , Yue, W. , Gullapalli, A. , Whitehurst, C. , Liu, R. , Pagano, J. S. , and Shackelford, J. ( 2009) Positive reciprocal regulation of ubiquitin C‐terminal hydrolase L1 and beta‐catenin/TCF signaling. PLoS One. 4, e5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Bheda, A. , Shackelford, J. , and Pagano, J. S. ( 2009) Expression and functional studies of ubiquitin C‐terminal hydrolase L1 regulated genes. PLoS One. 4, e6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Johnston, S. C. , Riddle, S. M. , Cohen, R. E. , and Hill, C. P. ( 1999) Structural basis for the specificity of ubiquitin C‐terminal hydrolases. EMBO J. 18, 3877–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Larsen, C. N. , Krantz, B. A. , and Wilkinson, K. D. ( 1998) Substrate specificity of deubiquitinating enzymes: ubiquitin C‐terminal hydrolases. Biochemistry 37, 3358–3368. [DOI] [PubMed] [Google Scholar]
- 171. Joenje, H. and Patel, K. J. ( 2001) The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2, 446–457. [DOI] [PubMed] [Google Scholar]
- 172. Sasaki, M. S. and Tonomura, A. ( 1973) A high susceptibility of Fanconi's anemia to chromosome breakage by DNA cross‐linking agents. Cancer Res. 33, 1829–1836. [PubMed] [Google Scholar]
- 173. Jacquemont, C. and Taniguchi, T. ( 2007) The Fanconi anemia pathway and ubiquitin. BMC Biochem. 8, S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Zhi, G. , Wilson, J. B. , Chen, X. , Krause, D. S. , Xiao, Y. , Jones, N. J. , and Kupfer, G. M. ( 2009) Fanconi anemia complementation groupFANCD2 protein serine 331 phosphorylation is important for Fanconi anemia pathway function and BRCA2 interaction. Cancer Res. 69, 8775–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Oestergaard, V. H. , Langevin, F. , Kuiken, H. J. , Pace, P. , Niedzwiedz, W. , Simpson, L. J. , Ohzeki, M. , Takata, M. , Sale, J. E. , and Patel, K. J. ( 2007) Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol. Cell 28, 798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Kim, J. M. , Parmar, K. , Huang, M. , Weinstock, D. M. , Ruit, C. A. , Kutok, J. L. , and D'Andrea, A. D. ( 2009) Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev. Cell 16, 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Dang, L. C. , Melandri, F. D. , and Stein, R. L. ( 1998) Kinetic and mechanistic studies on the hydrolysis of ubiquitin C‐terminal 7‐amido‐4‐methylcoumarin by deubiquitinating enzymes. Biochemistry 37, 1868–1879. [DOI] [PubMed] [Google Scholar]
- 178. Horton, R. A. , Strachan, E. A. , Vogel, K. W. , and Riddle, S. M. ( 2007) A substrate for deubiquitinating enzymes based on time‐resolved fluorescence resonance energy transfer between terbium and yellow fluorescent protein. Anal Biochem. 360, 138–143. [DOI] [PubMed] [Google Scholar]
- 179. Ratia, K. , Pegan, S. , Takayama, J. , Sleeman, K. , Coughlin, M. , Baliji, S. , Chaudhuri, R. , Fu, W. , Prabhakar, B. S. , Johnson, M. E. , Baker, S. C. , Ghosh, A. K. , and Mesecar, A. D. ( 2008) A noncovalent class of papain‐like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. USA 105, 16119–16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Ghosh, A. K. , Takayama, J. , Aubin, Y. , Ratia, K. , Chaudhuri, R. , Baez, Y. , Sleeman, K. , Coughlin, M. , Nichols, D. B. , Mulhearn, D. C. , Prabhakar, B. S. , Baker, S. C. , Johnson, M. E. , and Mesecar, A. D. ( 2009) Structure‐based design, synthesis, and biological evaluation of a series of novel and reversible inhibitors for the severe acute respiratory syndrome‐coronavirus papain‐like protease. J. Med. Chem. 52, 5228–5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Mermerian, A. H. , Case, A. , Stein, R. L. , and Cuny, G. D. ( 2007) Structure‐activity relationship, kinetic mechanism, and selectivity for a new class of ubiquitin C‐terminal hydrolase‐L1 (UCH‐L1) inhibitors. Bioorg. Med. Chem. Lett. 17, 3729–3732. [DOI] [PubMed] [Google Scholar]
- 182. Daviet, L. and Colland, F. ( 2008) Targeting ubiquitin specific proteases for drug discovery. Biochimie 90, 270–283. [DOI] [PubMed] [Google Scholar]


