Summary
The zoonotic introduction of an animal pathogen into the human population and the subsequent extension or alteration of its host range leading to the successful maintenance of the corresponding pathogen by human‐to‐human transmission pose a serious risk for world‐wide health care. Such a scenario occurred for instance by the introduction of simian immunodeficiency viruses into the human population resulting in the human immunodeficiency viruses (HIV) and the subsequent AIDS pandemic or the proposed recent host range switch of the SARS coronavirus from a presently unknown animal species to humans. The occurrence of zoonotic transmissions of animal viruses to humans is a permanent threat to human health and is even increased by changes in the human lifestyle. In this review, the potential of the zoonotic transmission of bovine, feline and equine foamy retroviruses will be discussed in the light of well‐documented cases of zoonotic transmissions of different simian foamy viruses to humans.
Introduction
Foamy viruses (FV), also designated spuma‐ or spumaretroviruses are the least studied group within the family of the Retroviridae (Rethwilm, 2003). During the last few years, interest in FVs increased for certain reasons. First, FVs are considered promising vectors for the targeted delivery and expression of therapeutic genes or antigens, e.g. for the transduction of human haematopoietic stem cells or as vaccine vectors (Vassilopoulos et al., 2001, 2003a; Schwantes et al., 2003). Secondly, the FVs of wild‐ranging and captive non‐human primates, the simian FVs (SFV), have repeatedly been shown to cross host‐range barriers resulting in zoonotic transmission to humans (Schweizer et al., 1997; Heneine et al., 1998). Finally, various features of the FV replication pathway, morphogenesis and particle structure are different from those of the other retroviruses (Linial, 1999; Lecellier and Saib, 2000; Linial and Eastman, 2003; Rethwilm, 2003). These FV‐specific differences even resulted in a novel systematic placement within the family of the Retroviridae that is now subdivided into two major subfamilies: the Spumavirinae and the Orthoretrovirinae which comprise all other known retroviruses including among others the highly pathogenic human immunodeficiency viruses (HIV), the human T‐cell leukaemia viruses (HTLV) and animal pathogens like equine infectious anemia virus, feline and bovine leukaemia viruses, immunodeficiency viruses of cattle and cats, and the ovine enzootic nasal tumour virus (Rethwilm, 2003).
Among the known FVs, the most thoroughly studied member of this group of viruses is the so‐called human foamy virus (HFV), which was the first retrovirus discovered in men, even before the detection and isolation of HTLV‐1 or HIV‐1 (Achong et al., 1971). After intense studies and a long‐lasting debate, it is generally accepted that the prototypic HFV isolate is of chimpanzee origin and that all documented FV infections of men are most likely zoonotically derived from non‐human primates (NHP), (Herchenröder et al., 1994; Heneine et al., 1998). Whereas the zoonotic potential of SFVs from NHPs is under current investigation, little is presently known about the zoonotic potential of FVs from farm and live‐stock animals and pets. As especially in the northern hemisphere contact to the bovine, feline and equine FVs (BFV, FFV, EFV) is far more likely, the potential of corresponding zoonoses will be discussed after a brief summary of the biology of FVs.
Molecular Biology of Foamy Viruses
Like HIV and HTLV, FV are complex retroviruses in terms of their genetic make‐up, their strategies of gene expression and their interactions with the infected hosts and the host cells (Fig. 1, Löchelt, 2003). However, besides these apparent similarities, FVs have several unique features that set them clearly apart from the classical oncoretroviruses and the lentiviruses and they even show striking similarities to the replication strategy of reverse‐transcriptase (RT)‐encoding hepadnaviruses, such as the hepatitis B virus (Rethwilm, 2003). The genome of FVs is a linear plus‐strand RNA ranging from 11 to 13 kb and thus one of the largest among retroviruses. In addition to the gag, pol and env genes and the regulatory elements located in both LTRs which are common to all retroviruses, the FV genome contains up to three additional open reading frames (bel1 to bel3) that are expressed predominantly by the internal promoter (IP) localized in the env gene (Fig. 1, summarized in Löchelt, 2003). The presence of a functionally active and essential IP is a distinguishing feature of FVs (Löchelt et al., 1993). Both FV promoters, the LTR‐promoter and the IP require the expression of the Bel1 post‐transcriptional transactivator for high‐level gene expression and infectivity. The IP has, in comparison with the LTR promoter, a higher basal activity and a higher affinity for Bel1. Thus, after integration of the FV provirus into the host cell genome, the basal activity of the IP leads to the expression of Bel1 that in turn trans‐activates the IP. When the level of Bel1 reaches a critical threshold, transactivation of the LTR‐promoter occurs, leading to the expression of the viral structural and enzymatic proteins and the synthesis of the viral genomic RNA (Meiering and Linial, 2002). This switch in promoter utilization allows a temporal shift in viral gene expression of early regulatory and late structural proteins. In HIV and HTLV, this temporal shift in protein expression is because of the regulatory proteins Rev and Rex, respectively (Cullen, 1992).
Figure 1.
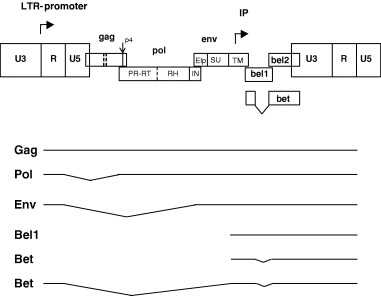
Genetic map and transcription pattern of FVs shown schematically for FFV. The FFV genes gag, pol, env, bel1, and bel2 are depicted as boxes. Sites of proteolytic processing of the precursor proteins are marked by solid and dashed lines. FV PR‐mediated processing results in the Gag p4 peptide and the Pol‐derived PR‐RT, RNase H (RH), and integrase (IN) proteins. The Env precursor is processed by cellular proteases into the novel N‐terminal Env leader protein (Elp) and the SU and TM products. The regulatory long terminal repeats (LTR) are subdivided into the U3, R, and U5 regions (not in scale). The bel 3 gene unique for HFV is not shown. The promoter in the 5′ LTR and the internal promoter (IP) are marked by rectangular arrows pointing into the direction of transcription. The Bet protein consists of Bel1 and Bel2 domains as schematically shown. Below, the LTR‐derived Gag‐, Pol‐, Env‐ and Bet‐encoding transcripts and the IP‐directed Bel1 and Bet mRNAs are schematically shown. Note that a spliced Pol mRNA and the IP transcripts are unique features of FVs. Splicing of the RNAs is indicated by thin broken lines.
Whereas the activities of Bel1 are well established, the function of the abundantly expressed, cytoplasmic Bet protein that consists of bel1 and bel2 sequences (Fig. 1) is not fully understood. Bet is essential for FFV infectivity by unknown mechanisms (Alke et al., 2001). Bet is expressed in FFV‐infected cats in different cells of several organs (J. Weikel, U. Truyen and M. Löchelt, unpublished observations) and antibodies against Bet are considered to have diagnostic value, at least for HFV and the SFVs (Hahn et al., 1994). A bel3 open reading frame is unique to HFV, the functions of Bel2 and Bel3 (if present) are unknown.
Comparison of the FV gag sequences with other RVs showed that the two hallmarks of retroviral Gag proteins are lacking (Fig. 1). These are the major homology regions (MHR) in the capsid (CA) domain and the cysteine‐histidine box(es) in the nucleo‐capsid (NC) domain. In addition, FV Gag‐processing differs from all other retroviruses as the different Gag domains in released particles are not separated by the viral protease, only a C‐terminal cut removing a 4 kDa‐peptide is clearly established (Enssle et al., 1997; Zemba et al., 1998). Thus, only two large Gag proteins are detectable in infected cells and released virions paralleling the situation in hepadnaviruses but not in the other retroviruses. The functions of internal, low efficiency cuts in Gag are presently unclear (Fig. 1; Pfrepper et al., 1999).
Unlike other retroviruses, the Pol proteins are expressed independently of Gag from a spliced, sub‐genomic mRNA (Fig. 1; Bodem et al., 1996; Enssle et al., 1996; Yu et al., 1996). As no Gag‐Pol fusion protein is formed, Pol must be specifically incorporated into newly formed virions (Löchelt and Flügel, 1996). Furthermore, the FV Pol precursor is incompletely cleaved leading to the unique situation in FVs of a protease (PR)– RT fusion protein detectable in infected cells and virions (Pfrepper et al., 1998).
Another unique feature is the onset of reverse transcription of the genomic RNA during the viral life cycle that occurs within the virus‐producing cells before release rather than after entry of the new host cell (Moebes et al., 1997; Yu et al., 1999). Therefore, a substantial portion of released, infectious particles already contains proviral DNA genomes. However, substantial DNA synthesis is detectable directly after infection of new target cells indicating that only part of the viral genome is already proviral DNA (Delelis et al., 2003).
As FVs are considered promising vectors, it might be important to inhibit an uncontrolled vector replication in treated patients/animals. Thus, different nucleoside analogs have been analyzed for their anti‐viral activity. Azidothymidine (AZT), a broadly used RT‐inhibitor, is presently the only known drug effective against HFV, other dideoxynucleosides like dideoxyinosine and dideoxycytidine showed a much lower inhibitory effect for HFV compared with HIV (Rosenblum et al., 2001; Yvon‐Groussin et al., 2001).
In contrast to the other known retroviruses, FV Env proteins are absolutely required for particle budding (Baldwin and Linial, 1998; Pietschmann et al., 1999) most probably because of a highly specific interaction between a distinct structure formed by the N‐terminal Gag sequences, the FV MA‐layer and the novel Env leader protein that is a component of released FV particles (Fig. 2a–c.; Wilk et al., 2001).
Figure 2.
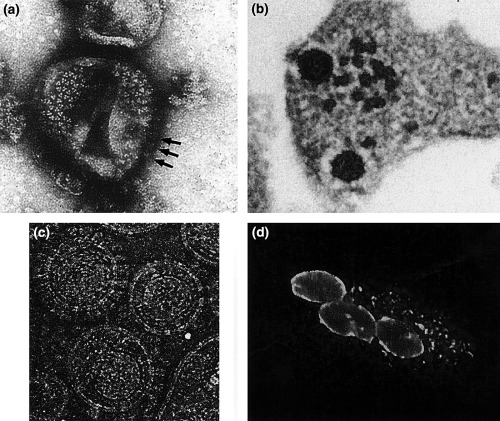
FV morphology and FV‐specific cytopathic effects. (a) Electron microscopy (EM) image of negative‐stained trimeric Env glycoprotein complexes (arrows indicate lateral views) arranged in hexameric rings on the surface of released HFV particles. (b) Thin‐section EM image of FFV particles budding from infected CRFK cells. The electron‐dense preformed capsids contact Env glycoprotein‐complexes in the cellular membrane before particle budding. (c) Cryo‐EM image of purified FFV particles displaying (from the centre to the periphery of the particle) the polygonal capsid, the MA‐layer following the shape of the capsid, the viral lipid membrane, and the outer Env surface glycoproteins (for comparison see also Wilk et al., 2001). (d) Small, early syncytium of FFV‐infected and bystander CRFK cells. FFV Gag protein located at the nuclear membrane and in cytoplasmic aggregates was visualized by indirect immunofluorescence using a Gag‐specific antiserum. Images were kindly provided by Thomas Wilk (a), Matthias Floetenmeyer (b), John Briggs and Brent Gowen (c), and Astrid Schwantes (d).
Sequence analysis of genomes of different FV revealed that pol is the most conserved gene, while gag diverges more than env (Tobaly‐Tapiero et al., 2000). In all other retroviruses, env is less conserved than gag (Wang and Mulligan, 1999). Within an FV species, the genomes are highly conserved (even after zoonotic transmission) showing only minute genetic variation (Schweizer et al., 1999; Phung et al., 2001). Only for FFV two distinct serotypes are known (Winkler et al., 1998). Most of the serotype‐specific differences are confined to the carboxy‐terminal part of the external Env surface (SU) domain.
Biology of Foamy Viruses
FVs were shown to have a wide tissue tropism and can be propagated in vitro in a variety of cell types from different species (Hill et al., 1999). The large range of cells permissive for FV infections probably reflects the ubiquity of the viral receptor. In cell cultures FVs are often highly cytopathic/lytic (Fig. 2d.), however, in defined cells also persistent, continuous replication without destruction of the host cells has been described (Meiering et al., 2001). In naturally infected hosts, FVs cause a life‐long persistent infection without obvious disease symptoms (Saib, 2003). Due to this apparently benign persistence in the naturally infected host, FVs are considered a‐pathogenic. An association of FFV infection with renal failure in cats has been discussed, however, the relevance of this finding is presently unclear. Although apparently a‐pathogenic under natural conditions, FVs possess a significant disease potential as determined in transgenic mice (Aguzzi, 1993). Strong cellular pathology in the form of syncytia development in the brain and neurological symptoms were consistently detectable in these mice (Tschopp et al., 1996). However, transgenic animals can be only considered as an experimental system that may not reflect the authentic situation.
Experimental or naturally occurring FV infections are known to induce a strong humoral immune response against different structural and non‐structural proteins. Reactivities against Gag, Pol, Env and Bet start within weeks after infection, increase with time and remain relatively stable at high levels over long periods indicating that the immune system is constantly or periodically challenged by all viral antigens (Alke et al., 2000). This pattern of reactivity is indicative of a constant viral gene expression probably associated with the continuous production of viral infectivity. In contrast, humoral immunity against lentiviruses is initially directed against Env, and antibodies specific for other viral antigens appear much later. In line with the idea of a productive FV persistence and not a classical viral latency associated with a more or less restricted viral gene expression, free‐ranging, naturally FFV‐infected cats were always positive for FFV‐specific DNA, antibodies and viral infectivity detected by co‐cultivation (Winkler et al., 1998).
Although neutralizing, isolate‐specific antibodies are clearly induced in the infected host, there is no sign of antigenic drift of the virus as the genetic heterogeneity within a FV species or the two known FFV sero‐types is minimal (Winkler et al., 1998). Whereas the humoral immunity against FVs is partially characterized and reactivity against Gag and Bet are considered as diagnostic markers (Alke et al., 2000; Hussain et al., 2003), the contribution of the cellular immune system in controlling persistence and the induction of a mucosal immunity are discussed but currently unknown (Schwantes et al., 2003).
Zoonotic Potential of Simian Foamy Viruses from NHPs
After isolation of the prototypic HFV isolate from an African naso‐pharynx carcinoma patient (Achong et al., 1971), an intensive search for HFV infection in different sick and healthy human populations was initiated (Schweizer et al., 1995; Ali et al., 1996). At present, no consistent and generally accepted association of the detection of HFV or HFV‐specific markers of infection and a defined disease is known although different disorders like Grave's disease, thyreoiditis and others had initially been suggested (Lagaye et al., 1992; Saib et al., 1994; Schweizer et al., 1994). In addition, it has been established by now that the prevalence of the prototypic HFV isolate in the general human population is extremely low in contrast to initial findings implicating geographic differences in its distribution with areas of relatively high incidence. Re‐evaluation of these initial findings did not establish those patterns (Schweizer et al., 1994, 1995; Ali et al., 1996).
During sequencing of FVs from NHPs, especially those obtained from chimpanzees, the so‐called SFVcpz, it became apparent that the HFV isolate and different SFVcpz are almost identical (Herchenröder et al., 1994). It was thus assumed that this single HFV isolate is simply the result of a chimpanzee‐derived FV contamination of the cell cultures used to propagate the HFV isolate. This interpretation was premature as extensive serological studies using sera from animal caretakers being occupationally exposed to NHPs showed that about 2–4% had clear reactivity against HFV whereas markers for zoonotic infection with other simian retroviruses were absent or extremely rare (Heneine et al., 1998; Sandstrom et al., 2000; Brooks et al., 2002). In these and other studies it was shown that zoonotic transmission was possible not only with SFVcpz but also with SFVs from African green monkeys, baboons, gorillas, mandrills and macaques (Heneine et al., 1998; Sandstrom et al., 2000; Brooks et al., 2002; Wolfe et al., 2003). Especially the finding that repeated transmission events of different, genetically and antigenetically distinct SFVs had occurred, clearly settled the point that SFVs possess a considerable zoonotic potential, significantly higher than that of the other NHP retroviruses tested (Heneine et al., 1998). From the known cases it appears that deep bites and possibly infectious saliva from the animals, bush‐meat preparation and transplantation of baboon organs resulted in SFV transmissions (Allan et al., 1998; Heneine et al., 1998; Wolfe et al., 2003). The acquired SFVs induce a persistent infection in men with persisting SFV‐specific antibodies and the propagation of intact and defective proviral genomes (Callahan et al., 1999). In some cases, even infectious virus had been re‐isolated (Schweizer et al., 1997; Boneva et al., 2003).
In none of the known SFV/HFV infected individuals, any symptoms of an SFV‐induced disease were detectable, despite the evidence of a long‐lasting SFV infection (summarized in Heneine et al., 2003). Whereas the initially infected person clearly showed all markers of FV infection, neither family members including the spouse nor blood‐product recipients scored SFV‐positive using different tests (Boneva et al., 2002, 2003). It thus appears that zoonotically infected humans are dead‐end hosts. However, care should be taken before drawing conclusions as the number of well studied cases is about 20, a number that does not allow to determine the frequency of further transmission events and that is too small to estimate the disease potential in men.
The reagents used for the detection of zoonotically acquired SFVs are primarily based on proteins (for ELISA and immuno‐blotting) and DNA sequences (PCR amplification) from the HFV isolate (Schweizer et al., 1995; Heneine et al., 1998). Thus, zoonotic infection with distantly related SFVs might have escaped detection; only a recently established immuno‐blot addresses this question by using a combination of HFV‐ and SFV‐derived antigens (Hussain et al., 2003). In this context it is worth mentioning that in African hunters, markers of SFV infections have recently been detected indicating that bush meat hunting and butchering may also result in zoonotic transmission of SFVs (Wolfe et al., 2003). Considering the wide‐spread use and consumption of bush meat in Central Africa, this part of the world harbours an increased risk of emerging human FVs. Importantly, the entry of HIV into the human population followed almost the same mechanisms and co‐infection with this or other immuno‐suppressive pathogens may even increase the zoonotic risk of SFVs (Weiss, 2001).
Potential of Zoonotic Transmission of Bovine, Feline and Equine Foamy Viruses
At present, three non‐primate FVs have been studied and characterized in vitro and, to a much lesser extent, in vivo: the bovine (BFV), equine (EFV) and feline foamy virus (FFV) (summarized in Saib, 2003). FVs from other species like sea lions and hamsters are not further considered as only limited information are presently available.
In the developed so called First World countries, exposure to NHP and the risk of zoonotic SFV transmission is almost entirely restricted to few laboratory workers and animal (NHP) caretakers. In contrast, in these countries, exposure to the pathogens of companion (horses and cats) or life‐stock animals (cattle) is much more likely and potentially affects almost the whole population. The zoonotic risk arises either by contacting FFV‐, BFV‐ and EFV‐infected animals or, alternatively, by food or medical products directly or indirectly derived from the FV‐infected hosts. BFV and FFV are highly prevalent in their natural host (between 30 and 70% of the outgrown animals are sero‐positive) making contacts between FV‐infected animals and human very likely (summarized in Saib, 2003). Presently, no corresponding data for EFV are available.
However, the risk of zoonotic inter‐species transmission events is not only determined by the extent and mode of exposure to the heterologous pathogen but also by the make‐up of the virus. Whereas the virus‐encoded proteins are the primary targets of the adaptive, humoural and cellular immune systems, the structures determined by the host, for instance the glycosylation pattern and host‐derived, virion‐associated proteins, are directly identified by mechanisms of innate immunity or by passively adapted cross‐reacting immune mechanisms (Burton, 2002). Thus, the risk of zoonotic transmission directly increases with the genetic relatedness of the authentic and novel host. This makes, for instance because of conserved glycosylation patterns, simian to human transmissions far more likely than those from distant species like cat, cattle and horse (Saib, 2003).
For FFV, direct contact to infected cats appears to be the most reasonable way for a potential zoonotic transmission. As infectious FFV can be reproducibly recovered from oral swabs of infected cats (Alke et al., 2000), cat bites intrinsically carry the risk of FFV transmission by breaking the intact skin barrier and allowing access to susceptible cells, for instance leukocytes. However, also non‐aggressive, social behaviour like licking has been considered to be a route of cat‐to‐cat spread (Winkler et al., 1999). Two studies enrolling veterinarians and others occupationally exposed to cats did not reveal any hint for a zoonotic transmission of FFV (Winkler et al., 1997; Butera et al., 2000). However, this apparent resistance against transmission may be not as effective in children with a developing immune system or in immuno‐compromised individuals. The juvenile exposure to cats and their pathogens has been considered as a risk factor of developing multi‐factorial, complex diseases or disease complexes in humans, for instance schizophrenia (Torrey and Yolken, 1995).
Similarly, potential zoonotic infection with EFV appears to be limited to those having direct contact to horses. Whether EFV is present in the oral cavity of horses, a prerequisite of infection by bites, is not known. A second potential route of EFV exposure may be by butchering and meat consumption.
With BFV, the above‐mentioned and additional modes of zoonotic transmission can be anticipated. First, spread of BFV is considered to take place predominantly via spread of BFV‐containing saliva (summarized in Saib, 2003). For the transmission of BFV, non‐aggressive social contacts and inhalation of aerosols (licking or sneezing) have been demonstrated (Johnson et al., 1988) indicating that BFV spread does not depend on aggressive behaviour and biting, thus increasing the probability of zoonotic transmissions. Secondly, milk has been shown to contain FV‐like particles and beef and other cattle‐derived foodstuff may also contain infectious BFV (summarized in Saib, 2003). In addition, butchering with the associated risk of skin injuries or aerosol production and inhalation may increase the exposure to BFV. Finally, medical products derived from cell cultures supplemented with bovine serum (e.g. viral vectors) or directly cattle‐derived therapeutics pose a risk of BFV contamination and thus careful testing and inactivation appears highly mandatory. Similar routes of transmission have been suggested for the transmission of the agent of bovine spongiform encephalopathy to humans (Ludwig et al., 2003).
Finally, FV vectors derived from the prototypic HFV and SFVs are currently constructed for use in humans and FFV‐derived vectors showed their potential to efficiently vaccinate cats (Vassilopoulos et al., 2001, 2003b; Mergia and Heinkelein, 2003; Schwantes et al., 2003). Considering the upcoming application of FV‐based vectors in farm and life‐stock animals, the zoonotic risk has to be carefully evaluated.
In conclusion, the issues summarized in Table 1 should be addressed to determine the potential and possible consequences of FFV, BFV and EFV zoonoses.
Table 1.
Determination of the zoonotic potential of feline, bovine and equine foamy virus
| 1 | Establishing species‐specific and group‐specific screening systems for high‐throughput analysis of human serum samples (e.g. enzyme‐ linked immuno‐sorbent assay, ELISA) |
| 2 | Establishing confirmatory, species‐specific serological test (e.g. immuno‐blotting tests) |
| 3 | Establishing independent, PCR‐based detection systems |
| 4 | Testing of samples from individuals with increased exposition probability |
| 5 | Testing of samples from patients where exposition to cat, bovine, or equine viruses has been implicated in disease |
Summary and Outlook
At present, no diagnostic and experimental data support the idea of intensive zoonotic transmission of FFV, BFV, or EFV into humans. However, recent zoonoses have significantly raised the public and health‐system awareness for such scenarios with unpredictable and even fatal consequences. It is generally accepted that zoonoses will become a novel challenge for the public and international health systems and a scientific preparedness is a prerequisite to find appropriate solutions for these threats (Ludwig et al., 2003). The emerging insights into the zoonotic potential of different primate FVs, the substantial changes in lifestyle and medical technologies and the availability of data and reagents for the setup of novel and sensitive detection systems make it possible to adequately address these issues of scientific and medical interest.
Acknowledgements
We thank Silke Bachmann (University of Heidelberg), Nadine Kirchner and Roman Wirtz (DKFZ) for critically reading the manuscript and Harald zur Hausen for continuous support. Work in the corresponding author's lab is supported by ‘The Stanley Medical Research Institute’, Bethesda, USA.
References
- Achong, B. G. , Mansell P. W., Epstein M. A., and Clifford P., 1971: An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Cancer Inst. 46, 299–307. [PubMed] [Google Scholar]
- Aguzzi, A. , 1993: The foamy virus family: molecular biology, epidemiology and neuropathology. Biochim. Biophys. Acta 1155, 1–24. [DOI] [PubMed] [Google Scholar]
- Ali, M. , Taylor G. P., Pitman R. J., Parker D., Rethwilm A., Cheingsong‐Popov R., Weber J. N., Bieniasz P. D., Bradley J., and McClure M. O., 1996: No evidence of antibody to human foamy virus in widespread human populations. AIDS Res. Hum. Retroviruses 12, 1473–1483. [DOI] [PubMed] [Google Scholar]
- Alke, A. , Schwantes A., Zemba M., Flügel R. M., and Löchelt M., 2000: Characterization of the humoral immune response and virus replication in cats experimentally infected with feline foamy virus. Virology 275, 170–176. [DOI] [PubMed] [Google Scholar]
- Alke, A. , Schwantes A., Kido K., Flötenmeyer M., Flügel R. M., and Löchelt M., 2001: The bet gene of feline foamy virus is required for virus replication. Virology 287, 310–320. [DOI] [PubMed] [Google Scholar]
- Allan, J. S. , Broussard S. R., Michaels M. G., Starzl T. E., Leighton K. L., Whitehead E. M., Comuzzie A. G., Lanford R. E., Leland M. M., Switzer W. M., and Heneine W., 1998: Amplification of simian retroviral sequences from human recipients of baboon liver transplants. AIDS Res. Hum. Retroviruses 14, 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, D. N. , and Linial M. L., 1998: The roles of Pol and Env in the assembly pathway of human foamy virus. J. Virol. 72, 3658–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodem, J. , Löchelt M., Winkler I., Flower R. P., Delius H., and Flügel R. M., 1996: Characterization of the spliced pol transcript of feline foamy virus: the splice acceptor site of the pol transcript is located in gag of foamy viruses. J. Virol. 70, 9024–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneva, R. S. , Grindon A. J., Orton S. L., Switzer W. M., Shanmugam V., Hussain A. I., Bhullar V. B., Chamberland M. E., Heneine W., Folks T. M., and Chapman L. E., 2002: Simian foamy virus infection in a blood donor. Transfusion. 42, 886–891. [DOI] [PubMed] [Google Scholar]
- Boneva, R. S. , Switzer W. M., Spira T. J., Shanmugam V., Bhullar V. B., Lam L., Hussain A. I., Cummins J. E., Heneine W., Folks T. M., and Chapman L. E., 2003: Interim Results of a prospective cohort study of persons infected with simian foamy virus (SFV) 11th International Conference on Human Retrovirology: HTLV and Related Viruses, San Francisco. [Google Scholar]
- Brooks, J. I. , Rud E. W., Pilon R. G., Smith J. M., Switzer W. M., and Sandstrom P. A., 2002: Cross‐species retroviral transmission from macaques to human beings. Lancet 360, 387–388. [DOI] [PubMed] [Google Scholar]
- Burton, D. R. , 2002: Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2, 706–713. [DOI] [PubMed] [Google Scholar]
- Butera, S. T. , Brown J., Callahan M. E., Owen S. M., Matthews A. L., Weigner D. D., Chapman L. E., and Sandstrom P. A., 2000: Survey of veterinary conference attendees for evidence of zoonotic infection by feline retroviruses. J. Am. Vet. Med. Assoc. 217, 1475–1479. [DOI] [PubMed] [Google Scholar]
- Callahan, M. E. , Switzer W. M., Matthews A. L., Roberts B. D., Heneine W., Folks T. M., and Sandstrom P. A., 1999: Persistent zoonotic infection of a human with simian foamy virus in the absence of an intact orf‐2 accessory gene. J. Virol. 73, 9619–9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, B. R. , 1992: Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol. Rev. 56, 375–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delelis, O. , Saib A., and Sonigo P., 2003: Biphasic DNA synthesis in spumaviruses. J. Virol. 77, 8141–8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enssle, J. , Jordan I., Mauer B., and Rethwilm A., 1996: Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc. Natl. Acad. Sci. USA 93, 4137–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enssle, J. , Fischer N., Moebes A., Mauer B., Smola U., and Rethwilm A., 1997: Carboxy‐terminal cleavage of the human foamy virus Gag precursor molecule is an essential step in the viral life cycle. J. Virol. 71, 7312–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, H. , Baunach G., Brautigam S., Mergia A., Neumann‐Haefelin D., Daniel M., McClure M. O., and Rethwilm A., 1994: Reactivity of primate sera to foamy virus Gag and Bet proteins. J. Gen. Virol. 75, 2635–2644. [DOI] [PubMed] [Google Scholar]
- Heneine, W. , Switzer W. M., Sandstrom P., Brown J., Vedapuri S., Schable C. A., Khan A., Lerche N. W., Schweizer M., Neumann‐Haefelin D., Chapman L. E., and Folks T. M., 1998: Identification of a human population infected with simian foamy viruses. Nat. Med. 4, 403–407. [DOI] [PubMed] [Google Scholar]
- Heneine, W. , Schweizer M., Sandstrom P., and Folks T., 2003: Human infection with foamy viruses. Curr. Top. Microbiol. Immunol. 277, 182–196. [DOI] [PubMed] [Google Scholar]
- Herchenröder, O. , Renne R., Loncar D., Cobb E. K., Murthy K. K., Schneider J., Mergia A., and Luciw P., 1994: Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (SFVcpz): high homology to human foamy virus (HFV). Virology 201, 187–199. [DOI] [PubMed] [Google Scholar]
- Hill, C. L. , Bieniasz P. D., and McClure M. O., 1999: Properties of human foamy virus relevant to its development as a vector for gene therapy. J. Gen. Virol. 80, 2003–2009. [DOI] [PubMed] [Google Scholar]
- Hussain, A. I. , Shanmugam V., Bhullar V. B., Beer B. E., Vallet D., Gautier‐Hion A., Wolfe N. D., Karesh W. B., Kilbourn A. M., Tooze Z., Heneine W., and Switzer W. M., 2003: Screening for simian foamy virus infection by using a combined antigen Western blot assay: evidence for a wide distribution among Old World primates and identification of four new divergent viruses. Virology 309, 248–257. [DOI] [PubMed] [Google Scholar]
- Johnson, R. H. , De J. La Rosa, I. Abher, I. G. Kertayadnya, K. W. Entwistle, G. Fordyce, and Holroyd R. G., 1988: Epidemiological studies of bovine spumavirus. Vet. Microbiol. 16, 25–33. [DOI] [PubMed] [Google Scholar]
- Lagaye, S. , Vexiau P., Morozov V., Guenebaut‐Claudet V., Tobaly‐Tapiero J., Canivet M., Cathelineau G., Peries J., and Emanoil‐Ravier R., 1992: Human spumaretrovirus‐related sequences in the DNA of leukocytes from patients with Graves disease. Proc. Natl. Acad. Sci. U S A 89, 10070–10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier, C. H. , and Saib A., 2000: Foamy viruses: between retroviruses and pararetroviruses. Virology 271, 1–8. [DOI] [PubMed] [Google Scholar]
- Linial, M. L. , 1999: Foamy viruses are unconventional retroviruses. J. Virol. 73, 1747–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial, M. L. , and Eastman S. W., 2003: Particle assembly and genome packaging. Curr. Top. Microbiol. Immunol. 277, 89–110. [DOI] [PubMed] [Google Scholar]
- Löchelt, M. , 2003: Foamy virus transactivation and gene expression. Curr. Top. Microbiol. Immunol. 277, 27–61. [DOI] [PubMed] [Google Scholar]
- Löchelt, M. , and Flügel R. M., 1996: The human foamy virus pol gene is expressed as a Pro‐Pol polyprotein and not as a Gag‐Pol fusion protein. J. Virol. 70, 1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löchelt, M. , Muranyi W., and Flügel R. M., 1993: Human foamy virus genome possesses an internal, Bel‐1‐dependent and functional promoter. Proc. Natl. Acad. Sci. USA 90, 7317–7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, B. , Kraus F. B., Allwinn R., Doerr H. W., and Preiser W., 2003: Viral zoonoses – a threat under control? Intervirology 46, 71–78. [DOI] [PubMed] [Google Scholar]
- Meiering, C. D. , and Linial M. L., 2002: Reactivation of a complex retrovirus is controlled by a molecular switch and is inhibited by a viral protein. Proc. Natl. Acad. Sci. USA 99, 15130–15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiering, C. D. , Rubio C., May C., and Linial M. L., 2001: Cell‐type‐specific regulation of the two foamy virus promoters. J. Virol. 75, 6547–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergia, A. , and Heinkelein M., 2003: Foamy virus vectors. Curr. Top. Microbiol. Immunol. 277, 131–159. [DOI] [PubMed] [Google Scholar]
- Moebes, A. , Enssle J., Bieniasz P. D., Heinkelein M., Lindemann D., Bock M., McClure M. O., and Rethwilm A., 1997: Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71, 7305–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrepper, K. I. , Rackwitz H. R., Schnölzer M., Heid H., Löchelt M., and Flügel R. M., 1998: Molecular characterization of proteolytic processing of the Pol proteins of human foamy virus reveals novel features of the viral protease. J. Virol. 72, 7648–7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrepper, K. I. , Rackwitz H. R., Schnölzer M., Heid H., Löchelt M., and Flügel R. M., 1999: Molecular characterization of proteolytic processing of the Pol proteins of human spumavirus. J. Virol. 73, 7907–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung, H. T. , Ikeda Y., Miyazawa T., Nakamura K., Mochizuki M., Izumiya Y., Sato E., Nishimura Y., Tohya Y., Takahashi E., and Mikami T., 2001: Genetic analyses of feline foamy virus isolates from domestic and wild feline species in geographically distinct areas. Virus. Res. 76, 171–181. [DOI] [PubMed] [Google Scholar]
- Pietschmann, T. , Heinkelein M., Heldmann M., Zentgraf H., Rethwilm A., and Lindemann D., 1999: Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73, 2613–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethwilm, A. , 2003: The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 277, 2–26. [DOI] [PubMed] [Google Scholar]
- Rosenblum, L. L. , Patton G., Grigg A. R., Frater A. J., Cain D., Erlwein O., Hill C. L., Clarke J. R., and McClure M. O., 2001: Differential susceptibility of retroviruses to nucleoside analogues. Antivir. Chem. Chemother. 12, 91–97. [DOI] [PubMed] [Google Scholar]
- Saib, A. , 2003: Non‐primate foamy viruses. Curr. Top. Microbiol. Immunol. 277, 197–211. [DOI] [PubMed] [Google Scholar]
- Saib, A. , Canivet M., Giron M. L., Bolgert F., Valla J., Lagaye S., Peries J., and de The H., 1994: Human foamy virus infection in myasthenia gravis. Lancet 343, 666. [DOI] [PubMed] [Google Scholar]
- Sandstrom, P. A. , Phan K. O., Switzer W. M., Fredeking T., Chapman L., Heneine W., and Folks T. M., 2000: Simian foamy virus infection among zoo keepers. Lancet 355, 551–552. [DOI] [PubMed] [Google Scholar]
- Schwantes, A. , Truyen U., Weikel J., Weiss C., and Löchelt M., 2003: Application of chimeric feline foamy virus‐based retroviral vectors for the induction of antiviral immunity in cats. J. Virol. 77, 7830–7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, M. , Turek R., Reinhardt M., and Neumann‐Haefelin D., 1994: Absence of foamy virus DNA in Graves’ disease. AIDS Res. Hum. Retroviruses 10, 601–605. [DOI] [PubMed] [Google Scholar]
- Schweizer, M. , Turek R., Hahn H., Schliephake A., Netzer K. O., Eder G., Reinhardt M., Rethwilm A., and Neumann‐Haefelin D., 1995: Markers of foamy virus infections in monkeys, apes, and accidentally infected humans: appropriate testing fails to confirm suspected foamy virus prevalence in humans. AIDS Res. Hum. Retroviruses 11, 161–1670. [DOI] [PubMed] [Google Scholar]
- Schweizer, M. , Falcone V., Gange J., Turek R., and Neumann‐Haefelin D., 1997: Simian foamy virus isolated from an accidentally infected human individual. J. Virol. 71, 4821–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer, M. , Schleer H., Pietrek M., Liegibel J., Falcone V., and Neumann‐Haefelin D., 1999. Genetic stability of foamy viruses: long‐term study in an African green monkey population. J. Virol. 73, 9256–9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobaly‐Tapiero, J. , Bittoun P., Neves M., Guillemin M., Lecellier C. H., Puvion‐Dutilleul F., Gicquel B., Zientara S., Giron M. L., de The H., and Saib A., 2000: Isolation and characterization of an equine foamy virus. J. Virol. 74, 4064–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey, E. F. , and Yolken R. H., 1995: Could schizophrenia be a viral zoonosis transmitted from house cats? Schizophr. Bull. 21, 167–171. [DOI] [PubMed] [Google Scholar]
- Tschopp, R. R. , Brandner S., Marino S., Bothe K., Horak I., Rethwilm A., and Aguzzi A., 1996: Analysis of the determinants of neurotropism and neurotoxicity of HFV in transgenic mice. Virology 216, 338–346. [DOI] [PubMed] [Google Scholar]
- Vassilopoulos, G. , Trobridge G., Josephson N. C., and Russell D. W., 2001: Gene transfer into murine hematopoietic stem cells with helper‐free foamy virus vectors. Blood 98, 604–609. [DOI] [PubMed] [Google Scholar]
- Vassilopoulos, G. , Josephson N. C., and Trobridge G., 2003a: Development of foamy virus vectors. Methods Mol. Med. 76, 545–564. [DOI] [PubMed] [Google Scholar]
- Vassilopoulos, G. , Wang P. R., and Russell D. W., 2003b: Transplanted bone marrow regenerates liver by cell fusion. Nature 422, 901–904. [DOI] [PubMed] [Google Scholar]
- Wang, G. , and Mulligan M. J., 1999. Comparative sequence analysis and predictions for the envelope glycoproteins of foamy viruses. J. Gen. Virol. 80, 245–254. [DOI] [PubMed] [Google Scholar]
- Weiss, R. A. , 2001: Gulliver's travels in HIVland. Nature 410, 963–967. [DOI] [PubMed] [Google Scholar]
- Wilk, T. , Geiselhart V., Frech M., Fuller S. D., Flügel R. M., and Löchelt M., 2001: Specific interaction of a novel foamy virus Env leader protein with the N‐terminal Gag domain. J. Virol. 75, 7995–8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, I. G. , Löchelt M., Levesque J. P., Bodem J., Flügel R. M., and Flower R. L., 1997: A rapid streptavidin‐capture ELISA specific for the detection of antibodies to feline foamy virus. J. Immunol. Methods 207, 69–77. [DOI] [PubMed] [Google Scholar]
- Winkler, I. G. , Flügel R. M., Löchelt M., and Flower R. L., 1998: Detection and molecular characterisation of feline foamy virus serotypes in naturally infected cats. Virology 247, 144–151. [DOI] [PubMed] [Google Scholar]
- Winkler, I. G. , Löchelt M., and Flower R. L., 1999: Epidemiology of feline foamy virus and feline immunodeficiency virus infections in domestic and feral cats: a seroepidemiological study. J. Clin. Microbiol 37, 2848–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, N. D. , Switzer W. M., Carr J. K., Bhullar V. B., Shanmugam V., Tamoufe U., Prosser A. T., Torimiro J. N., Mpoudi‐Ngole E., McCutchan F. E., Birx D. L., Folks T. M., Burke D. S., and Heneine W., 2003: Naturally acquired simian foamy virus (SFV) infection anong Central African hunters 11th International Conference on Human Retrovirology: HTLV and Related Viruses, San Francisco. [Google Scholar]
- Yu, S. F. , Baldwin D. N., Gwynn S. R., Yendapalli S., and Linial M. L., 1996: Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271, 1579–1582. [DOI] [PubMed] [Google Scholar]
- Yu, S. F. , Sullivan M. D., and Linial M. L., 1999: Evidence that the human foamy virus genome is DNA. J. Virol. 73, 1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvon‐Groussin, A. , Mugnier P., Bertin P., Grandadam M., Agut H., Huraux J. M., and Calvez V., 2001: Efficacy of dideoxynucleosides against human foamy virus and relationship to its reverse transcriptase amino acid sequence and structure. J. Virol. 75, 7184–7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemba, M. , Wilk T., Rutten T., Wagner A., Flügel R. M., and Löchelt M., 1998: The carboxy‐terminal p3Gag domain of the human foamy virus Gag precursor is required for efficient virus infectivity. Virology 247, 7–13. [DOI] [PubMed] [Google Scholar]


