Abstract
Summary: Natural killer (NK) cells are large granular lymphocytes of the innate immune system that participate in the early control of microbial infections and cancer. NK cells can induce the death of autologous cells undergoing various forms of stress, recognizing and providing non‐microbial ‘danger’ signals to the immune system. NK cells are widely distributed in lymphoid and non‐lymphoid organs. NK cell precursors originate from the bone marrow and go through a complex maturation process that leads to the acquisition of their effector functions, to changes in their expression of integrins and chemotactic receptors, and to their redistribution from the bone marrow and lymph nodes to blood, spleen, liver, and lung. Here, we describe the tissue localization of NK cells, using NKp46 as an NK cell marker, and review the current knowledge on the mechanisms that govern their trafficking in humans and in mice.
Keywords: innate immunity, anatomy of the immune system, natural killer cells
Introduction
Natural killer (NK) cells are lymphocytes of the innate immune system that can induce the death of allogeneic cells and autologous cells undergoing various forms of stress, such as upon microbial infection and malignant transformation (1, 2). NK cells express an array of activating and inhibitory receptors, whose engagement allows them to discriminate between target and non‐target cells (3, 4, 5). The repertoire of NK cell receptors complements that of other innate sensors, such as scavanger receptors, Toll‐like receptors (TLRs) or nucleotide oligomerization domain (NOD) proteins. The strategies of NK cell recognition thus broaden the detection of pathogenic situations where microbial ‘danger signals’ are missing in vivo, such as in the case of poorly immunogenic tumors.
Consistent with their role in immune surveillance, NK cells are widely distributed in the body. It is unclear, however, whether this wide distribution is due to their recirculation, due to the existence of NK subsets with different homing capacities, or due to their development at multiples sites. NK cells can also be recruited in various tissues upon inflammation. However, in contrast to B and T cells, the mechanisms governing NK cell trafficking remain poorly dissected.
NK cellwide tissue distribution
It has been long appreciated that NK cells are widely distributed in mammals (6). However, few studies have precisely addressed this distribution. An early study in rat showed that the frequency of large granular lymphocytes (LGLs), including NK and T‐cell subsets, was high in the lung and peripheral blood, superior to that in the spleen, peritoneal exudates, and lymph nodes. LGLs were also found to be absent in the thymus and bone marrow (7). Subsequent studies in the mouse have shown the presence of NK cells, defined as NK1.1+CD3− or DX5+CD3− by flow cytometry, in various organs but never focused on their distribution (8). We revisited this question and measured the percentage and number of mouse NK cells in various organs. Our results confirmed the wide distribution of NK cells in lymphoid and non‐lymphoid organs (Fig. 1). The frequency of NK cells in lymphocytes was found to be the highest in non‐lymphoid organs such as the lung and liver. A similar phenomenon has also been observed for effector memory CD8+ T cells (9), suggesting similar mechanisms of trafficking for these two cell types and the existence of a niche for interleukin‐15 (IL‐15)‐dependent lymphocytes in non‐lymphoid organs. The order for NK cell frequency is lung>liver>peripheral blood mononuclear cells (PBMCs)>spleen>bone marrow (BM)>lymph node (LN)>thymus, where NK cells are almost undetectable. For comparison, NK1.1+ T cells predominate in the liver, while T cells prevail in blood and LN (Fig. 1). Human NK cells also appear to be frequent in non‐lymphoid organs (10, 11).
Figure 1.

Tissue distribution of natural killer (NK), NK1.1+ T, and T cells. Lymphocyte populations were isolated from the indicated organs of 6‐week‐old C57BL/6 female mice, as previously described (33). (A) The percentage of NK1.1+CD3− (NK cells), NK1.1+CD3+ (NK1.1+ T cells), and NK1.1−CD3+ (mature T cells) cells was measured by flow cytometry. (B) Cell numbers of the indicated subsets were obtained by multiplying the respective frequency of each subset by the total number of lymphocytes in the organ. For blood and lymph nodes, we used an estimate number of 10 million cells in peripheral blood mononuclear cells and 100 million cells in LNs. Results show the mean±standard deviation (SD) of six mice for each organ.
The largest number of mouse NK cells, 2–3 million, can be found in the spleen. Significant reservoirs of NK cells may also be found in all other organs tested excluding the thymus (Fig. 1). In humans, it was found that lymph node NK cells outnumber blood NK cells by 10:1 (12), whereas an estimated 1:1 ratio was observed in the mouse. The reason for this discrepancy could be linked to the expression of CCR7 on a subset of human but not on mouse NK cells, as discussed later.
The presence of NK cells in epithelial tissues has been poorly investigated. NK cell numbers are massively increased in the uterus during pregnancy both in humans and mice (13). In humans, CD3–CD56+ NK cells have been detected in the skin of healthy donors (14) and in the lymph draining this tissue (15). An infiltration of NK cells has been reported in lesional atopic dermatitis skin, in both epidermis and dermis, after Malassezia exposure (16), and in the inflammatory skin during the elicitation phase of contact hypersensitivity in a mouse model of allergic contact dermatitis (17). Furthermore, several groups have reported an accumulation of cells expressing an NK cell phenotype in skin psoriatic lesions (18, 19, 20). However, further investigations are required to clearly determine whether infiltrating cells are ‘true’ NK cells or rather NK‐like T cells. In a normal human intestine, CD3− intraepithelial leukocytes expressing NK cell markers such as CD122, CD161, CD2, CD94, CD56, or CD16 have been described (21, 22, 23, 24). Finally, CD3−NK1.1+ cells have been recently identified in cell suspensions prepared from mouse vaginal tissue (25). Moreover, IL‐15−/− and/or RAG2−/−γc −/− mice are more sensitive to genital herpes simplex virus 2 (HSV‐2) infection, suggesting a potential implication of NK cells in the control of infection spreading in mucosal tissues (26, 27).
NK cells in sinuses
In situ visualization of NK cells has been hampered by the lack of specific reagents. Previous attempts to identify NK cells in situ were based on adoptive transfers of fluorescently labeled cells or staining with anti‐NK1.1, anti‐Ly49G2, or anti‐CD49b (28, 29, 30, 31, 32). However, none of the aforementioned antibodies are NK cell specific (NK1.1, CD49b) or expressed on all NK cells (Ly49G2, CD49b). We recently described that the cell surface expression of the activating NK cell receptor NKp46 (CD336) defines at best NK cells across mammalian species, providing the opportunity to look at NK cells in situ on tissue sections using anti‐NKp46 antibodies (33).
In the spleen, NK cells are mostly found in the red pulp at a steady state, thus excluded from the T and B lymphocyte‐rich area (defined by CD3 and CD19 staining) (Fig. 2A).
Figure 2.
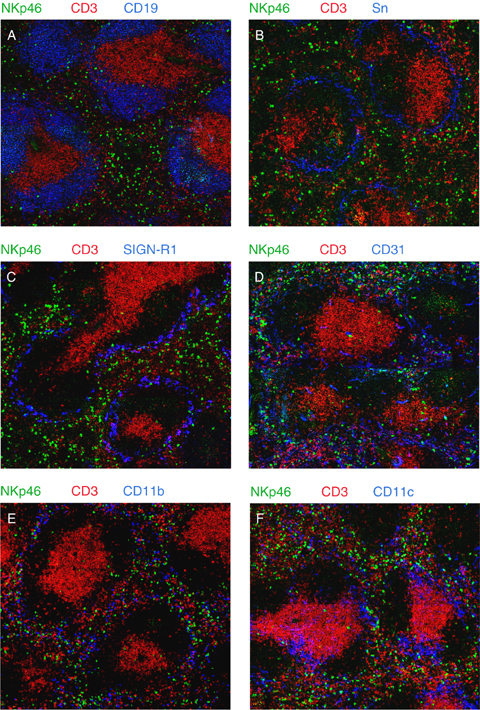
Localization of natural killer (NK) cells in the spleen. Frozen sections of spleen were fixed with acetone and stained with fluorescently coupled antibodies or biotinylated antibodies revealed with fluorochrome‐coupled streptavidin. Anti‐CD3 (145‐2C11), anti‐CD19 (1D3), anti‐CD31 (MEC13‐3), anti‐CD11b (M1/70), and anti‐CD11c (HL3) monoclonal antibodies were from BD Pharmingen (San Diego, CA, USA). Anti‐sialoadhesin (MOMA‐1 for metallophilic macrophages) and anti‐Sign‐R1 (ER‐TR9 for marginal zone macrophages) monoclonal antibodies were obtained from AbD Serotec (Raleigh, NC, USA) and BMA Biomedicals (Augst, Switzerland), respectively. Polyclonal goat anti‐NKp46 (R&D Systems, Minneapolis, MN, USA) was revealed with donkey‐anti‐goat antibody (Invitrogen, Carlsbad, CA, USA), Sections were visualized by confocal microscopy (Zeiss LSM 510 META, Iena, Germany). Panels A–F show representative images for the indicated staining.
Staining for metallophilic macrophages [sialoadhesin (Sn)] (Fig. 2B) and marginal zone macrophages (SignR1) (Fig. 2C) reveals that a few NK cells are also present in the marginal zone but do not go beyond this zone toward the white pulp. Antibodies to NKp46 and CD31 [platelet‐endothelial cell adhesion molecule‐1 (PECAM‐1), an endothelial marker] stain the same regions, suggesting that most splenic NK cells are in fact located inside blood sinuses (Fig. 2D). Occasional NKp46+ cells are found in the white pulp, but staining for CD31 suggests that many of them are in fact located within sinuses or vessels (Fig. 2D). Many other cell types can be found in the vicinity of the ‘NK cell zone’. In particular, most CD11bhigh cells (including macrophages) (Fig. 2E) are found in the red pulp and the marginal zone, whereas many CD11chigh cells (including dendritic cells) (Fig. 2F) are found not only in the white pulp but also in the marginal zone and the red pulp. Cell–cell contacts between NK cells and macrophages (either of the red pulp or of the marginal zone) can be seen on these static images. These observations support the existence of interactions between NK cells and macrophages, as well as NK cells and dendritic cells. Such interactions have been extensively documented in vitro (34, 35, 36, 37) and lead to the mutual regulation of these different cell types in the orchestration of immune responses.
At a steady state, NK cells are also found in the peripheral LN, mostly excluded from T‐ and B‐cell zones (33, authors' unpublished data), consistent with previous studies (29). NK cells are found in perifollicular regions, in the paracortex, and especially in the medulla zone within lymphatic sinuses (29). In the perifollicular region, NK cells are found again in sinuses surrounding the follicles and especially between these follicles, at the site where presumably dendritic cells migrating from tissues arrive to the draining LN through an afferent lymph (29).
Within secondary lymphoid organs, the three types of lymphocytes (T, B, and NK cells) thus localize in distinct compartments. NK cells are preferentially found inside vessels or sinuses, either blood or lymphatics. The same conclusion could also be reached for the liver, as it is known that most hepatic lymphocytes are present in the sinusoids (38) and are absent from the parenchymal space.
NK cell development and maturation
Compelling evidence points to the BM as the primary site of NK cell development in adults at a steady state (6). Moreover, recent articles suggested that LN and thymus could be alternative sources of NK cells (39, 40). The relative contribution of these organs to the pool of NK cells is not known but is expected to be low at a steady state. Indeed, this extramedullary ‘NK‐poiesis’ appears to produce phenotypically distinct NK cells, expressing the α chain of the IL‐7 receptor (40). Such NK cells make up <5% of the total NK cells. Moreover, a careful examination of thymic NK cells reveals that many of them express CD3 intracellularly and share phenotypic and functional features with NK1.1+ T cells, suggesting that they are in fact NK‐like T cells (41). However, in mice where T‐cell development is blocked by genetic means, a very high number of NK cells develop in the thymus, showing the ability of T‐cell precursors to develop into NK cells under particular conditions (40). Furthermore, the normal liver contains a substantial number of immature NK cells, suggesting that NK cell precursors originating in the BM could seed the periphery and develop in situ (42). Thus, it is possible that extramedullary NK cell development may take place under lymphopenic or other conditions that remain to be dissected.
Upon commitment to the NK cell lineage, NK cells go through a complex maturation process that leads to the gradual acquisition of effector functions. Three stages of NK cell maturation can be defined in the mouse based on the expression of CD11b and CD27: CD27highCD11bdull (abbreviated as CD11bdull, the most immature), double‐positive CD11bhighCD27high (abbreviated as DP), and CD11bhigh CD27dull (abbreviated as CD27dull, the most mature) (43, 44, 45). DP and CD27dull NK cells display stronger effector functions than CD11bdull NK cells (113). The repartition of the three NK cell subsets varies with the tissue distribution. Whereas CD11bdull NK cells predominate in BM and LN, DP and especially CD27dull NK cells prevail in the blood, liver, spleen, and lung (44). As discussed later, the expression of different chemotactic receptors participates in the distinct homing capabilities of the NK cell subsets. In human, two subsets of NK cells have been described, CD56dim and CD56bright NK cells, that share many characteristics with CD11bdull and DP/CD27dull NK cells, respectively (45). This resemblance and other observations (12, 46) suggest that CD56bright could be precursors of CD56dim NK cells. Like their mouse counterparts, human CD56bright NK cells are preferentially found in the LN, whereas CD56dim cells are more abundant in the blood and spleen (12).
It is still unknown how and where NK cells complete their maturation. As the three NK cell subsets are found in all organs, it is likely that NK cell precursors or immature NK cells (CD11bdull stage) are seeded from the BM to the periphery, where they further develop in situ under the influence of micro‐environmental factors.
NK cell recirculation
Few data are available on the recirculation of NK cells between organs. Do NK cells traffic through peripheral organs? Do they exit lymphoid and non‐lymphoid organs, or do they reside in tissues? An early study (47) showed that upon intravenous transfer into recipient rats, radiolabeled LGLs preferentially homed into the alveolar walls of the lung and the red pulp of the spleen. LGLs were found, however, to be absent from the thoracic duct and LN (47). Later, adoptive transfer experiments in mouse showed that splenic NK cells can home to the spleen, liver, and BM of recipient mice (48, 49, 50). To gain an insight into NK cell trafficking, we performed an intravenous adoptive transfer of carboxyfluorescein succinimidyl ester (CFSE)‐labeled splenocytes into syngeneic, non‐irradiated recipient mice. Twenty‐four hours post‐transfer, the percentage of NK cells within lymphocytes was measured in the blood, spleen, LN, BM, lung, and liver. The results showed that the distribution of transferred NK cells paralleled that of recipient NK cells (Fig. 3), as did the distribution of NK1.1+ T cells. Moreover, the relative proportion of NK cell subsets (CD11bdull to CD27dull) was similar between recipient and donor NK cells in every organ (data not shown). This simple experiment shows that (i) splenic NK cells are not programmed to home to the spleen but instead recirculate through all NK cell‐containing organs and (ii) NK cell recirculation appears to be subset‐specific: LN and BM are preferentially repopulated by CD11bdull NK cells and the blood, spleen and lung by CD27dull NK cells.
Figure 3.

Recirculation of natural killer (NK) cells. C57BL/6 spleen cells were labeled with 3 μM 5‐(and 6‐)‐carboxyfluorescein succinimidyl ester (CFSE) and injected retro‐orbitally to C57BL/6 recipient mice. One day after transfer, lymphocyte populations were isolated from the indicated organs. The percentage of NK1.1+CD3− (NK cells) and NK1.1+CD3+ (NK1.1+ T cells) in gated CFSE+ (transferred) and CFSE− (recipient) cells was measured by flow cytometry. Results are the mean of six transferred mice in two independent experiments.
The relative contribution of the factors that regulate tissue homing and egress, such as chemotactic receptors or homeostatic mechanisms, in the distribution of NK cells, remains to be addressed. Nevertheless, multiple approaches have been used to address the expression of chemotactic receptors in NK cells. First, several studies have measured the expression of chemotactic receptors on NK cells by flow cytometry, when antibodies were available (51, 52, 53, 54, 55, 56, 57). Second, several strains of mice were created in which a green fluorescent protein cDNA was knocked in to genes encoding chemokine receptors (58, 59). Using such mice, we found that CXCR6 is only expressed by a fraction of CD11bdull NK cells, whereas CX3CR1 is acquired with maturation, almost selectively expressed by CD27dull NK cells (Fig. 4).
Figure 4.
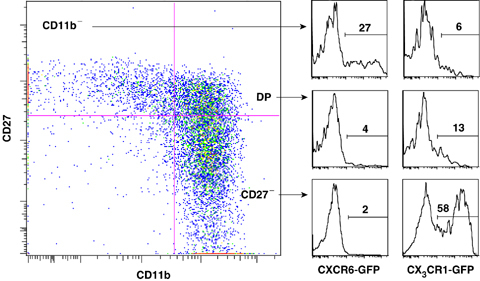
Expression of CXCR6 and CX3CR1 by mouse natural killer (NK) cell subsets. Spleen cells from CXCR6gfp/gfp and CX3CR1gfp/gfp knockin mice were stained for CD3, NK1.1, CD11b, and CD27 and analyzed by flow cytometry. Left panel shows the representative expression of CD11b and CD27 in gated NK1.1+CD3− NK cells. NK cell subsets were gated as indicated and CXCR6‐GFP or CX3CR1‐GFP was measured. Results shown are representative of three mice in each group.
Third, microarray experiments have been performed to measure gene expression at the pan‐genomic level in human (60, 61) and mouse NK cell subsets (Table 1). The findings obtained with these different approaches reveal several trends (Fig. 5).
Table 1.
Relative levels of selectin and integrin transcripts in mouse natural killer (NK) cell subsets
| other name | CD11b− | DP | CD27− | |
|---|---|---|---|---|
| Integrin subunits | ||||
| Itga1 | CD49a | +++ | +++ | +++ |
| Itga2 | CD49b (DX5) | ++ | +++ | +++ |
| Itga3 | CD49c | +++ | +++ | +++ |
| Itga4 | CD49d | ++ | +++ | +++ |
| Itga5 | CD49e | +++ | +++ | +++ |
| Itga6 | CD49f | ++ | ++ | +++ |
| Itga7 | +++ | +++ | +++ | |
| Itgae | CD103 | ++ | ++ | +++ |
| Itgal | CD11a | +++ | +++ | +++ |
| Itgam | CD11b | + | ++ | +++ |
| Itgav | CD51 | +++ | ++ | ++ |
| Itgax | CD11c | +++ | +++ | ++ |
| Itgb1 | VLA‐4 b | ++ | +++ | +++ |
| Itgb2 | CD18 | ++ | ++ | +++ |
| Itgb3 | CD61 | +++ | ++ | + |
| Itgb5 | +++ | +++ | ++ | |
| Itgb7 | ++ | ++ | +++ | |
| Selectins | ||||
| L‐selectin | CD62L | +++ | +++ | +++ |
| P‐selectin ligand | PSGL1 | +++ | +++ | +++ |
Spleen cells from C57BL/6 mice were stained for NK1.1, CD3, CD27 and CD11b expression. NK cell subsets were sorted by flow cytometry, and total RNA was extracted using Qiagen RNAmicro kit (Valencia, CA, USA). The quality of total RNA was assessed using an Agilent Bioanalyzer (Santa Clara, CA, USA). Biotinylated antisense cRNA was prepared by using two cycles of in vitro amplification according to the Affymetrix Small Sample Labeling Protocol II (Affymetrix, Santa Clara, CA, USA). Biotinylated cRNA (15 μg) was fragmented and hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 arrays. All data analyses were performed by using Bioconductor version 1.5 for the statistical software R. Expression values were background corrected, normalized, and summarized by using the default settings of the gcrma package. Accession numbers: The complete microarray data set is available on the CIML website (Vivier lab: http://www.ciml.univ-mrs.fr/Lab/Vivier/Resource.htm). Changes in the level of expression between subsets are highlighted in bold.
Figure 5.

Expression of chemotactic receptors by natural killer (NK) cells. The pattern of expression of chemotactic receptors displayed by human (top) and mouse (bottom) NK cell subsets is based on the reported expression at the protein level by flow cytometry or at the mRNA level using microarray experiments (see references in text). Chemokine aliases: CCL19: ELC, CCL21: SLC, CCL3: MIP1‐α, CCL4: MIP1‐β, CCL5: RANTES, CCL2: MCP1, CCL8: MCP2, CCL7: MCP3, CCL13: MCP4, CXCL12: SDF1, CXCL9: MIG, CXCL10: IP10, CXCL11: I‐TAC, CXCL8: IL‐8, CXCL6: GCP‐2, CX3CL1: fractalkine.
First, a strong similarity in the expression pattern of chemokine receptors on human and mouse NK cell subsets emphasizes the homology between mouse CD11bdull NK cells and CD56bright NK cells, on the one hand, and mouse DP/CD27dull and human CD56dim NK cells on the other. There are, however, important differences. In particular, CCR7 is expressed by human CD56bright NK cells, but no Ccr7 transcripts are detected in mouse CD11bdull NK cells. This difference might account for the distinct representation of NK cells in LN in humans (5% of lymphocytes) and in mice (0.5% of lymphocytes). Second, NK cells are poised to be rapidly recruited on sites of inflammation, as they express receptors for a broad range of inflammatory chemokines (CCR2, CCR5, CXCR3, CX3CR1). Third, the expression of chemotactic receptors switches over maturation in NK cells. In particular, CD27dull/CD56dim NK cells lose CXCR3 (and also CXCR4 in mouse) but acquire CX3CR1 (and also ChemR23 in human) expression, an array of receptors that are anticipated to influence their recirculation and their recruitment upon inflammation.
NK cell trafficking at a steady state
How do NK cells leave the bone marrow?
A role for CXCR4 in human NK cell homing to the BM of reconstituted NOD (nonobese diabetic)/SCID (severe combined immunodeficient) mice was reported (62), suggesting that loss of CXCR4 could contribute to the export of newly produced NK cells. However, CXCR4 seems to be uniformly expressed by NK cells from all organs. Moreover, a recent study showed that mouse treatment with a selective inhibitor of CXCR4 did not induce NK cell mobilization from the BM (63). Another study showed that NK cells were virtually absent from the periphery of CXCR3 knockout mice (64). However, three other studies did not report any defect in the distribution of NK cells in these mice (65, 66, 67), questioning the role of CXCR3 in the emigration of NK cells from BM. More recently, we found that NK cell maturation correlated with the acquisition of S1P5, one of the five sphingosine‐phosphate (S1P) G‐protein‐coupled receptors (113). S1P is a secreted lysophospholipid bound extensively to albumin and other plasma proteins. Coordinated activities of biosynthetic (sphingosine kinases) and biodegradative (sphingosine lyase and phosphatases) enzymes maintain S1P gradients in vivo, with high S1P concentrations in extracellular fluids and low S1P concentrations in tissues (68, 69). In S1p 5‐deficient mice, a drastic decrease in peripheral NK cell counts is observed in the blood, spleen, and lung. This defective homing is NK cell‐intrinsic and correlates with an increased number of NK cells in the BM and LN. S1P5 operates in NK cells as a chemotactic receptor for S1P in vitro, promoting NK cell homing in the blood, spleen, and lung in vivo. The extent of NK cell accumulation in BM and LN correlates with the level of S1P5 expression (113). Altogether, these observations suggest that S1P5 provides an egress signal to NK cells, allowing both their export from the BM and their exit from LN (Fig. 6). Interestingly, Id2 −/− E2a −/− mice have normal BM NK cells but very few mature NK cells in the spleen (70), prompting an investigation of whether these transcription factors control S1P5 expression and/or function.
Figure 6.

Model of mouse natural killer (NK) cell circulation at steady state. NK cells develop mostly in the bone marrow (BM) and, for the CD127+ fraction, also in lymph node (LN) and thymus. NK cells mature from the CD11bdull stage to the double positive CD11bhighCD27high (DP) and further to the CD27dull stage in all organs, starting in the BM (vertical dotted arrows). Upon maturation, they acquire S1P5 expression and exit the BM in a S1P5‐dependent manner. In this model, the more NK cells express S1P5, the more they exit the BM. Once in the periphery, they may return to BM and LN, through a CD62L‐dependent mechanism for the LN.
How do NK cells reach lymphoid organs?
Like other lymphocytes, NK cells enter LNs through high endothelial venules (HEVs) and the spleen through the marginal sinus. CD56bright human NK cells express L‐selectin (CD62L) that allows interaction with glycosylated L‐selectin ligands on HEVs (71). CD62L is expressed at a similar level on mouse NK cell subsets isolated from the LN (44) and is required for mouse NK cell entry in LNs (72) (Fig. 6). Whether CD62L is sufficient for the entry is not known. As discussed previously, most mouse NK cells localize in the sinusoids around T and B‐cell areas, a location reminiscent of that in spleen where NK cells are found in the red pulp surrounding the white pulp. One possibility is that this area is reached by default, in the absence of CCR7 expression. Consistent with this model, human LN CD56 bright NK cells that do express CCR7 are localized, at least in part, in the T‐cell cortex (73). Recently, it was shown that CCR7 is induced on CD56dim cells by IL‐18 in vitro (74), suggesting that under certain conditions, these cells could also traffic to LN.
Transfer experiments have shown that peripheral NK cells may also return to the BM. Central memory T cells preferentially home in this tissue by a mechanism that depends both on CXCL12 (CXCR4 ligand) and on E/P/L selectins (75). Whether this is also the case for NK cells requires further investigation.
How do NK cells reach non‐lymphoid organs?
The distribution of NK cell subsets in the blood, spleen, liver, and lung is very similar, with a majority of CD27dull NK cells. S1P5 deficiency affects NK cells from these compartments in the same way (113). This finding suggests, after their S1P5‐dependent export to the periphery, that NK cells are carried by the blood flow in a S1P5‐independent manner (Fig. 6). A fraction of CD11bdull NK cells expressing CXCR6 is enriched in the liver. Similar to what was shown for NKT cells, these CXCR6 NK cells could patrol hepatic sinusoids through the interaction of CXCR6 with endothelium‐bound CXCL16 (58).
NK cell trafficking under inflammatory conditions
Upon inflammation, NK mouse cells may be recruited in various organs such as the LN, lung, liver, or central nervous system (CNS), and can then extravasate in the parenchyma or cavities (6). The migration of leukocytes from the vascular lumen to tissues depends on a series of sequential molecular interactions between leukocytes and endothelial cells, involving selectins, integrins, and chemokine receptors.
Selectins
In the mouse, L‐selectin and P‐selectin glycoprotein ligand‐1 (PSGL‐1) are expressed at similar levels on NK cell subsets (Table 1). L‐selectin is required for mouse NK cell homing and recruitment to the LN (72). In human, CD56bright but not mature CD56dimCD16+ NK cells also express L‐selectin (71, 76). Sialyl stage‐specific embryonic antigen 1 and an uncharacterized sLex‐bearing receptor may serve as E‐selectin ligands (77, 78). Although PSGL‐1 is expressed on freshly isolated human NK cells, only a minor population of NK cells binds P‐selectin‐immunoglobulin (Ig) (78, 79). Human NK cell differentiation is accompanied by the cell surface expression of a mucin‐like glycoprotein bearing an NK cell‐restricted keratan sulfate‐related lactosamine, the PEN5 epitope (80). The PEN5 carbohydrate decorates PSGL‐1, creating a unique binding site for L‐selectin, which is independent of PSGL‐1 tyrosine sulfation (76). By analogy to the ability of rolling neutrophils to capture free‐flowing neutrophils in a PSGL‐1:L‐selectin‐dependent manner (81), the PEN5 epitope might allow NK cells to interact with other L‐selectin+ leukocytes (e.g. neutrophils, monocytes, or T/B lymphocytes) attached to the inflammatory endothelium to amplify the immune response.
Chemotaxis
The role of chemokine receptors in mouse NK cell recruitment to inflammatory sites has been studied using knockout strains and blocking antibodies (25, 64, 65, 66, 67, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92). In humans, the expression of chemokines and their receptors at inflammatory sites has also provided some information (20, 57, 74, 93, 94, 95, 96) (Table 2).
Table 2.
Experimental models of natural killer (NK) cell trafficking
| Stimulus | Organ where NK cells are recruited | Mechanisms | Reference |
|---|---|---|---|
| Human | |||
| Granulomas in TAP2‐deficient patients | Skin lung | CCR2? | Hanna et al. (93) |
| Oral lichen planus | Oral mucosa | ChemR23 | Parolini et al. (57) |
| Hemolytic uremic syndrome | Kidney | CX3CR1 | Ramos et al. (95) |
| Psoriasis | Skin | CXCR3/CCR5 | Ottaviani et al. (20) |
| Menstruation | Uterus | CXCR3? | Sentman et al. (96) |
| Invasive trophoblasts | Decidua | CXCL12/CXCR4 | Hanna et al. (94) |
| IL‐18 | LN? | CCR7 | Mailliard et al. (74) |
| Mouse | |||
| TLR7/8 ligands, injection sc | Draining LN | CXCR3 | Martin‐Fontecha et al. (87) |
| ConA‐induced hepatitis | Blood, mobilization from spleen | CXCR3 | Wald et al. (65) |
| ConA‐induced hepatitis | Liver | CCR1 | Wald et al. (91) |
| MCMV, i.p. | Liver | MCP1/CCR2 | Hokeness et al. (83) |
| MCMV or poly(I:C), i.p. | Liver | MIP1‐a/CCR5? | Salazar Mather et al. (89) |
| Toxoplasma gondii | Liver, spleen | CCR5 | Khan et al. (85) |
| Dengue virus | Liver | CXCL10/CXCR3? | Chen et al. (82) |
| Bordetella bronchiseptica | Lung | CXCR3 | Widney et al. (66) |
| Bleomycin‐induced lung fibrosis | Lung | CXCR3 | Jiang et al. (64) |
| Intracerebral coronavirus infection | CNS | CXCL10/CXCR3? | Stiles et al. (90) |
| MOG (s.c.)‐induced EAE | CNS | CX3CR1 not CXCR3 | Huang et al., Liu et al. (67, 84) |
| None | Lung | CX3CR1 | Yu et al. (92) |
| B16‐F10 | Lung | CX3CR1 | Yu et al. (92) |
| Invasive aspergillosis | Lung | MCP1/CCR2 | Morrison et al. (88) |
| EL4 tumor cells, s.c. | Tumor site | CX3CR1 | Lavergne et al. (86) |
| HSV2 genital infection | CNS | CCR5 | Thapa et al. (25) |
Compilation of data from the literature describing the recruitment of NK cells in various organs in response to the indicated stimuli. In each case, the receptor involved is indicated in bold.
HSV, herpes simplex virus; CNS, central nervous system; MCMV, mouse cytomegalovirus; LN, lymph node.
Four receptors appear to play a key role in mouse NK cell recruitment following an inflammatory stimulus: CCR2, CCR5, CXCR3, and CX3CR1. These receptors allow NK cells to respond to a large array of inflammatory chemokines such as CCL2, CCL3, CCL5, CCL7, CCL8, CCL9, CCL11, CCL13, CXCL9‐11 and CX3CL1. This broad responsiveness could warrant NK cell recruitment in situations where limited sets of chemokines are expressed. Thus, NK cell recruitment in the same tissue can be mediated by different chemokine receptors. For example, human NK cell recruitment to the epithelia appears to be dependent on CCR2, CCR5, CXCR3, or ChemR23, depending on the inflammatory conditions (20, 57, 93). Not only may NK cells respond to different chemokines, but it also seems that chemokines may act in concert to recruit them. CCR2 and CCR5 are both required for NK cell recruitment to the liver of mouse cytomegalovirus (MCMV)‐infected mice (83, 89). A similar phenomenon had been observed previously for monocytes (97). Importantly, NK cell subsets could also be differentially recruited, depending on the stimulus. Indeed, CX3CR1 but not CXCR3 is required for NK cell recruitment in the CNS in a model of experimental autoimmune encephalomyelitis (67, 84). Conversely, CXCR3 has a central role in NK cell recruitment to the inflammatory LN (87). As CXCR3 and CX3CR1 are expressed in a quasi‐mutually exclusive fashion on NK cell subsets (Fig. 5), this finding suggests that different NK cell subsets may be independently recruited in distinct inflammatory settings. Such a division of labor has already been shown for other types of leukocytes, including monocytes (98) and memory CD8+ T cells (99). Besides chemokines, NK cells have been shown to respond to various other chemotactic compounds that are either induced or augmented upon inflammation. These include lysophosphatidic acid (100), N‐formyl‐methionyl‐leucyl‐phenylalanine (f‐MLP) (101), leukotrienes (102), and C5a (103). The roles of these inflammatory mediators and of their receptors in NK cell recruitment in vivo remain to be thoroughly investigated.
Integrins
Upon activation by chemokines, NK cells interact firmly with the endothelium through integrins. The role of LFA1 (leukocyte function‐associated antigen‐1), as for other leukocytes, has been shown to be central in NK cell extravasation (104). Interestingly, NK cells sequentially express different integrins over development and maturation (43). To gain an insight into this phenomenon, we examined integrin expression at the mRNA level in sorted NK cells subsets, using microarrays. All NK cells express many transcripts of integrins involved in extravasation (Table 1). As reported previously, we found that CD49b and CD11b were acquired while CD51 was decreased upon NK cell maturation. Our data also show that CD11c and especially CD61 (β3 integrin) are decreased upon maturation. The significance of this observation is unclear, but different sets of integrins may contribute to the differential recruitment of NK cell subsets upon inflammation. Besides integrins, NK cell adhesion to the endothelium could also be mediated by CX3CR1. Indeed, it was reported previously that CX3CL1 mediates the rapid capture, integrin‐independent firm adhesion, and activation of circulating leukocytes under flowthrough CX3CR1 (105).
Recruitment of NK cells to the inflammatory liver
NK cells migrate to the liver in response to a variety of stimuli, such as injection of maleic anhydride divinyl ether (MVE‐2), Corynebacterium parvum (106), poly(I:C) (89, 107, 108), MCMV (109), hepatitis B (110), or concanavalin‐A (Con‐A) (111). Concomitant to the increase in liver NK cells, NK cell counts decrease in the spleen and BM, suggesting that these organs serve as reservoirs of NK cells in case of inflammation (65). Various chemokine receptors may orchestrate this recruitment. CXCR3 is partly required in the mobilization of spleen NK cells, while CCR1 is required for the accumulation of NK cells in the liver (65). To gain an insight into the mechanism of spleen NK cell mobilization, we performed immunofluorescence experiments on frozen sections of spleens obtained from mice intravenously injected with lipopolysaccharide (LPS) or Con‐A. A redistribution of splenic NK cells was observed in response to Con‐A injection (Fig. 7).
Figure 7.

Relocalization of natural killer (NK) cells in the spleen in response to Con‐A injection. C57BL/6 mice were treated with indicated stimuli [Con‐A, Sigma (St. Louis, MO, USA): 300 μg i.v., lipopolysaccharide (LPS), Sigma (St. Louis, MO, USA), 25 μg i.v.] for the indicated times. Frozen sections of spleen were fixed with acetone and stained with anti‐CD3, anti‐CD19, (panel A, C) or anti‐CD31 and goat anti‐NKp46 antibodies (panel B, D), and goat anti‐NKp46 antibodies, as described in Fig. 2. Sections were visualized by confocal microscopy.
NK cells appear to migrate from the red pulp to the marginal zone. This relocalization of NK cells was not observed when mice were treated with LPS, although this stimulus induces a massive redistribution of dendritic cells (Fig. 7A and data not shown). After Con‐A injection, NK cells are thus found in close contact with marginal zone macrophages (Fig. 8). It will be of interest to test whether CXCR3 is involved in this process. This phenomenon is transient, as 48 h after Con‐A injection, NK cell distribution in the spleen is normal (Fig. 8).
Figure 8.

Kinetics of natural killer (NK) cell response to Con‐A injection. C57BL/6 mice were treated with Con‐A (300 μg) and sacrificed at the indicated times. Frozen sections of spleen were fixed with acetone and stained with anti‐sialoadhesin and goat anti‐NKp46 antibodies, as described in Fig. 2. Sections were visualized by confocal microscopy.
Recruitment of NK cells to the inflammatory LN
A massive recruitment of NK cells is observed in the draining LN in response to a footpad injection of Leishmania major (29), TLR 7/8 ligands, or LPS‐activated dendritic cells (87). This recruitment is dependent on CXCR3 (87). CXCR3 ligands such as CXCL9 are expressed on the surface of inflammatory HEVs (112). Under inflammatory conditions, most NK cells are recruited preferentially to the T‐cell zone of the LN, near HEVs, in close proximity to dendritic cells (29). NK cell recruitment is required for T‐helper 1 cell polarization of naive T cells activated within the LN (87). In another system, it was found that CD62L was also required for the recruitment on the inflammatory LN in response to a subcutaneous injection of tumor cells (72).
Concluding remarks
The precise knowledge of the anatomy of the immune system is obviously critical for our understanding of immunity. In contrast to T, B, and dendritic cells, the trafficking of NK cells in normal and disease conditions is poorly characterized. Nevertheless, it appears that NK cells patrol lymphoid and non‐lymphoid organs. At a steady state, NK cells are present at a high frequency in the circulation, ready to extravasate to tissues under inflammatory conditions. Besides the roles of S1P5 and chemokine receptors, the set of molecules that govern NK cells trafficking in vivo remain to be identified. The dissection of the mechanisms that regulate NK cell migration may likely provide new perspectives for the manipulation of these cells for therapeutic purposes, as exemplified by the use of the S1P agonist, FTY720, as a T‐ and B‐cell immunosuppressant.
Acknowledgements
The authors thank the mouse functional genomics platform of the Marseille‐Nice Genopole® for immunohistochemistry and Marc Barad et Mathieu Fallet for help with confocal microscopy. The E. V. lab is supported by European Union FP6, LSHB‐CT‐2004‐503319‐Allostem, Ligue Nationale contre le Cancer (‘Equipe labellisée La Ligue’), Agence Nationale de la Recherche (‘Réseau Innovation Biotechnologies’ and ‘Microbiologie Immunologie – Maladies Emergentes’), INSERM, CNRS, and Ministère de l'Enseignement Supérieur et de la Recherche. C. L. is supported by Fondation pour la Recherche Médicale.
References
- 1. Moretta A, Bottino C, Mingari MC, Biassoni R, Moretta L. What is a natural killer cell? Nat Immunol 2002;3:6–8. [DOI] [PubMed] [Google Scholar]
- 2. Raulet DH. Interplay of natural killer cells and their receptors with the adaptive immune response. Nat Immunol 2004;5:996–1002. [DOI] [PubMed] [Google Scholar]
- 3. Cerwenka A, Lanier LL. Ligands for natural killer cell receptors: redundancy or specificity. Immunol Rev 2001;181:158–169. [DOI] [PubMed] [Google Scholar]
- 4. Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 2002;20:217–251. [DOI] [PubMed] [Google Scholar]
- 5. Vivier E, Nunès JA, Vély F. Natural killer cell signaling pathways. Science 2004;306:1517–1519. [DOI] [PubMed] [Google Scholar]
- 6. Trinchieri G. Biology of natural killer cells. Adv Immunol 1989;47:187–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reynolds CW, Timonen T, Herberman RB. Natural killer (NK) cell activity in the rat. I. Isolation and characterization of the effector cells. J Immunol 1981;127:282–287. [PubMed] [Google Scholar]
- 8. Colucci F, Caligiuri MA, Di Santo JP. What does it take to make a natural killer? Nat Rev Immunol 2003;3:413–425. [DOI] [PubMed] [Google Scholar]
- 9. Masopust D, Vezys V, Marzo AL, Lefrancois L. Preferential localization of effector memory cells in nonlymphoid tissue. Science 2001;291:2413–2417. [DOI] [PubMed] [Google Scholar]
- 10. Crispe IN, Mehal WZ. Strange brew: T cells in the liver. Immunol Today 1996;17:522–525. [DOI] [PubMed] [Google Scholar]
- 11. Norris S, et al Resident human hepatic lymphocytes are phenotypically different from circulating lymphocytes. J Hepatol 1998;28:84–90. [DOI] [PubMed] [Google Scholar]
- 12. Ferlazzo G, et al The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig‐like receptors and become cytolytic. J Immunol 2004;172:1455–1462. [DOI] [PubMed] [Google Scholar]
- 13. Moffett‐King A. Natural killer cells and pregnancy. Nat Rev Immunol 2002;2:656–663. [DOI] [PubMed] [Google Scholar]
- 14. Ebert LM, Meuter S, Moser B. Homing and function of human skin {gamma}{delta} T cells and NK cells: relevance for tumor surveillance. J Immunol 2006;176:4331–4336. [DOI] [PubMed] [Google Scholar]
- 15. Hunger RE, Yawalkar N, Braathen LR, Brand CU. The HECA‐452 epitope is highly expressed on lymph cells derived from human skin. Br J Dermatol 1999;141:565–569. [DOI] [PubMed] [Google Scholar]
- 16. Buentke E, et al Natural killer and dendritic cell contact in lesional atopic dermatitis skin – Malassezia‐influenced cell interaction. J Invest Dermatol 2002;119:850–857. [DOI] [PubMed] [Google Scholar]
- 17. O'Leary JG, Goodarzi M, Drayton DL, Von Andrian UH. T cell‐ and B cell‐independent adaptive immunity mediated by natural killer cells. Nat Immunol 2006;7:507–516. [DOI] [PubMed] [Google Scholar]
- 18. Cameron AL, Kirby B, Fei W, Griffiths CE. Natural killer and natural killer‐T cells in psoriasis. Arch Dermatol Res 2002;294:363–369. [DOI] [PubMed] [Google Scholar]
- 19. Gilhar A, et al Psoriasis is mediated by a cutaneous defect triggered by activated immunocytes: induction of psoriasis by cells with natural killer receptors. J Invest Dermatol 2002;119:384–391. [DOI] [PubMed] [Google Scholar]
- 20. Ottaviani C, Nasorri F, Bedini C, De Pita O, Girolomoni G, Cavani A. CD56brightCD16(−) NK cells accumulate in psoriatic skin in response to CXCL10 and CCL5 and exacerbate skin inflammation. Eur J Immunol 2006;36:118–128. [DOI] [PubMed] [Google Scholar]
- 21. Eiras P, et al Intestinal intraepithelial lymphocytes contain a CD3− CD7+ subset expressing natural killer markers and a singular pattern of adhesion molecules. Scand J Immunol 2000;52:1–6. [DOI] [PubMed] [Google Scholar]
- 22. Leon F, Roldan E, Sanchez L, Camarero C, Bootello A, Roy G. Human small‐intestinal epithelium contains functional natural killer lymphocytes. Gastroenterology 2003;125:345–356. [DOI] [PubMed] [Google Scholar]
- 23. Tagliabue A, Befus AD, Clark DA, Bienenstock J. Characteristics of natural killer cells in the murine intestinal epithelium and lamina propria. J Exp Med 1982;155:1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tagliabue A, Luini W, Soldateschi D, Boraschi D. Natural killer activity of gut mucosal lymphoid cells in mice. Eur J Immunol 1981;11:919–922. [DOI] [PubMed] [Google Scholar]
- 25. Thapa M, Kuziel WA, Carr DJ. Susceptibility of CCR5‐deficient mice to genital herpes simplex virus type 2 is linked to NK cell mobilization. J Virol 2007;81:3704–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ashkar AA, Rosenthal KL. Interleukin‐15 and Natural Killer and NKT Cells Play a Critical Role in Innate Protection against Genital Herpes Simplex Virus Type 2 Infection. J Virol 2003;77:10168–10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gill N, Rosenthal KL, Ashkar AA. NK and NKT cell‐independent contribution of interleukin‐15 to innate protection against mucosal viral infection. J Virol 2005;79:4470–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andrews DW, Farrell HE, Densley EH, Scalzo AA, Shellam GR, Degli‐Esposti MA. NK1.1+ cells and murine cytomegalovirus infection: what happens in situ? J Immunol 2001;166:1796–1802. [DOI] [PubMed] [Google Scholar]
- 29. Bajenoff M, et al Natural killer cell behavior in lymph nodes revealed by static and real‐time imaging. J Exp Med 2006;203:619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dokun AO, Chu DT, Yang L, Bendelac AS, Yokoyama WM. Analysis of in situ NK cell responses during viral infection. J Immunol 2001;167:5286–5293. [DOI] [PubMed] [Google Scholar]
- 31. Grundy MA, Sentman CL. Immunodeficient mice have elevated numbers of NK cells in non‐lymphoid tissues. Exp Cell Res 2006;312:3920–3926. [DOI] [PubMed] [Google Scholar]
- 32. Salazar‐Mather TP, Ishikawa R, Biron CA. NK cell trafficking and cytokine expression in splenic compartments after IFN induction and viral infection. J Immunol 1996;157:3054–3064. [PubMed] [Google Scholar]
- 33. Walzer T, et al Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci USA 2007;104:3384–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moretta A. Natural killer cells and dendritic cells: rendezvous in abused tissues. Nat Rev Immunol 2002;2:957–964. [DOI] [PubMed] [Google Scholar]
- 35. Walzer T, Dalod M, Robbins SH, Zitvogel L, Vivier E. Natural‐killer cells and dendritic cells: “l'union fait la force”. Blood 2005;106:2252–2258. [DOI] [PubMed] [Google Scholar]
- 36. Degli‐Esposti MA, Smyth MJ. Close encounters of different kinds: dendritic cells and NK cells take centre stage. Nat Rev Immunol 2005;5:112–124. [DOI] [PubMed] [Google Scholar]
- 37. Newman KC, Riley EM. Whatever turns you on: accessory-cell-dependent activation of NK cells by pathogens. Nat Rev Immunol 2007;7:279–291. [DOI] [PubMed] [Google Scholar]
- 38. Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology 2006;43:S54–S62. [DOI] [PubMed] [Google Scholar]
- 39. Freud AG, et al A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity 2005;22:295–304. [DOI] [PubMed] [Google Scholar]
- 40. Vosshenrich CA, et al A thymic pathway of mouse natural killer cell development characterized by expression of GATA‐3 and CD127. Nat Immunol 2006;7:1217–1224. [DOI] [PubMed] [Google Scholar]
- 41. Stewart CA, Walzer T, Robbins SH, Malissen B, Vivier E, Prinz I. Germ‐line and rearranged Tcrd transcription distinguish bona fide NK cells and NK‐like gammadelta T cells. Eur J Immunol 2007;37:1442–1452. [DOI] [PubMed] [Google Scholar]
- 42. Takeda K, et al TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood 2005;105:2082–2089. [DOI] [PubMed] [Google Scholar]
- 43. Kim S, et al In vivo developmental stages in murine natural killer cell maturation. Nat Immunol 2002;3:523–528. [DOI] [PubMed] [Google Scholar]
- 44. Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol 2006;176:1517–1524. [DOI] [PubMed] [Google Scholar]
- 45. Hayakawa Y, Huntington ND, Nutt SL, Smyth MJ. Functional subsets of mouse natural killer cells. Immunol Rev 2006;214:47–55. [DOI] [PubMed] [Google Scholar]
- 46. Romagnani C, et al CD56brightCD16‐ killer Ig‐like receptor‐ NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol 2007;178:4947–4955. [DOI] [PubMed] [Google Scholar]
- 47. Rolstad B, Herberman RB, Reynolds CW. Natural killer cell activity in the rat. V. The circulation patterns and tissue localization of peripheral blood large granular lymphocytes (LGL). J Immunol 1986;136:2800–2808. [PubMed] [Google Scholar]
- 48. Cooper MA, et al In vivo evidence for a dependence on interleukin 15 for survival of natural killer cells. Blood 2002;100:3633–3638. [DOI] [PubMed] [Google Scholar]
- 49. Prlic M, Blazar BR, Farrar MA, Jameson SC. In vivo survival and homeostatic proliferation of natural killer cells. J Exp Med 2003;197:967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ranson T, Vosshenrich CA, Corcuff E, Richard O, Muller W, Di Santo JP. IL‐15 is an essential mediator of peripheral NK‐cell homeostasis. Blood 2003;101:4887–4893. [DOI] [PubMed] [Google Scholar]
- 51. Campbell JJ, et al Unique subpopulations of CD56+ NK and NK‐T peripheral blood lymphocytes identified by chemokine receptor expression repertoire. J Immunol 2001;166:6477–6482. [DOI] [PubMed] [Google Scholar]
- 52. Imai T, et al Identification and molecular characterization of fractalkine receptor CX3CR1, which mediates both leukocyte migration and adhesion. Cell 1997;91:521–530. [DOI] [PubMed] [Google Scholar]
- 53. Inngjerdingen M, Damaj B, Maghazachi AA. Human NK cells express CC chemokine receptors 4 and 8 and respond to thymus and activation‐regulated chemokine, macrophage‐derived chemokine, and I‐309. J Immunol 2000;164:4048–4054. [DOI] [PubMed] [Google Scholar]
- 54. Inngjerdingen M, Damaj B, Maghazachi AA. Expression and regulation of chemokine receptors in human natural killer cells. Blood 2001;97:367–375. [DOI] [PubMed] [Google Scholar]
- 55. Johnston B, Kim CH, Soler D, Emoto M, Butcher EC. Differential chemokine responses and homing patterns of murine TCR alpha beta NKT cell subsets. J Immunol 2003;171:2960–2969. [DOI] [PubMed] [Google Scholar]
- 56. Nishimura M, et al Dual functions of fractalkine/CX3C ligand 1 in trafficking of perforin+/granzyme B+cytotoxic effector lymphocytes that are defined by CX3CR1 expression. J Immunol 2002;168:6173–6180. [DOI] [PubMed] [Google Scholar]
- 57. Parolini S, et al The role of chemerin in the colocalization of NK and dendritic cell subsets into inflamed tissues. Blood 2007;109:3625–3632. [DOI] [PubMed] [Google Scholar]
- 58. Geissmann F, et al Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol 2005;3:e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jung S, et al Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 2000;20:4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hanna J, Bechtel P, Zhai Y, Youssef F, McLachlan K, Mandelboim O. Novel insights on human NK cells' immunological modalities revealed by gene expression profiling. J Immunol 2004;173:6547–6563. [DOI] [PubMed] [Google Scholar]
- 61. Wendt K, Wilk E, Buyny S, Buer J, Schmidt RE, Jacobs R. Gene and protein characteristics reflect functional diversity of CD56dim and CD56bright NK cells. J Leukoc Biol 2006;80:1529–1541. [DOI] [PubMed] [Google Scholar]
- 62. Beider K, et al Involvement of CXCR4 and IL‐2 in the homing and retention of human NK and NK T cells to the bone marrow and spleen of NOD/SCID mice. Blood 2003;102:1951–1958. [DOI] [PubMed] [Google Scholar]
- 63. Abraham M, et al Enhanced unique pattern of hematopoietic cell mobilization induced by the CXCR4 antagonist 4F‐benzoyl‐TN14003. Stem Cells 2007; in press. [DOI] [PubMed] [Google Scholar]
- 64. Jiang D, et al Regulation of pulmonary fibrosis by chemokine receptor CXCR3. J Clin Invest 2004;114:291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wald O, et al IFN‐gamma acts on T cells to induce NK cell mobilization and accumulation in target organs. J Immunol 2006;176:4716–4729. [DOI] [PubMed] [Google Scholar]
- 66. Widney DP, et al CXCR3 and its ligands participate in the host response to Bordetella bronchiseptica infection of the mouse respiratory tract but are not required for clearance of bacteria from the lung. Infect Immun 2005;73:485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu L, et al Severe disease, unaltered leukocyte migration, and reduced IFN‐gamma production in CXCR3−/− mice with experimental autoimmune encephalomyelitis. J Immunol 2006;176:4399–4409. [DOI] [PubMed] [Google Scholar]
- 68. Cyster JG. Chemokines, sphingosine‐1‐phosphate, and cell migration in secondary lymphoid organs. Annu Rev Immunol 2005;23:127–159. [DOI] [PubMed] [Google Scholar]
- 69. Goetzl EJ, Rosen H. Regulation of immunity by lysosphingolipids and their G protein‐coupled receptors. J Clin Invest 2004;114:1531–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue‐inducing cell development requires Id2‐mediated suppression of E protein activity. J Exp Med 2007;204:1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Frey M, et al Differential expression and function of L‐selectin on CD56bright and CD56dim natural killer cell subsets. J Immunol 1998;161:400–408. [PubMed] [Google Scholar]
- 72. Chen S, Kawashima H, Lowe JB, Lanier LL, Fukuda M. Suppression of tumor formation in lymph nodes by L‐selectin‐mediated natural killer cell recruitment. J Exp Med 2005;202:1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ferlazzo G, et al Distinct roles of IL‐12 and IL‐15 in human natural killer cell activation by dendritic cells from secondary lymphoid organs. Proc Natl Acad Sci USA 2004;101:16606–16611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mailliard RB, et al IL‐18‐induced CD83+CCR7+ NK helper cells. J Exp Med 2005;202:941–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mazo IB, et al Bone marrow is a major reservoir and site of recruitment for central memory CD8+ T cells. Immunity 2005;22:259–270. [DOI] [PubMed] [Google Scholar]
- 76. Andre P, et al Modification of P‐selectin glycoprotein ligand‐1 with a natural killer cell‐restricted sulfated lactosamine creates an alternate ligand for L‐selectin. Proc Natl Acad Sci USA 2000;97:3400–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ohmori K, et al Differentiation‐dependent expression of sialyl stage‐specific embryonic antigen‐1 and I‐antigens on human lymphoid cells and its implications for carbohydrate‐mediated adhesion to vascular endothelium. Blood Article 1993;81:101–111. [PubMed] [Google Scholar]
- 78. Yago T, et al IL‐12 promotes the adhesion of NK cells to endothelial selectins under flow conditions. J Immunol 1998;161:1140–1145. [PubMed] [Google Scholar]
- 79. Snapp KR, Ding H, Atkins K, Warnke R, Luscinskas FW, Kansas GS. A novel P‐selectin glycoprotein ligand‐1 monoclonal antibody recognizes an epitope within the tyrosine sulfate motif of human PSGL‐1 and blocks recognition of both P‐ and L‐selectin. Blood 1998;91:154–164. [PubMed] [Google Scholar]
- 80. Vivier E, et al Developmental regulation of a mucinlike glycoprotein selectively expressed on natural killer cells. J Exp Med 1993;178:2023–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Walcheck B, Moore KL, McEver RP, Kishimoto TK. Neutrophil–neutrophil interactions under hydrodynamic shear stress involve L‐selectin and PSGL‐1. A mechanism that amplifies initial leukocyte accumulation of P‐selectin in vitro. J Clin Invest 1996;98:1081–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chen JP, et al Dengue virus induces expression of CXC chemokine ligand 10/IFN‐gamma‐inducible protein 10, which competitively inhibits viral binding to cell surface heparan sulfate. J Immunol 2006;177:3185–3192. [DOI] [PubMed] [Google Scholar]
- 83. Hokeness KL, Kuziel WA, Biron CA, Salazar‐Mather TP. Monocyte chemoattractant protein‐1 and CCR2 interactions are required for IFN‐alpha/beta‐induced inflammatory responses and antiviral defense in liver. J Immunol 2005;174:1549–1556. [DOI] [PubMed] [Google Scholar]
- 84. Huang D, et al The neuronal chemokine CX3CL1/fractalkine selectively recruits NK cells that modify experimental autoimmune encephalomyelitis within the central nervous system. FASEB J 2006;20:896–905. [DOI] [PubMed] [Google Scholar]
- 85. Khan IA, et al CCR5 is essential for NK cell trafficking and host survival following Toxoplasma gondii infection. PLoS Pathog 2006;2:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lavergne E, et al Fractalkine mediates natural killer‐dependent antitumor responses in vivo. Cancer Res 2003;63:7468–7474. [PubMed] [Google Scholar]
- 87. Martin‐Fontecha A, et al Induced recruitment of NK cells to lymph nodes provides IFN‐gamma for T(H)1 priming. Nat Immunol 2004;5:1260–1265. [DOI] [PubMed] [Google Scholar]
- 88. Morrison BE, Park SJ, Mooney JM, Mehrad B. Chemokine‐mediated recruitment of NK cells is a critical host defense mechanism in invasive aspergillosis. J Clin Invest 2003;112:1862–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Salazar‐Mather TP, Orange JS, Biron CA. Early murine cytomegalovirus (MCMV) infection induces liver natural killer (NK) cell inflammation and protection through macrophage inflammatory protein 1alpha (MIP‐1alpha)‐dependent pathways. J Exp Med 1998;187:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Stiles LN, Hardison JL, Schaumburg CS, Whitman LM, Lane TE. T cell antiviral effector function is not dependent on CXCL10 following murine coronavirus infection. J Immunol 2006;177:8372–8380. [DOI] [PubMed] [Google Scholar]
- 91. Wald O, Weiss ID, Galun E, Peled A. Chemokines in hepatitis C virus infection: pathogenesis, prognosis and therapeutics. Cytokine 2007; in press. [DOI] [PubMed] [Google Scholar]
- 92. Yu YR, Fong AM, Combadiere C, Gao JL, Murphy PM, Patel DD. Defective antitumor responses in CX3CR1‐deficient mice. Int J Cancer 2007;121:316–322. [DOI] [PubMed] [Google Scholar]
- 93. Hanna J, et al Functional aberrant expression of CCR2 receptor on chronically activated NK cells in patients with TAP‐2 deficiency. Blood 2005;106:3465–3473. [DOI] [PubMed] [Google Scholar]
- 94. Hanna J, et al CXCL12 expression by invasive trophoblasts induces the specific migration of CD16− human natural killer cells. Blood 2003;102:1569–1577. [DOI] [PubMed] [Google Scholar]
- 95. Ramos MV, et al Involvement of the fractalkine pathway in the pathogenesis of childhood hemolytic uremic syndrome. Blood 2007;109:2438–2445. [DOI] [PubMed] [Google Scholar]
- 96. Sentman CL, Meadows SK, Wira CR, Eriksson M. Recruitment of uterine NK cells: induction of CXC chemokine ligands 10 and 11 in human endometrium by estradiol and progesterone. J Immunol 2004;173:6760–6766. [DOI] [PubMed] [Google Scholar]
- 97. Tacke F, et al Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J Clin Invest 2007;117:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tacke F, Randolph GJ. Migratory fate and differentiation of blood monocyte subsets. Immunobiology 2006;211:609–618. [DOI] [PubMed] [Google Scholar]
- 99. Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999;401:708–712. [DOI] [PubMed] [Google Scholar]
- 100. Jin Y, Knudsen E, Wang L, Maghazachi AA. Lysophosphatidic acid induces human natural killer cell chemotaxis and intracellular calcium mobilization. Eur J Immunol 2003;33:2083–2089. [DOI] [PubMed] [Google Scholar]
- 101. Pohajdak B, Gomez J, Orr FW, Khalil N, Talgoy M, Greenberg AH. Chemotaxis of large granular lymphocytes. J Immunol 1986;136:278–284. [PubMed] [Google Scholar]
- 102. Pilaro AM, Sayers TJ, McCormick KL, Reynolds CW, Wiltrout RH. An improved in vitro assay to quantitate chemotaxis of rat peripheral blood large granular lymphocytes (LGL). J Immunol Methods 1990;135:213–223. [DOI] [PubMed] [Google Scholar]
- 103. Bottazzi B, Introna M, Allavena P, Villa A, Mantovani A. In vitro migration of human large granular lymphocytes. J Immunol 1985;134:2316–2321. [PubMed] [Google Scholar]
- 104. Bianchi G, et al Migration of natural killer cells across endothelial cell monolayers. J Immunol 1993;151:5135–5144. [PubMed] [Google Scholar]
- 105. Fong AM, et al Fractalkine and CX3CR1 mediate a novel mechanism of leukocyte capture, firm adhesion, and activation under physiologic flow. J Exp Med 1998;188:1413–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wiltrout RH, et al Augmentation of organ‐associated natural killer activity by biological response modifiers. Isolation and characterization of large granular lymphocytes from the liver. J Exp Med 1984;160:1431–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Twilley TA, Mason L, Talmadge JE, Wiltrout RH. Increase in liver‐associated natural killer activity by polyribonucleotides. Nat Immun Cell Growth Regul 1987;6:279–290. [PubMed] [Google Scholar]
- 108. Wang J, Xu JW, Zhang WC, Wei HM, Tian ZG. TLR3 ligand‐induced accumulation of activated splenic natural killer cells into liver. Cell Mol Immunol 2005;2:449–453. [PubMed] [Google Scholar]
- 109. Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar‐Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol 1999;17:189–220. [DOI] [PubMed] [Google Scholar]
- 110. Chen Y, Wei H, Gao B, Hu Z, Zheng S, Tian Z. Activation and function of hepatic NK cells in hepatitis B infection: an underinvestigated innate immune response. J Viral Hepat 2005;12:38–45. [DOI] [PubMed] [Google Scholar]
- 111. Takeda K, Hayakawa Y, Van Kaer L, Matsuda H, Yagita H, Okumura K. Critical contribution of liver natural killer T cells to a murine model of hepatitis. Proc Natl Acad Sci USA 2000;97:5498–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Guarda G, et al L‐selectin‐negative CCR7(−) effector and memory CD8(+) T cells enter reactive lymph nodes and kill dendritic cells. Nat Immunol 2007;8:743–752. [DOI] [PubMed] [Google Scholar]
- 113. Walzer T, et al Natural cell trafficking in vivo requires a dedicated sphingosine 1‐phosphate receptor. Nat Immunol; in press. [DOI] [PubMed] [Google Scholar]


