Summary
Bats are considered as the reservoirs of several emerging infectious disease, and novel viruses are continually found in bats all around the world. Studies conducted in southern China found that bats carried a variety of viruses. However, few studies have been conducted on bats in northern China, which harbours a diversity of endemic insectivorous bats. It is important to understand the prevalence and diversity of viruses circulating in bats in northern China. In this study, a total of 145 insectivorous bats representing six species were collected from northern China and screened with degenerate primers for viruses belonging to six families, including coronaviruses, astroviruses, hantaviruses, paramyxoviruses, adenoviruses and circoviruses. Our study found that four of the viruses screened for were positive and the overall detection rates for astroviruses, coronaviruses, adenoviruses and circoviruses in bats were 21.4%, 15.9%, 20% and 37.2%, respectively. In addition, we found that bats in northern China harboured a diversity of novel viruses. Common Serotine (Eptesicus serotinu), Fringed long‐footed Myotis (Myotis fimriatus) and Peking Myotis (Myotis pequinius) were investigated in China for the first time. Our study provided new information on the ecology and phylogeny of bat‐borne viruses.
Keywords: adenoviruses, astroviruses, bats, China, circoviruses, coronaviruses
Impacts.
We conducted a survey of viruses in bats in northern China; a total of 145 insectivorous bats representing six species were screened with degenerate primers for viruses belonging to six families, including coronaviruses, astroviruses, hantaviruses, paramyxoviruses, adenoviruses and circoviruses.
Bats in northern China harboured a diversity of novel viruses that might be of public health significance.
To our knowledge, this represented the first documentations of viruses in Common Serotine (Eptesicus serotinu), Fringed long‐footed Myotis (Myotis fimriatus) and Peking Myotis (Myotis pequinius) in China.
1. Introduction
The majority of emerging infectious diseases (EIDs) of humans are zoonoses originating from wildlife (Lloyd‐Smith et al., 2009; Wolfe, Dunavan, & Diamond, 2007). As the second largest order of mammals, bats are considered as the reservoirs of several emerging infectious disease, such as Ebola, Marburg, SARS, MERS and Henipavirus disease (Han et al., 2015).
Since the outbreak of SARS in 2003, interest in bat‐borne viruses has been increasing and novel viruses are continually found in bats all around the world. A number of studies reported that bats, especially from southern China, carried a variety of viruses (Chu, Poon, Guan, & Peiris, 2008; Ge et al., 2011, 2012, 2016; Hu et al., 2014; Li et al., 2010; Wu et al., 2012; Xu et al., 2015; Yang et al., 2013; Yuan et al., 2014). However, so far, few studies have been conducted on bats in northern China, which harbours a diversity of endemic insectivorous bats. To ascertain the prevalence and diversity of viruses circulating in bats in northern China, insectivorous bat representing six species were collected and screened for viruses belonging to six families including coronaviruses, astroviruses, hantaviruses, paramyxoviruses, adenoviruses and circoviruses.
2. Materials and Methods
2.1. Sample collection
Bats were collected in various habitats, including karst caves, city sewers and human houses in Mengyin County, Shandong Province of China, from March to October, 2015. Bat species was molecularly identified by amplification and sequencing of the cytochrome B (cytB) gene with bat liver DNA as described previously (Ishii et al., 2014; Linacre & Lee, 2005). Captured bats were euthanized by intramuscular injection of overdose chloral hydrate. Thoracic and abdominal organs were collected and stored at −80°C until analysis.
This study was carried out in accordance with the Guidelines of Regulations for the Administration of Laboratory Animals (Decree No. 2 of the State Science and Technology Commission of the People's Republic of China, 1988). The collection of bats for this study was approved by the Ethics Committee of Prevention Medicine of Shandong University (No. 20150501), and all efforts were made to minimize discomfort to animals.
2.2. Preparation of samples and PCR screening for viruses
RNA and DNA were extracted separately from intestinal and spleen samples using AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany), and RNA was extracted from kidney and lung tissue with RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. PCR screening for viruses was performed using broadly reactive consensus PCR assays targeting RNA viruses including coronaviruses (Woo et al., 2005), astroviruses (Chu et al., 2008), hantaviruses (Klempa et al., 2006), paramyxoviruses (Tong, Chern, Li, Pallansch, & Anderson, 2008), DNA viruses including adenoviruses (Wellehan et al., 2004) and circoviruses (Lima et al., 2015). Paramyxoviruses were screened with one‐step RT‐PCR with Access RT‐PCR System (Promega, Madison, WI), while screening for coronaviruses, astroviruses and hantaviruses, cDNA was first generated by using reverse transcription system (Promega) with random primers. Detailed information of primer sequences and amplicon lengths is listed in Table S1.
PCR products were analyzed using 1.2% agarose gel electrophoresis and detected with ethidium bromide staining under UV light. PCR products of expected size were excised from gels and extracted with Gel Extraction Kit (Promega) and were then cloned into pMD 19‐T vectors (TaKaRa, Shiga, Japan) for sequencing. Three recombined plasmids for each PCR product were sequenced on both strands.
2.3. Phylogenetic analysis
Sequence chromatograms and sequence analysis were examined with Chromas 2.6 (http://technelysium.com.au/wp/chromas) and blast program (http://blast.ncbi.nlm.nih.gov/Blast.cgi), respectively. Sequences were aligned and trimmed with mega 7.0. Phylogenetic trees were constructed with mega 7.0 (http://www.megasoftware.net) by using the neighbour‐joining method with an amino acid percentage distance substitution model and complete deletion option, and bootstrap values were calculated with 1,000 replicates.
3. Results
3.1. Molecular detection of viruses in bats
A total of 145 insectivorous bats were collected for microbiological studies from Mengyin County of Shandong Province, China. They were molecularly identified into six species, with four from the family Vespertilionidae and two from the family Rhinolophidae (Table 1). The partial cytB gene for the six bat species was deposited in GenBank with accession numbers KX655809, KX655810, KX655817, KX655826, KX655831, KX655837 and KX655840.
Table 1.
Prevalence of viruses in bats from northern China
| Family | Species | Common name | No. sampled | AstVs | CoVs | PMVs | HTVs | AdVs | CVs |
|---|---|---|---|---|---|---|---|---|---|
| Vespertilionidae | Eptesicus serotinus | Common Serotine | 26 | 8 | 11 | 0 | 0 | 2 | 1 |
| Myotis fimbriatus | Fringed long‐footed Myotis | 34 | 14 | 6 | 0 | 0 | 18 | 29 | |
| Myotis ricketti | Rickett's big‐footed Myotis | 10 | 1 | 2 | 0 | 0 | 5 | 6 | |
| Myotis pequinius | Peking Myotis | 57 | 8 | 3 | 0 | 0 | 4 | 15 | |
| Rhinolophidae | Rhinolophus ferrumequinum | Greater Horseshoe Bat | 4 | 0 | 0 | 0 | 0 | 0 | 1 |
| Rhinolophus pusillus | Least Horseshoe Bat | 14 | 0 | 1 | 0 | 0 | 0 | 2 | |
| 145 | 31 | 22 | 0 | 0 | 29 | 51 |
AstVs, astroviruses; CoVs, coronaviruses; PMVs, paramyxoviruses; HTVs, hantaviruses; AdVs, adenoviruses; CVs, circoviruses.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
None of the bats screened were positive for hantaviruses or paramyxoviruses, despite previous studies reporting their existence in bats. The overall detection rates for astroviruses, coronaviruses, adenoviruses and circoviruses in bats were 21.4% (31/145), 15.9% (23/145), 20% (29/145) and 37.2% (54/145), respectively. Four species from the family Vespertilionidae were all positive for astroviruses, coronaviruses, adenoviruses and circoviruses, while two species from the family Rhinolophidae were both positive for circoviruses, with one species positive for coronaviruses and neither positive for astroviruses and adenoviruses. The highest positive rates for astroviruses, adenoviruses and circoviruses were observed in Fringed long‐footed Myotis (Myotis fimbriatus) and for coronaviruses in Common Serotine (Eptesicus serotinu) (Table 1).
3.2. Phylogenetic analysis
3.2.1. Astroviruses
Phylogenetic analysis based on the partial RdRp gene sequences of astroviruses showed that most bat astroviruses derived from this study along with previously described bat astroviruses formed a separate bat astroviruses lineage. However, different from previous findings, several bat astroviruses detected in this study clustered with porcine/murine/rat astroviruses (Figure 1).
Figure 1.
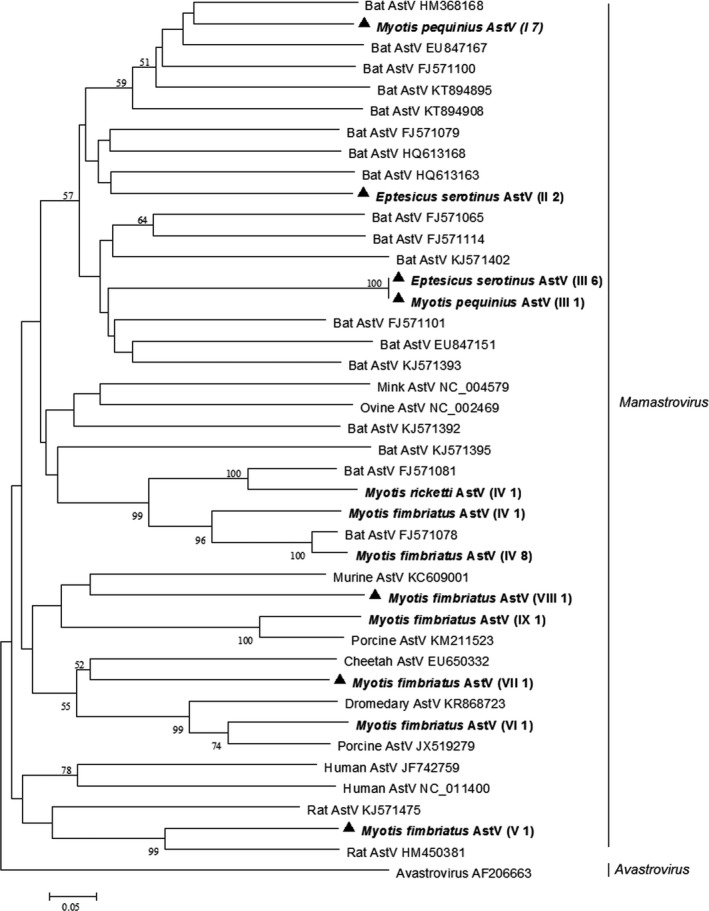
Phylogenetic analysis of the RdRp gene of astroviruses (AstVs) detected from bats in northern China. The tree was constructed with mega 7.0, by using neighbour‐joining method with an amino acid p‐distance substitution model and complete deletion option, and 1,000 bootstrap reiterations. Bootstrap values (≥50) are shown next to the branches. Astroviruses detected in this study are shown in boldface with the Latin name of the bat species, followed by parentheses, in which the Roman numerals represented astrovirus genotypes and Arabic numerals were the number of positive bats
Phylogenetic analysis also revealed that bat astroviruses detected in this study exhibited a definite host specificity (Figure 1). First, genotype I astroviruses were only detected in Peking Myotis (Myotis pequinius), genotype II and III astroviruses were almost exclusively found in Common Serotine (E. serotinus), and genotype IV astroviruses were only detected in Fringed long‐footed Myotis (Myotis fimriatus). Second, no cross‐species transmission was observed in bats collected from the same habitat on the same day. For example, Fringed long‐footed Myotis (M. fimriatus) and Rickett's big‐footed Myotis (Myotis rickettis) were collected simultaneously from a sewer; however, astroviruses detected in Fringed long‐footed bat (M. fimriatus) were absolutely negative in Rickett's big‐footed Myotis (M. rickettis).
Genotype I, II III, V, VII and VIII astroviruses detected in this study shared 69%–84% nucleotide identity and 70%–88% amino acid identity with the most closely related known astroviruses, indicating that they represented novel bat astroviruses. In addition, it was noteworthy that Fringed long‐footed Myotis (M. fimriatus) could harbour much more diverse astroviruses than bats of the other five species studied (Figure 1). Partial RdRp gene sequences of bat astroviruses of this study were deposited in GenBank with accession numbers KX702337–KX702367.
3.2.2. Coronaviruses
Phylogenetic analysis based on the 440‐bp pol gene sequences of coronaviruses identified five coronavirus genotypes (Figure 2). Three genotypes (genotype III, IV and V) clustered in the Alphacoronavirus (α‐CoV) genus and two (genotype I and II) clustered in the Betacoronavirus (β‐CoV) genus. Genotype III in the α‐CoV genus shared <81% nucleotide identity with the most closely related known coronaviruses and may represent a novel coronavirus. Significantly, genotype I and II in the β‐CoV genus clustered with lineage C β‐CoV and shared 85%–88% nucleotide identity with MERS‐CoV, which was the causative agent of the Middle East Respiratory Diseases (MERS). Partial pol gene sequences of bat coronaviruses of this study were deposited in GenBank with accession numbers KY009612–KY009634.
Figure 2.
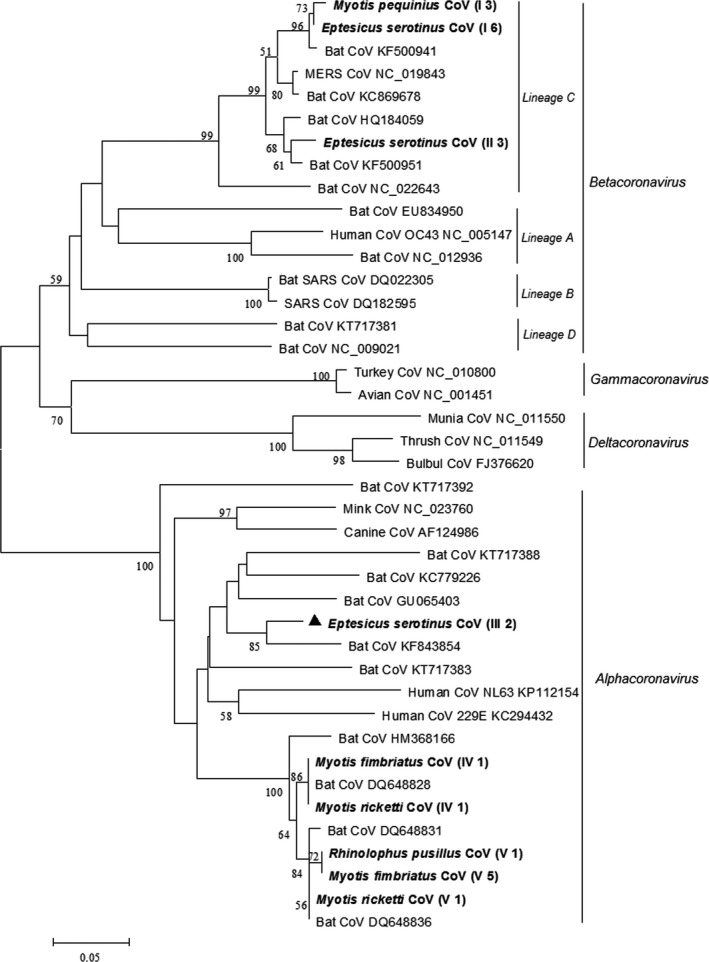
Phylogenetic analysis of the pol gene of coronaviruses (CoVs) detected from bats in northern China. The tree was constructed with mega 7.0, by using neighbour‐joining method with an amino acid p‐distance substitution model and complete deletion option and 1,000 bootstrap reiterations. Bootstrap values (≥50) are shown next to the branches. Coronaviruses detected in this study are shown in boldface with the Latin name of the bat species, followed by parentheses, in which the Roman numerals represented coronavirus genotypes and Arabic numerals were the number of positive bats
3.2.3. Adenoviruses
Phylogenetic analysis of bat adenoviruses based on the partial pol gene sequences identified six bat adenovirus genotypes in this study, which clustered within the Mastadenovirus genus (Figure 3). Genotype IV, V and VI adenoviruses from Rickett's big‐footed Myotis (M. ricketti) and Peking Myotis (M. pequinius) shared only 69%–75% nucleotide identity and 76%–82% amino acid identity with the most closely related known adenoviruses and therefore may represent novel adenoviruses. Genotype III AdV identified in a single Fringed long‐footed Myotis (M. fimbriatus) shared 97% nucleotide identity and 99% amino acid identity with adenoviruses detected in rat and shrew from southern China (Zheng et al., 2016), indicating possible cross‐species transmission. Moreover, bat adenoviruses with adenoviruses of other mammals formed a mixed cluster, instead of a separated group like bat astroviruses, which also indicate cross‐species jump events.
Figure 3.

Phylogenetic analysis of the pol gene of adenoviruses (AdVs) detected from bats in northern China. The tree was constructed with mega 7.0, by using neighbour‐joining method with an amino acid p‐distance substitution model and complete deletion option, and 1,000 bootstrap reiterations. Bootstrap values (≥50) are shown next to the branches. Adenoviruses detected in this study are shown in boldface with the Latin name of the bat species, followed by parentheses, in which the Roman numerals represented adenovirus genotypes and Arabic numerals were the number of positive bats
Phylogenetic analysis revealed that adenoviruses detected in bats in this study showed host specificity, because bats of different species collected from different habitats harboured different adenovirus genotypes, while bats in the same habitat carried the same genotypes. Common Serotine (E. serotinu) were collected from a human dwelling and carried genotype I adenoviruses, Fringed long‐footed Myotis (M. fimriatus) and Rickett's big‐footed Myotis (M. rickettis) were collected simultaneously from a sewer and were infected with genotype II, III and IV adenoviruses, while Peking Myotis (M. pequinius), which were sampled from a cave, carried genotype V adenoviruses. Partial pol gene sequences of bat adenoviruses of this study were deposited in GenBank with accession numbers KY009635–KY009663.
3.2.4. Circoviruses
Phylogenetic analysis of the partial rep gene sequences of circoviruses showed that circoviruses detected in this study could be divided into 10 genotypes, with nine genotypes in the Circovirus genus and one genotype in Cyclovirus genus (Figure 4). Most circoviruses detected in this study shared 65%–88% nucleotide identity with the most closely related known circoviruses and therefore might represent novel circoviruses. Partial rep gene sequences of bat circoviruses of this study were deposited in GenBank with accession numbers KX834465–KX834507.
Figure 4.
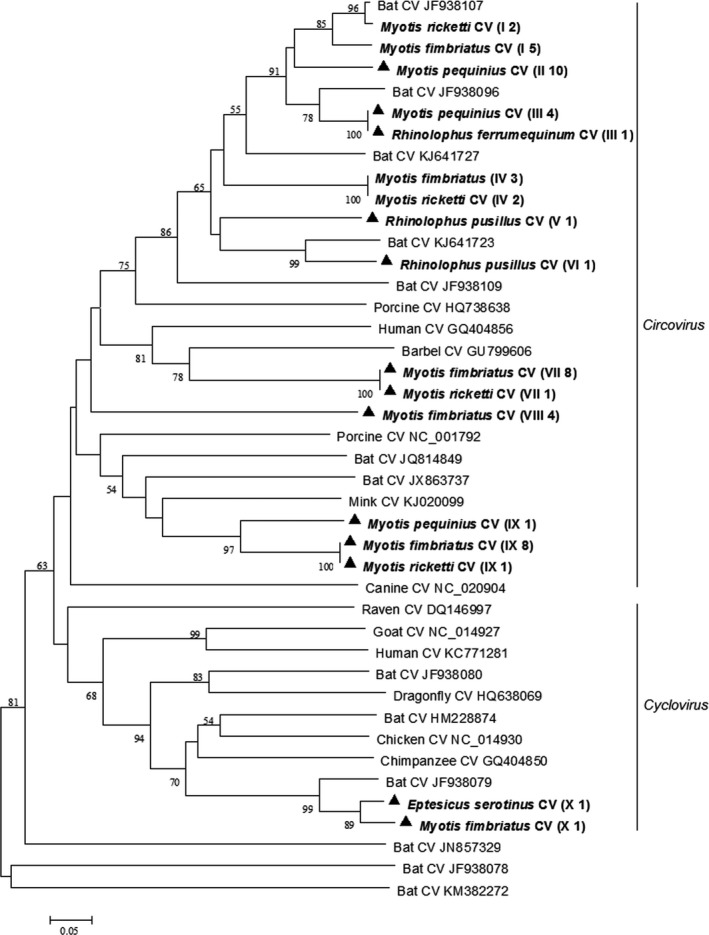
Phylogenetic analysis of the rep gene of circoviruses (CVs) detected from bats in northern China. The tree was constructed with mega 7.0, by using neighbour‐joining method with an amino acid p‐distance substitution model and complete deletion option, and 1,000 bootstrap reiterations. Bootstrap values (≥50) are shown next to the branches. Circoviruses detected in this study are shown in boldface with the Latin name of the bat species, followed by parentheses, in which the Roman numerals represented circoviruses genotypes and Arabic numerals were the number of positive bats
4. Discussion
4.1. Astroviruses
The Astroviridae is a family of small, single‐stranded positive‐sense RNA viruses, which are important causative pathogens of human and animal acute gastroenteritis. It comprises two genera: Avastrovius and Mamastrovirus, which infect birds and mammals, respectively (Guix, Bosch, & Pintó, 2013).
Consistent with previous studies, our study also revealed that bats carried genetically diverse astroviruses, which showed a definite host specificity for astroviruses in bat species (Fischer et al., 2016; Zhu et al., 2009). However, some bat astroviruses detected in this study clustered with porcine/murine/rat astroviruses, implying cross‐species transmission between different mammalian hosts. Several studies also showed the existence of cross‐species transmission among mamastroviruses. For example, some turkey flocks had antibody against chicken astroviruses (Baxendale & Mebatsion, 2004), several astroviruses isolated from brown rats were most closely related to human astrovirus MLB1 (Chu et al., 2010) and bat astroviruses were phylogenetically related to astroviruses of mink, ovine and human (Xiao et al., 2011).
Phylogenetic analysis of astroviruses in this study was based on the partial RdRp gene (ORF1b) sequence; however, as capsid protein (ORF2) was also a key protein for astrovirus classification according to ICTV's criteria (Martella et al., 2014), a further analysis based on ORF2 might provide a more accurate understanding of the phylogenetic relationship of the astroviruses detected in this study, and a better assessment of the cross‐species potential.
In conclusion, we detected a genetic diversity of astroviruses in bats of several species in northern China, and many astroviruses detected in this study were novel. With previous reports (Chu et al., 2008; Xiao et al., 2011; Zhu et al., 2009), it was proposed that bats may be important natural reservoirs of astroviruses. Despite the definite host specificity, there were implications for cross‐species transmission of bat astroviruses between mammals. Whether these novel astroviruses detected in bats in this study had any public significance needed to be further investigated.
4.2. Coronaviruses
Coronaviruses are enveloped, positive‐sense RNA viruses that belong to the subfamily Coronavirinae, family Coronaviridae. Coronaviruses are classified into four genera: Alphacoronavirus (α‐CoV), Betacoronavirus (β‐CoV), Gammacoronavirus (γ‐CoV) and Deltacoronavirus (δ‐CoV), with the genus β‐CoV further divided into four genetic lineages, namely A–D (de Groot et al., 2012). SARS‐CoV and MERS‐CoV are causative agents of severe human respiratory infections SARS and MERS and belong to the lineage B and lineage C of β‐CoV, respectively (Zaki, van Boheemen, Bestebroer, Osterhaus, & Fouchier, 2012).
Molecular analysis was based on the 440‐bp RdRp gene sequence of coronaviruses, although it was proposed to extend the 440‐bp RdRp fragment to 816 bp to obtain better phylogenetic resolution (Drexler et al., 2010). Despite the short sequences, we were able to identify diverse coronaviruses in bats in China. We documented two lineage C β‐CoV genotypes in Fringed long‐footed Myotis (M. fimbriatus) and Common Serotine (E. serotinus). So far, genotypes of lineage C β‐CoV in bats have been reported in Common Serotine (E. serotinus), Kuhl's pipistrelle (Pipistrellus kuhlii), Pipistrelle bat (Pipistrellus spp.), Common noctule (Nyctalus noctula) and Savi's pipistrelle (Hypsugo savii) in Italy (De Benedictis et al., 2014; Lelli et al., 2013), broad‐eared bat (Nyctinomops laticaudatus) and Davy's naked‐backed bat (Pteronotus davyi) in Mexico (Anthony et al., 2013; Goes et al., 2013), Zulu Pipistrelle bat (Neoromicia zuluensisin) in South Africa (Ithete et al., 2013), Egyptian tomb bat (Taphozous perforatus) in Saudi Arabia (Memish et al., 2013), Gambian Slit‐faced bat (Nycteris gambiensis) in Ghana (Annan et al., 2013), Common Pipistrelle (Pipistrellus pipistrellus) in the Netherlands (Reusken et al., 2010) and Asian Particolored bat (Vespertilio superans) in China (Yang et al., 2014). Compared with the previously reported lineage C β‐CoV in Asian Particolored bats (Vespertilio superans) in China (Yang et al., 2014), which shared 75.7% nucleotide identity with that of human MERS‐CoV, the genotypes of lineage C β‐CoV detected in this study were more closely related to MERS‐CoV with 85%–88% nucleotide identity.
Our study was the first to report the discovery of lineage C β‐CoV in Fringed long‐footed Myotis (M. fimbriatus) and Common Serotine (E. serotinus) in China. Notably, this may be of public health concern, as both species are in close proximity to human beings: Fringed long‐footed Myotis (M. fimbriatus) was collected from a city sewer, and Common Serotine (E. serotinus) was captured from rural homes. Despite previous reports of SARS‐like coronaviruses in Rhinolophus bats in China (Chen et al., 2016; He et al., 2014; Li et al., 2005), no SARS‐like coronaviruses were detected in Rhinolophus bats in this study, which may be due to the limited sample size of Rhinolophus bats.
This was also the first documentation of coronaviruses in Peking Myotis (M. pequinius) and Fringed long‐footed Myotis (M. fimbriatus), expanding our understanding of the diversity and ecology of coronaviruses in bats.
4.3. Adenoviruses
Adenoviruses are linear, non‐segmented, double‐stranded DNA viruses belonging to the family Adenoviridae, which includes five genera: Mastadenovirus, Aviadenovirus, Atadenovirus, Siadenovirus and Ichtadenovirus (King, Adams, Carstens, & Lefkowitz, 2012). Adenoviruses are prevalent in human and can cause human infections ranging from respiratory disease, conjunctivitis to gastroenteritis (Kojaoghlanian, Flomenberg, & Horwitz, 2003).
A previous study found that bat adenoviruses in different bat species displayed greater genetic diversity (Li et al., 2010). However, our study found that adenoviruses showed limited genetic diversity among bats collected from the same location, which may be due to the lower mutation rate of DNA viruses compared with that of RNA viruses (Domingo‐Calap & Sanjuan, 2011). This was the first report of adenoviruses in Fringed long‐footed Myotis (M. fimbriatus), Common Serotine (E. serotinus) and Peking Myotis (M. pequinius) in China.
A previous study reported that an adenovirus, which was responsible for the respiratory disease of a pygmy marmoset, was genetically related to bat and canine adenoviruses, indicating a cross‐species jump event (Gal et al., 2013). Cross‐species transmission of adenoviruses might result in as severe outcomes such as the highly pathogenic canine adenoviruses in dogs, which were genetically related to bat adenoviruses (Kohl et al., 2012). Despite the short pol gene sequences, our study also showed the existence of possible cross‐species transmission of bat adenoviruses. Genotype III adenovirus identified in a single Fringed long‐footed Myotis (M. fimbriatus) was closely related to adenoviruses detected in rats and shrews from southern China and bat adenoviruses formed a mixed cluster with other mammalian adenoviruses. So far, the pathogenesis of these bat adenoviruses is not yet clear and we cannot rule out the possibility of cross‐species transmission with further investigations needed.
4.4. Circoviruses
Circoviruses are non‐enveloped, small, circular, single‐stranded DNA viruses that belong to the genus Circovirus and the recently proposed genus Cyclovirus in the family Circoviridae (Lima et al., 2015). So far, several circoviruses can be related to severe clinical conditions in animals. Porcine circovirus 2 (PCV2) is the primary etiological agent of post‐weaning multisystemic wasting syndrome (PMWS), which may be responsible for significant economic losses (Firth, Charleston, Duffy, Shapiro, & Holmes, 2009). Beak feather disease virus (BFDV) is responsible for fatal disease in parrots called Psittacine beak and feather disease (PBFD) (Fogell, Martin, & Groombridge, 2016). A novel circovirus was reported to be the causative agent of a long‐established epidemic of Mink enteritis in China (Lian et al., 2014). In addition, a new cyclovirus was identified in the cerebrospinal fluid of Malawi paraplegia patients with acute central nervous system infection of unknown aetiology, indicating the possibility of disease association, although it has yet to be proven (Smits et al., 2013).
Based on the partial rep gene sequences, we found that bats in northern China carried genetically diverse novel circoviruses, which showed no obvious host specificity. Six bat species tested in this study were all positive for circoviruses with a high prevalence of 37.2%. So far, the pathogenesis of these novel circoviruses detected is not yet clear and future investigations are needed to understand the relationships between these circoviruses and bats, as well as potential effects on other mammals.
In conclusion, we reported the molecular detection of viruses belonging to six families in insectivorous bat representing six species in northern China. Four of six viruses screened were positive and bats in northern China harboured a diversity of novel viruses. Common Serotine (E. serotinu), Fringed long‐footed Myotis (M. fimriatus) and Peking Myotis (M. pequinius) were investigated in China for the first time, which provided new information on the ecology and phylogeny of bat‐borne viruses.
Supporting information
Acknowledgements
This study was supported by The Shandong Province Science and Technology Development Program (grant number 2014GSF121004) and The National Natural Science Funds of China (grant numbers 31570167, 81102171 and 31402173).
Han H‐J, Wen H‐L, Zhao L, et al. Novel coronaviruses, astroviruses, adenoviruses and circoviruses in insectivorous bats from northern China. Zoonoses Public Health. 2017;64:636–646. 10.1111/zph.12358
[Correction added on 5 Dec 2017, after first online publication]
References
- Annan, A. , Baldwin, H. J. , Corman, V. M. , Klose, S. M. , Owusu, M. , Nkrumah, E. E. , … Drexler, J. F. (2013). Human betacoronavirus 2c EMC/2012‐related viruses in bats, Ghana and Europe. Emerging Infectious Diseases, 19, 456–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony, S. J. , Ojeda‐Flores, R. , Rico‐Chavez, O. , Navarrete‐Macias, I. , Zambrana‐Torrelio, C. M. , Rostal, M. K. , … Lipkin, W. I. (2013). Coronaviruses in bats from Mexico. The Journal of General Virology, 94, 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxendale, W. , & Mebatsion, T. (2004). The isolation and characterisation of astroviruses from chickens. Avian Pathology, 33, 364–370. [DOI] [PubMed] [Google Scholar]
- Chen, Y. N. , Phuong, V. N. , Chen, H. C. , Chou, C. H. , Cheng, H. C. , & Wu, C. H. (2016). Detection of the severe acute respiratory syndrome‐related coronavirus and alphacoronavirus in the bat population of Taiwan. Zoonoses and Public Health, 63, 608–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, D. K. , Chin, A. W. , Smith, G. J. , Chan, K. H. , Guan, Y. , Peiris, J. S. , & Poon, L. L. (2010). Detection of novel astroviruses in urban brown rats and previously known astroviruses in humans. The Journal of General Virology, 91, 2457–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, D. K. , Poon, L. L. , Guan, Y. , & Peiris, J. S. (2008). Novel astroviruses in insectivorous bats. Journal of Virology, 82, 9107–9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedictis, P. , Marciano, S. , Scaravelli, D. , Priori, P. , Zecchin, B. , Capua, I. , … Cattoli, G. (2014). Alpha and lineage C betaCoV infections in Italian bats. Virus Genes, 48, 366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo‐Calap, P. , & Sanjuan, R. (2011). Experimental evolution of RNA versus DNA viruses. Evolution; International Journal of Organic Evolution, 65, 2987–2994. [DOI] [PubMed] [Google Scholar]
- Drexler, J. F. , Gloza‐Rausch, F. , Glende, J. , Corman, V. M. , Muth, D. , Goettsche, M. , … Drosten, C. (2010). Genomic characterization of severe acute respiratory syndrome‐related coronavirus in European bats and classification of coronaviruses based on partial RNA‐dependent RNA polymerase gene sequences. Journal of Virology, 84, 11336–11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth, C. , Charleston, M. A. , Duffy, S. , Shapiro, B. , & Holmes, E. C. (2009). Insights into the evolutionary history of an emerging livestock pathogen: Porcine circovirus 2. Journal of Virology, 83, 12813–12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, K. , Zeus, V. , Kwasnitschka, L. , Kerth, G. , Haase, M. , Groschup, M. H. , & Balkema‐Buschmann, A. (2016). Insectivorous bats carry host specific astroviruses and coronaviruses across different regions in Germany. Infection, Genetics and Evolution, 37, 108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogell, D. J. , Martin, R. O. , & Groombridge, J. J. (2016). Beak and feather disease virus in wild and captive parrots: An analysis of geographic and taxonomic distribution and methodological trends. Archives of Virology, 161, 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal, J. , Hornyak, A. , Mandoki, M. , Bakonyi, T. , Balka, G. , Szeredi, L. , … Farkas, S. L. (2013). Novel mastadenovirus infection and clinical disease in a pygmy marmoset (Callithrix [Cebuella] pygmaea). Veterinary Microbiology, 167, 695–699. [DOI] [PubMed] [Google Scholar]
- Ge, X. , Li, J. , Peng, C. , Wu, L. , Yang, X. , Wu, Y. , Zhang, Y. , & Shi, Z. (2011). Genetic diversity of novel circular ssDNA viruses in bats in China. The Journal of General Virology, 92, 2646–2653. [DOI] [PubMed] [Google Scholar]
- Ge, X. , Li, Y. , Yang, X. , Zhang, H. , Zhou, P. , Zhang, Y. , & Shi, Z. (2012). Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. Journal of Virology, 86, 4620–4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, X. Y. , Wang, N. , Zhang, W. , Hu, B. , Li, B. , Zhang, Y. Z. , … Shi, Z. L. (2016). Coexistence of multiple coronaviruses in several bat colonies in an abandoned mineshaft. Virologica Sinica, 31, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goes, L. G. , Ruvalcaba, S. G. , Campos, A. A. , Queiroz, L. H. , de Carvalho, C. , Jerez, J. A. , … Dominguez, S. R. (2013). Novel bat coronaviruses, Brazil and Mexico. Emerging Infectious Diseases, 19, 1711–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot, R. J. , Baker, S. , Baric, R. , Enjuanes, L. , Gorbalenya, A. E. , Holmes, K. V. , … Ziebuhr, J. (2012). Family coronaviridae (pp. 806–820). Oxford, UK: Elsevier. [Google Scholar]
- Guix, S. , Bosch, A. , & Pintó, R. M. (2013). Astrovirus taxonomy In Schultz‐Cherry S. (Ed.), Astrovirus research: Essential Ideas, everyday impacts, future directions. New York, NY: Springer New York. [Google Scholar]
- Han, H. J. , Wen, H. L. , Zhou, C. M. , Chen, F. F. , Luo, L. M. , Liu, J. W. , & Yu, X. J. (2015). Bats as reservoirs of severe emerging infectious diseases. Virus Research, 205, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, B. , Zhang, Y. , Xu, L. , Yang, W. , Yang, F. , Feng, Y. , … Tu, C. (2014). Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome‐like coronavirus from bats in China. Journal of Virology, 88, 7070–7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Chmura, A. A. , Li, J. , Zhu, G. , Desmond, J. S. , Zhang, Y. , … Shi, Z. (2014). Detection of diverse novel astroviruses from small mammals in China. The Journal of General Virology, 95, 2442–2449. [DOI] [PubMed] [Google Scholar]
- Ishii, A. , Ueno, K. , Orba, Y. , Sasaki, M. , Moonga, L. , Hang'ombe, B. M. , … Sawa, H. (2014). A nairovirus isolated from African bats causes haemorrhagic gastroenteritis and severe hepatic disease in mice. Nature Communications, 5, 5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ithete, N. L. , Stoffberg, S. , Corman, V. M. , Cottontail, V. M. , Richards, L. R. , Schoeman, M. C. , … Preiser, W. (2013). Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerging Infectious Diseases, 19, 1697–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, A. M. Q. , Adams, M. J. , Carstens, E. B. , & Lefkowitz, E. J. (2012). Family – Adenoviridae. Virus taxonomy. San Diego: Elsevier. [Google Scholar]
- Klempa, B. , Fichet‐Calvet, E. , Lecompte, E. , Auste, B. , Aniskin, V. , Meisel, H. , … Kruger, D. H. (2006). Hantavirus in African wood mouse, Guinea. Emerging Infectious Diseases, 12, 838–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl, C. , Vidovszky, M. Z. , Muhldorfer, K. , Dabrowski, P. W. , Radonic, A. , Nitsche, A. , … Harrach, B. (2012). Genome analysis of bat adenovirus 2: Indications of interspecies transmission. Journal of Virology, 86, 1888–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojaoghlanian, T. , Flomenberg, P. , & Horwitz, M. S. (2003). The impact of adenovirus infection on the immunocompromised host. Reviews in Medical Virology, 13, 155–171. [DOI] [PubMed] [Google Scholar]
- Lelli, D. , Papetti, A. , Sabelli, C. , Rosti, E. , Moreno, A. , & Boniotti, M. B. (2013). Detection of coronaviruses in bats of various species in Italy. Viruses, 5, 2679–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Ge, X. , Zhang, H. , Zhou, P. , Zhu, Y. , Zhang, Y. , … Shi, Z. (2010). Host range, prevalence, and genetic diversity of adenoviruses in bats. Journal of Virology, 84, 3889–3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Shi, Z. , Yu, M. , Ren, W. , Smith, C. , Epstein, J. H. , … Wang, L. F. (2005). Bats are natural reservoirs of SARS‐like coronaviruses. Science (New York, N.Y.), 310, 676–679. [DOI] [PubMed] [Google Scholar]
- Lian, H. , Liu, Y. , Li, N. , Wang, Y. , Zhang, S. , & Hu, R. (2014). Novel circovirus from mink, China. Emerging Infectious Diseases, 20, 1548–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, F. E. , Cibulski, S. P. , Dos Santos, H. F. , Teixeira, T. F. , Varela, A. P. , Roehe, P. M. , … Franco, A. C. (2015). Genomic characterization of novel circular ssDNA viruses from insectivorous bats in Southern Brazil. PLoS One, 10, e0118070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linacre, A. , & Lee, J. C. (2005). Species determination: The role and use of the cytochrome b gene. Methods in Molecular Biology, 297, 45–52. [PubMed] [Google Scholar]
- Lloyd‐Smith, J. O. , George, D. , Pepin, K. M. , Pitzer, V. E. , Pulliam, J. R. , Dobson, A. P. , … Grenfell, B. T. (2009). Epidemic dynamics at the human‐animal interface. Science (New York, N.Y.), 326, 1362–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella, V. , Pinto, P. , Tummolo, F. , De Grazia, S. , Giammanco, G. M. , Medici, M. C. , … Banyai, K. (2014). Analysis of the ORF2 of human astroviruses reveals lineage diversification, recombination and rearrangement and provides the basis for a novel sub‐classification system. Archives of Virology, 159, 3185–3196. [DOI] [PubMed] [Google Scholar]
- Memish, Z. A. , Mishra, N. , Olival, K. J. , Fagbo, S. F. , Kapoor, V. , Epstein, J. H. , … Lipkin, W. I. (2013). Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerging Infectious Diseases, 19, 1819–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken, C. B. , Lina, P. H. , Pielaat, A. , de Vries, A. , Dam‐Deisz, C. , Adema, J. , … Kooi, E. A. (2010). Circulation of group 2 coronaviruses in a bat species common to urban areas in Western Europe. Vector Borne and Zoonotic Diseases, 10, 785–791. [DOI] [PubMed] [Google Scholar]
- Smits, S. L. , Zijlstra, E. E. , van Hellemond, J. J. , Schapendonk, C. M. , Bodewes, R. , Schurch, A. C. , … Osterhaus, A. D. (2013). Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010‐2011. Emerging Infectious Diseases, 19, 1511–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, S. , Chern, S. W. , Li, Y. , Pallansch, M. A. , & Anderson, L. J. (2008). Sensitive and broadly reactive reverse transcription‐PCR assays to detect novel paramyxoviruses. Journal of Clinical Microbiology, 46, 2652–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellehan, J. F. , Johnson, A. J. , Harrach, B. , Benko, M. , Pessier, A. P. , Johnson, C. M. , … Jacobson, E. R. (2004). Detection and analysis of six lizard adenoviruses by consensus primer PCR provides further evidence of a reptilian origin for the atadenoviruses. Journal of Virology, 78, 13366–13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, N. D. , Dunavan, C. P. , & Diamond, J. (2007). Origins of major human infectious diseases. Nature, 447, 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, P. C. , Lau, S. K. , Chu, C. M. , Chan, K. H. , Tsoi, H. W. , Huang, Y. , … Yuen, K. Y. (2005). Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. Journal of Virology, 79, 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z. , Ren, X. , Yang, L. , Hu, Y. , Yang, J. , He, G. , … Jin, Q. (2012). Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. Journal of Virology, 86, 10999–11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, J. , Li, J. , Hu, G. , Chen, Z. , Wu, Y. , Chen, Y. , … Chen, Q. (2011). Isolation and phylogenetic characterization of bat astroviruses in southern China. Archives of Virology, 156, 1415–1423. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Wu, J. , He, B. , Qin, S. , Xia, L. , Qin, M. , … Tu, C. (2015). Novel hantavirus identified in black‐bearded tomb bats, China. Infection, Genetics and Evolution, 31, 158–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, F. , Wang, Y. , Zheng, W. , He, B. , Jiang, T. , Li, Y. , … Tu, C. (2013). Metagenomic analysis of bat virome in several Chinese regions. Chinese Journal of Biotechnology, 29, 586–600. [PubMed] [Google Scholar]
- Yang, L. , Wu, Z. , Ren, X. , Yang, F. , Zhang, J. , He, G. , … Jin, Q. (2014). MERS‐related betacoronavirus in Vespertilio superans bats, China. Emerging Infectious Diseases, 20, 1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L. , Li, M. , Li, L. , Monagin, C. , Chmura, A. A. , Schneider, B. S. , … Chen, J. (2014). Evidence for retrovirus and paramyxovirus infection of multiple bat species in china. Viruses, 6, 2138–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki, A. M. , van Boheemen, S. , Bestebroer, T. M. , Osterhaus, A. D. , & Fouchier, R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. The New England Journal of Medicine, 367, 1814–1820. [DOI] [PubMed] [Google Scholar]
- Zheng, X. Y. , Qiu, M. , Ke, X. M. , Guan, W. J. , Li, J. M. , Huo, S. T. , … Chen, Q. (2016). Detection of novel adenoviruses in fecal specimens from rodents and shrews in southern China. Virus Genes, 52, 417–421. [DOI] [PubMed] [Google Scholar]
- Zhu, H. C. , Chu, D. K. , Liu, W. , Dong, B. Q. , Zhang, S. Y. , Zhang, J. X. , … Guan, Y. (2009). Detection of diverse astroviruses from bats in China. The Journal of General Virology, 90, 883–887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


