Summary
Faeces of 230 calves with and without diarrhoea collected during the winter period 2004/2005 in 100 Austrian farms (Styria and Lower Austria) were examined for viral, bacterial and parasitic enteropathogens. Torovirus‐specific nucleic acid confirmed by reverse transcriptase‐polymerase chain reaction was found in 12 of 230 calves (5.2%). Ten of these calves were clinically ill, several of them showing signs of dehydration and abnormal faecal consistency at the time of sampling. Computer assisted analysis of two nucleotide sequences obtained from Austrian bovine samples revealed 93% similarity to Breda strain, but only 71% or 52% similarity to Equine Berne or Porcine Markelo torovirus strains respectively. Phylogenetic analysis grouped Austrian torovirus samples into the Bovine torovirus cluster indicating the first detection of Bovine torovirus in Austria. In addition, the following agents were detected in bovine faecal samples: Bovine coronavirus, 25.7%; Escherichia coli, 17%; Cryptosporidium spp., 11.7%; Eimeria spp., 10.4%; Rotavirus, 9.1%; Clostridium perfringens, 9.1% and Giardia spp., 6.1%. Salmonella spp. was not detected.
Introduction
Neonatal calf diarrhoea is a major problem encountered in calf rearing and leads to losses attributable to growth retardation and death. The most common pathogens are Rotaviruses (RV), Bovine coronavirus (BCV), Escherichia coli K99, Salmonella spp. and Cryptosporidium spp. (Koopmans et al., 1991).
Bovine torovirus (BoTV) was first associated with enteritis of calves by Woode et al. (1982). These enveloped single‐stranded RNA viruses have a fringe of peplomers on their surface, measure 100–140 nm at their largest diameter and contain a tightly coiled nucleocapsid that generally assumes a torus shape in the virion (Jamieson et al., 1998). By electron microscopy, it is difficult to differentiate between BoTV and BCV. However, there is no antigenic relationship between these two viruses (Pohlenz et al., 1984).
Bovine torovirus fails to grow in cell culture. In order to obtain data about pathology and pathogenesis, gnotobiotic calves were infected orally with BoTV (Woode et al., 1982; Fagerland et al., 1986). The inoculated calves developed mild to severe diarrhoea with clinical signs of dehydration and weakness. Faecal shedding of BoTV lasted for 3–4 days in these calves.
Bovine torovirus was described in diarrhoeic calves in various countries (Liebler et al., 1992; Duckmanton et al., 1998; Matiz et al., 2002; Smits et al., 2003). The faecal prevalence of BoTV in calf diarrhoea ranges from 5% in Lower Saxony, Germany (Liebler et al., 1992) to 36.4% in southern Ontario, Canada (Duckmanton et al., 1998).
Seroepidemiological studies revealed a wide distribution of BoTV in cattle herds; 94.6% of adult cattle in the Netherlands and Germany (Koopmans et al., 1989), 88.5% of cattle in the USA (Woode et al., 1985) and 55% of cattle in the UK (Brown et al., 1987) were positive for antibodies against BoTV.
To the authors’ knowledge, BoTV has not yet been investigated in Austria. There are no data available about sero‐ or faecal‐prevalences. The aim of the study reported here was to determine the faecal prevalence of BoTV in Austrian calves (with and without diarrhoea) and to find out whether there is a significant difference in the occurrence of BoTV between calves suffering from diarrhoea and healthy calves. Furthermore, the faecal prevalences of the most important viral, bacterial and parasitic enteropathogens as co‐infecting agents to BoTV on the one hand and on the other as causative pathogens in neonatal calf diarrhoea were determined. The health status of cattle herds and single animals in relation to a variety of parameters like clinical symptoms, farm management and presence of pathogens were evaluated statistically.
Materials and Methods
Specimens
During the winter period (October–February) 2004/2005, a total of 100 farms in Styria and Lower Austria were visited. Predominantly milk or milk replacer fed calves from 1 to 42 days of age were included in this study. In 50 of these farms (‘diseased’ farms), one or more calves were suffering from acute diarrhoea at the time of the visit. Other 50 farms (‘healthy’ farms) with a similar structure and geographical distribution had been free from calf diarrhoea for at least 3 weeks and served as a standard of comparison (control group).
Each calf was examined clinically before collection of a faecal sample. In the ‘diseased’ farms, one to four sick calves and, if available, one to four healthy neighbour calves were selected randomly. In the corresponding ‘healthy’ farm, (if available) an equivalent number of calves was included in the study.
The health status of each calf was evaluated by clinical examination. Healthy calves had to be free from diarrhoea, whereas sick calves showed signs of dehydration and weakness and/or abnormal faecal consistency at the time of sampling or shortly before.
Farm management characteristics were evaluated for each farm, including parameters like farm size, animal housing, cleaning and disinfection, feeding of the calves and vaccinations of cows etc.
Virological examination
Faecal samples were stored at −80°C and then tested by reverse transcriptase‐polymerase chain reaction (RT‐PCR) for specific nucleic acid of BoTV, BCV and RV. A commercially available kit (QIAamp® viral RNA mini kit; Qiagen, Hilden, Germany) was used for extraction of viral RNA from faecal samples. The RT‐PCR reaction mixture was prepared following the protocol of a ready‐to‐use kit (QIAGEN® OneStep RT‐PCR kit; Qiagen). Primers were diluted to a final concentration of 20 pmol/μl.
To minimize the risk of cross‐contamination and DNA carry‐over from previous reactions, all procedures were strictly separated physically and carried out following good laboratory practice. Several negative controls of distilled water as well as one positive control were included in each assay. All working units and laboratory materials were decontaminated with disinfectants and/or ultraviolet light before use.
For the detection of specific nucleic acids of BoTV, BCV and RV, primers designed by Tsunemitsu et al. (1999); Hoet et al. (2002) and Schwarz (2002) were used respectively. All details are shown in Table 1.
Table 1.
Details of reverse transcriptase‐polymerase chain reactions (RT‐PCRs) for the detection of bovine torovirus‐, bovine coronavirus‐ and rotavirus‐specific nucleic acids
| Bovine torovirus | Bovine coronavirus | Rotavirus | |
|---|---|---|---|
| Amplification product | 741 bp | 407 bp | 433 bp |
| Upstream primer | 5′‐GTG TTA AGT TTG TGC AAA AAT G‐3′ | 5′‐GCC GAT CAG TCC GAC CAA TC‐3′ | 5′‐AAG TAG CTG GAT TTG ATT ATT C‐3′ |
| Downstream primer | 5′‐TGC ATG AAC TCT ATA TGG TGT‐3′ | 5′‐AGA ATG TCA GCC GGG GTA T‐3′ | 5′‐GAC TCA CAA ACT GCA GAT TCA A‐3′ |
| Volume template | 2.5 μl | 1.6 μl | 1.5 μl |
| Volume RT‐PCR mixture | 22.5 μl | 18.4 μl | 13.5 μl |
| Reverse transcription | 50°C, 30 min | 50°C, 30 min | 50°C, 30 min |
| Initial denaturation | 95°C, 15 min | 95°C, 15 min | 95°C, 15 min |
| Number of cycles | 35 | 35 | 40 |
| Temperature profile | 95°C, 1 min | 94°C, 30 s | 94°C, 1 min |
| 55°C, 1.5 min | 58°C, 1 min | 55°C, 1 min | |
| 72°C, 1.5 min | 72°C, 2 min | 72°C, 1 min | |
| Final extension | 72°C, 10 min | 72°C, 10 min | 72°C, 7 min |
Sequencing of PCR products
Amplified DNA was purified using a commercially available kit (NucleoSpin®Extract; Machery‐Nagel, Düren, Germany) following the manufacturer's instructions and served as a template for sequencing PCR. Two BoTV‐positive samples as well as one randomly chosen sample of BCV and RV each were sequenced in both directions using a commercial sequencing kit (DNA sequencing kit; Applied Biosystems, Warrington, UK) and corresponding PCR primers on ABI prism 310 Genetic Analyser (Applied Biosystems).
Computer‐assisted analysis
Nucleotide sequences of two BoTV‐specific amplified products and corresponding sequences from GenBank were compared using the ClustalW program. Evolutionary distances were calculated using the program dnadist, employing the Kimura two‐parameter method (Kimura, 1980). Phylogenetic analysis of 644 bp DNA fragments was performed using the neighbor program based on the Neighbour‐joining method (Saitou and Nei, 1987) from phylip inference package programs (Felsenstein, 1993). Statistical parameters of phylogenetic trees were determined by bootstrap analysis carried out on 1000 replicates using phylip programs seqboot and consence. The tree was drawn using the drawtree program from phylip.
Parasitological examination
Faecal samples were examined for the presence of parasites using a flotation technique with concentrated sugar solution.
Bacteriological examination
Faecal material was plated on McConkey and blood agar, and incubated aerobically at 37°C for 18–24 h. For the detection of Clostridium spp., samples were plated on blood agar and incubated anaerobically at 37°C for 48 h. For the detection of Salmonella spp., faecal material was incubated in 5 ml of peptone water at 37°C for 24 h, and then, subcultivated on brilliant green agar at 37°C for 24 h.
Statistical analysis
Data were statistically evaluated using the spss for Windows package version 11.5.1 (SPSS Inc., Chicago, IL, USA). First of all, possible associations between the health status of the single animal, respectively, of the farms and farm management characteristics like farm size, animal housing, cleaning and disinfection, feeding of the calves, vaccinations of the cows, etc., were investigated by the chi‐squared test. Whenever an association was hypothesized by an alpha error of P < 0.1, the respective variable was included in the statistical model evaluating the association between the distribution of enteropathogen positive/negative animals and the health status of the single animal.
For final evaluation, a logistic regression model was created, which included the infection status of the animal as dependent variable and the health status of the animal and associated variables as independent variables. The resulting odds ratios (OR), P‐values as well as the 95% confidence interval (CI) of the respective OR are listed in Table 4. Furthermore, the distribution of clinical symptoms in infected versus non‐infected animals was investigated for different pathogens by descriptive analysis.
Table 4.
Occurrence of BoTV and other enteropathogens
| Enteropathogen | BoTV | BCV | RV | Cryptosporidia | Giardia | E. b./z. | Other eim. | Escherichia coli | Clostridium perfringens | Salmonella |
|---|---|---|---|---|---|---|---|---|---|---|
| Pos. cal. (n = 230) Faecal prev. (%) | 12 5.2 | 59 25.7 | 21 9.1 | 27 11.7 | 14 6.1 | 9 3.9 | 15 6.5 | 39 17.0 | 21 9.1 | 0 0 |
| Diseased f. | 8 | 45 | 19 | 25 | 8 | 8 | 8 | 21 | 13 | 0 |
| Healthy f. | 1 | 14 | 2 | 2 | 6 | 1 | 7 | 18 | 8 | 0 |
| Sick calves | 10 | 36 | 17 | 23 | 4 | 7 | 6 | 17 | 9 | 0 |
| Healthy calves | 2 | 23 | 4 | 4 | 10 | 2 | 9 | 22 | 12 | 0 |
| P + significance | 0.022 (s) | 0.1 (ns) | <0.001 (s) | <0.001 (s) | 0.15 (ns) | 0.14 (ns) | 0.72 (ns) | 0.67 (ns) | 0.68 (ns) | nd |
| OR | 6.35 | 2.1 | 9.0 | 12.2 | 0.4 | 3.43 | 1.51 | 1.2 | 0.8 | nd |
| 95% CI of OR | 1.3–30.8 | 0.9–5.2 | 2.9–28.2 | 4.0–36.8 | 0.1–1.4 | 0.7–17.7 | 0.2–14.7 | 0.6–2.5 | 0.2–2.8 | nd |
| Single infections | 2 | 22 | 6 | 6 | 9 | 2 | 9 | 19 | 7 | 0 |
| S.i. + diarrhoea | 2 | 6 | 4 | 5 | 1 | 1 | 3 | 4 | 0 | 0 |
E. b./z., Eimeria bovis and zuernii; other eim., other eimeriae than bovis and zuernii; Pos. cal., calves positive for respective enteropathogen; faecal prev. (%), faecal prevalence (n = 230); diseased f., positive calves from ‘diseased’ farms; healthy f., positive calves from ‘healthy’ farms; sick calves, positive calves with diarrhoea; healthy calves, positive calves without diarrhoea; P + significance, P‐value (ns, not significant; s, significant); OR, odds ratio; 95% CI of OR, 95% confidence interval of OR; single infections, number of calves with single infections with respective enteropathogen; S.i. + diarrhoea, number of calves with single infections with respective enteropathogen suffering from diarrhoea; nd, not done.
Results
A total of 230 calves was included in the study. 125 calves originated from ‘diseased’ farms; 90 of these calves were clinically ill at the time of the visit and 35 calves were clinically healthy neighbour calves. On the 50 ‘healthy’ farms, 105 calves were examined.
Bovine torovirus
Reverse transcriptase‐PCR with BoTV‐specific primers provided an expected 741 bp DNA product originating from S gene with 12 of 230 faecal samples analysed. The specificity of our amplification system was confirmed by sequencing and computer‐assisted phylogenetic analysis of two positive samples (Aut5 and Aut6). When 644 nucleotide sequence stretches of Aut5 and Aut6 were aligned with torovirus sequences available in the GenBank, the analysis revealed 99% similarity between the two Austrian viruses, 93% similarity to Bovine Breda virus but only 71% identities to Equine Berne virus and 52% to Porcine Markelo virus (Table 2). Phylogenetic analysis (Fig. 1) clearly demonstrated that Aut5 and Aut6 were grouped with BoTV strain B145 originating from the Netherlands and Breda strain isolated in Iowa.
Table 2.
Nucleotide sequence similarity of 644 bp (S gene fragment) of two Austrian torovirus‐positive samples (Aut5 and Aut6), bovine Breda and B145, Porcine Markelo and Equine Berne strains
| Aut6 | B145 | Breda | Berne | Markelo | |
|---|---|---|---|---|---|
| 98.8 | 95.3 | 92.5 | 70.8 | 52.3 | Aut5 |
| 95.3 | 92.9 | 70.7 | 52.8 | Aut6 | |
| 91.0 | 70.5 | 52.6 | B145 | ||
| 72.7 | 53.1 | Breda | |||
| 60.0 | Berne |
Figure 1.
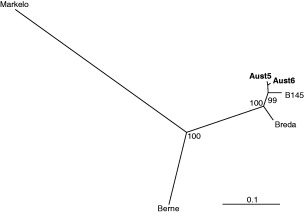
Unrooted phylogenetic tree of two Austrian toroviruses (Aut5, Aut6). The phylogenetic tree was prepared by analysis of a 644 nucleotide sequence fragment of the S gene using the Neighbor‐joining method employing Kimura‐2 parameter method. Numbers over branches represent bootstrap confidence resulting from analysis of 1000 replicates. The scale bar represents 0.1 nucleotide substitution per site. Nucleotide sequences extracted from the GenBank: B145 – accession no. AJ575373, Breda –AF076621, Berne – X52506 and Markelo –AF076621.
Twelve positive samples for BoTV RNA of 230 faecal samples examinated correspond to a virus prevalence of 5.2%. They were identified on nine different farms, eight of which were ‘diseased’ and one ‘healthy’. Ten of the BoTV‐positive calves were clinically ill while the other two showed no clinical symptoms of diarrhoea; one positive calf was a clinically healthy neighbour animal. Six BoTV‐positive calves were younger than 14 days, while the age of the other six ranged from 15 to 28 days. The statistical analysis showed a significant difference (P = 0.022) in the prevalence of BoTV in healthy and diseased calves. The calculated OR‐values indicate that calves shedding BoTV in their faeces are 6.35 times more likely to suffer from diarrhoea than calves not shedding BoTV (95% CI 1.3–30.8; P = 0.022).
On clinical examination, several BoTV‐positive calves showed signs of dehydration: skin elasticity was reduced in eight calves, bulbi were sunken and general behaviour was reduced in two calves. Three BoTV‐positive calves, two of them diarrhoeic, were suffering from additional lung diseases ranging from bronchitis to bronchopneumonia possibly also due to an additional BCV infection present in all three. One of these calves was further co‐infected with Eimeria bovis/zuernii and Klebsiella spp. Faecal consistency was abnormal in seven calves ranging from watery to mushy. Admixtures could be found in the faeces of 11 calves: mucus (nine calves) and blood (two calves). Single infection with BoTV was only found in two calves, both suffering from diarrhoea at the time of sampling. The other animals shed one or more other enteropathogens (Table 3).
Table 3.
Bovine torovirus‐positive calves: concomitant enteropathogens and clinical status
| Enteropathogen | Total | Diarrhoeal calves | Healthy calves |
|---|---|---|---|
| BoTV infections | |||
| BoTV | 2 | 2 | 0 |
| BoTV + BCV | 2 | 1 | 1 |
| BoTV + Clostridium perfringens | 1 | 1 | 0 |
| BoTV + Escherichia coli | 1 | 1 | 0 |
| BoTV + Giardia spp. | 1 | 1 | 0 |
| BoTV + BCV + RV | 1 | 1 | 0 |
| BoTV + BCV + cryptosporidia | 1 | 1 | 0 |
| BoTV + BCV + RV + C. perfringens | 1 | 1 | 0 |
| BoTV + cryptosporidia + C. perfringens + E. Coli | 1 | 0 | 1 |
| BoTV + BCV + Eimeria bovis/zuernii + Klebsiella spp. | 1 | 1 | 0 |
BoTV, Bovine torovirus; BCV, Bovine coronavirus; RV, rotavirus.
Other enteropathogens
The occurrence of other enteropathogens, the P‐value and the corresponding OR values are listed in Table 4. Rotaviruses and cryptosporidia were significantly more frequently detected in calves with diarrhoea with an OR of 9.0 (95% CI 2.9–28.2) and 12.2 (95% CI 4.0–36.8), respectively, (P < 0.001). Concerning BCV, the OR was 2.1 (95% CI 0.9–5.2) with an alpha error of P = 0.1, and therefore, at least showing a trend towards an association between the detection of BCV and diarrhoea.
The following farm management characteristics finally showed significant associations to the infection status with specific enteropathogens (the cited OR values are corrected for the influence of the health status of the calves).
Vaccination
The OR of calves born from cows vaccinated against rotavirus, coronavirus and E. coli to be positive for BoTV was 5.07 (95% CI 1.37–18.77; P = 0.015).
Calving box
The OR of calves from farms not having a calving box to be positive for RV was 0.35 (95% CI 0.12–0.96; P = 0.043).
Housing of the calves
The OR of single kept calves to be positive for RV was 3.80 (95% CI 1.49–9.66; P = 0.005).
Kind of farm
The OR of calves from dairy farms to be positive for cryptosporidia was 0.32 (95% CI 0.10–0.99; P = 0.049).
Discussion
This is the first study to document the presence of BoTV in Austria extending geographic regions where this virus has already been detected. Sequence and phylogenetic analyses of two Austrian BoTV‐positive samples (Fig. 1, Table 2) revealed the highest nucleotide identities with BoTV strain B145 (the Netherlands) and the North American strain Breda of 95% and 93% respectively. Phylogenetic analysis supported these data as Austrian BoTV clustered clearly within BoTV belonging to the family Coronaviridae.
The calculated percentage (5.2%) of BoTV RNA shedders in Austrian cattle herds is comparable with faecal prevalences reported from Lower Saxony (5%; Liebler et al., 1992) and the Netherlands (6%; Koopmans et al., 1991). In contrast, Duckmanton et al. (1998) reported a considerably higher faecal prevalence of 36.4% in southern Ontario. Compared with other enteropathogens, BoTV faecal prevalence is rather low. A possible explanation might be the short excretion period of only 3–4 days (Woode et al., 1982, 1985; Liebler et al., 1992). However, using the very sensitive RT‐PCR, Hoet et al. (2003a) were able to detect BoTV in faeces as early as 24 h post‐infection (p.i.) and up to 8–9 days p.i., with the most consistent range between days 2 and 6 p.i.
In our study, all animals with different histories and duration of diarrhoea were sampled only once. Therefore, it is very likely that not all BoTV infections have been detected. On the other hand, it has to be considered that the detected RNA does not necessarily represent infectious virus and acute infection (Hoet et al., 2003a).
According to previous studies (Koopmans et al., 1991; Duckmanton et al., 1998; Hoet et al., 2003b) we found BoTV significantly (P = 0.022) more frequently in calves with diarrhoea than in asymptomatic calves. Calves shedding BoTV in faeces were 6.35 times more likely to have diarrhoea than calves not shedding BoTV. These results indicate that BoTV, although not a major cause of disease, is of considerable importance in calf diarrhoea. However, it remains impossible to determine whether BoTV was the primary cause of diarrhoea in the symptomatic calves, which had concomitant infections with other enteropathogens. Even in the calves with BoTV single infections, the occurrence of other enteropathogens before the time of sampling cannot be excluded.
On clinical examination, the majority of BoTV‐positive calves showed signs of dehydration, and faecal consistency was abnormal in seven animals ranging from watery to mushy. Three calves were suffering from additional respiratory tract disease (RTD), ranging from bronchitis to bronchopneumonia. In all three calves co‐infected with BCV, a possible trigger and cause of RTD, was detected, the aetiological involvement of the two pathogens is impossible to determine. Observations by Vanopdenbosch et al. (1992) confirmed the pathogenicity of BoTV for the respiratory tract. Respiratory toroviral infections occur mainly during the first month of life and at the age of 4–6 months. It is possible that BoTV might play a role in RTD of young calves (Vanopdenbosch et al., 1991). As in this study, calves with RTD were not investigated for specific aetiological pathogens, BoTV might possibly have played a role as a cofactor in those diseases too. The role of BoTV in RTD has to be clarified by further studies with different sampling techniques.
In this study, the most frequently detected enteropathogen was BCV (25.7%), followed by various types of E. coli, cryptosporidia, rotavirus and Clostridium perfringens. Bovine coronavirus is estimated to account for 20–26% of all cases of calf diarrhoea (Anonymous, 1983), and BCV involvement in calf scours can go up to 48% (Doll, 2002). Rotavirus was detected with a faecal prevalence of 9.1%, which is rather low compared with investigations from Lower Saxony (Germany), where RV was detected in 31.5% of all collected samples (Liebler et al., 1992). In particular, for RV and BCV, the respective prevalences might be higher in our calves, as intermittent virus shedding has been reported in both healthy and diseased animals (Crouch and Acres, 1984; Schwers et al., 1984; Collins et al., 1987). Cryptosporidia faecal prevalence was 11.7%. Faecal prevalences can go up to 60% (Doll, 2002), with a peak shedding in calves 1–3 weeks old (Huetink et al., 2001). Giardia duodenalis was detected in 3–45% of calves older than 2 weeks (Doll, 2002), while in our study, it could be found in 6.1% of all calves. Eimeria bovis and zuernii and other eimeriae faecal prevalences were 3.9% and 6.5% respectively. Depending on animal housing and pasture management, infestation rates can range from 12% to 100% (Rommel, 2000). Concerning E. coli subtype, faecal prevalences range from 3% to 54% (Steiner et al., 1997; Doll, 2002), which corresponds to our results (17.0%). C. perfringens (faecal prevalence in our study: 9.1%) causes a number of diseases among humans and animals; information in literature goes up to a morbidity of 50% in single outbreaks (Klee, 2002).
Liebler et al. (1992) found RV, BCV and cryptosporidia significantly more often in calves with diarrhoea than in healthy calves. In our study, we found a significant difference only for RV and Cryptosporidia (P < 0.001), but not for BCV. However, OR values indicate a tendency that calves shedding BCV are 2.1 times (95% CI 0.9–5.2) more likely to develop diarrhoea.
Significant differences between clinically ill and healthy calves could neither be found for bacteria (E. coli, C. perfringens) nor for eimeriae and Giardia spp.
Concerning the significant associations of certain farm management characteristics with specific enteropathogens, a satisfying interpretation cannot be given; further investigations would need to be done.
In conclusion, we demonstrated for the first time at the genetic level that toroviruses circulate in Austrian cattle populations. Seroepidemiological studies are needed to gain more information about the distribution of these agents. Additionally, the recently described recombinations between different BoTV strains, as well as between BoTV and porcine torovirus, and most interesting with toroviruses of yet unknown origin (Smits et al., 2003) offer a wide field of future research. It remains to be elucidated whether such molecular events are present in the genomes of our torovirus isolates.
Acknowledgements
The authors want to thank the organization ‘Niederoesterreichischer Bauernbund’ for financial support. S. V. was supported by VEGA grant no. 1/2330/05 and SP51/0280800/0280803. Special thanks to Ms. Michaela Stolz for technical support.
References
- Anonymous , 1983: Laboratory Methods for Detecting Calf Diarrhea Viruses. Norden Laboratories, Lincoln, NE. [Google Scholar]
- Brown, D. , Beards G. M., and Flewett T. H., 1987: Detection of Breda virus antigen and antibody in humans and animals by enzyme immunoassay. J. Clin. Microbiol. 25, 637–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, J. K. , Riegel C. A., Olson J. D., and Fountain A., 1987: Shedding of enteric coronavirus in adult cattle. Am. J. Vet. Res. 48, 361–365. [PubMed] [Google Scholar]
- Crouch, C. F , and Acres S. D., 1984: Prevalence of rotavirus and coronavirus antigens in the feces of normal cows. Can. J. Comp. Med. 48, 340–342. [PMC free article] [PubMed] [Google Scholar]
- Doll, K. , 2002: Neugeborenendiarrhoe In: Dirksen G., Gründer H. ‐D., and Stöber M. (eds), Innere Medizin und Chirurgie des Rindes, 4. Auflage, pp. 561–572. Parey, Berlin. [Google Scholar]
- Duckmanton, L. , Carman S., Nagy E., and Petric M., 1998: Detection of bovine torovirus in fecal specimens of calves with diarrhea from Ontario farms. J. Clin. Microbiol. 36, 1266–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerland, J. A. , Pohlenz J. F. L., and Woode G. N., 1986: A morphological study of the replication of Breda virus (proposed family Toroviridae) in bovine intestinal cells. J. Gen. Virol. 67, 1293–1304. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. , 1993: PHYLIP (Phylogeny Inference Package), version 3.5c. Department of Genetics, University of Washington, Seattle, WA. [Google Scholar]
- Hoet, A. E. , Cho K. O., Chang K. O., Loerch S. C., Wittum T. E., and Saif L. J., 2002: Enteric and nasal shedding of bovine torovirus (Breda virus) in feedlot cattle. Am. J. Vet. Res. 63, 342–348. [DOI] [PubMed] [Google Scholar]
- Hoet, A. E. , Chang K. O., and Saif L. J., 2003a: Comparison of ELISA and RT‐PCR versus immune electron microscopy for detection of bovine torovirus (Breda virus) in calf fecal specimens. J. Vet. Diagn. Invest. 15, 100–106. [DOI] [PubMed] [Google Scholar]
- Hoet, A. E. , Smiley J., Thomas C., Nielsen P. R., Wittum T. E., and Saif L. J., 2003b: Association of enteric shedding of bovine torovirus (Breda virus) and other enteropathogens with diarrhea in veal calves. Am. J. Vet. Res. 64, 485–490. [DOI] [PubMed] [Google Scholar]
- Huetink, R. E. C. , Van Der Giessen J. W. B., Noordhuizen J. P. T. M., and Ploeger H. W., 2001: Epidemiology of Cryptosporidium spp. and Giardia duodenalis on a dairy farm. Vet. Parasitol. 102, 53–67. [DOI] [PubMed] [Google Scholar]
- Jamieson, F. B. , Wang E. E. L., Bain C., Good J., Duckmanton L., and Petric M., 1998: Human torovirus: a new nosocomial gastrointestinal pathogen. J. Infect. Dis. 178, 1263–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M. , 1980: A simple method for estimation of evolutionary rates of base substitution through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. [DOI] [PubMed] [Google Scholar]
- Klee, W. , 2002: Clostridiose (Enterotoxämie) In: Dirksen G., Gründer H. ‐D., and Stöber M. (eds), Innere Medizin und Chirurgie Des Rindes, 4. Auflage, pp. 591–593. Parey, Berlin. [Google Scholar]
- Koopmans, M. , Van den Boom U., Woode G., and Horzinek M. C., 1989: Seroepidemiology of Breda virus in cattle using ELISA. Vet. Microbiol. 19, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans, M. , Van Wuijckhuise‐Sjouke L., Schukken Y. H., Cremers H., and Horzinek M. C., 1991: Association of diarrhea in cattle with torovirus infections on farms. Am. J. Vet. Res. 52, 1769–1773. [PubMed] [Google Scholar]
- Liebler, E. M. , Klüver S., Pohlenz J., and Koopmans M., 1992: Zur Bedeutung des Bredavirus als Durchfallerreger in niedersächsischen Kälberbeständen. Dtsch. Tierarztl. Wochenschr. 99, 195–200. [PubMed] [Google Scholar]
- Matiz, K. , Kecskemeti S., Kiss I., Adam Z., Tanyi J., and Nagy B., 2002: Torovirus detection in faecal specimens of calves and pigs in Hungary: short communication. Acta Vet. Hung. 50, 293–296. [DOI] [PubMed] [Google Scholar]
- Pohlenz, J. F. L. , Cheville N. F., Woode G. N., and Mokresh A. H., 1984: Cellular lesions in intestinal mucosa of gnotobiotic calves experimentally infected with a new unclassified bovine virus (Breda virus). Vet. Pathol. 21, 407–417. [DOI] [PubMed] [Google Scholar]
- Rommel, M. , 2000: Protozoeninfektionen der Wiederkäuer In: Rommel M., Eckert J., Kutzer E., Körting W., and Schnieder T. (eds), Veterinärmedizinische Parasitologie, 5. Auflage, pp. 121–191. Parey, Berlin. [Google Scholar]
- Saitou, N. , and Nei M., 1987: The neighbor‐joining method: a new method for reconstructing phylogentic trees. Mol. Biol. Evol. 4, 406–409. [DOI] [PubMed] [Google Scholar]
- Schwarz, B. A. , 2002: Entwicklung einer Reversen Transkription‐Polymerase‐Kettenreaktion (RT‐PCR) zum Nachweis der Persistenz von Rotaviren beim Schwein. Dissertation, Universität Leipzig, Leipzig, Germany. [Google Scholar]
- Schwers, A. , Maenhoudt M., and Pastoret P. P., 1984: Repeated bovine rotavirus infection and excretion in calves. Vet. Rec. 21, 411. [DOI] [PubMed] [Google Scholar]
- Smits, S. L. , Lavazza A., Matiz K., Horzinek M. C., Koopmans M. P., and De Groot R. J., 2003: Phylogenetic and evolutionary relationships among torovirus field variants: evidence for multiple intertypic recombination events. J. Virol. 77, 9567–9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, L. , Busato A., Burnens A., and Gaillard C., 1997: Häufigkeiten und Ursachen von Kälberverlusten und Kälberkrankheiten in Mutterkuhbetrieben. II Mikrobiologische und Parasitologische Diagnosen bei Kälbern mit Durchfall. Dtsch. Tierarztl. Wochenschr. 104, 169–173. [PubMed] [Google Scholar]
- Tsunemitsu, H. , Smith D. R., and Saif L. J., 1999: Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT‐PCR. Arch. Virol. 144, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanopdenbosch, E. , Wellemans G., and Petroff K., 1991: Breda virus associated with respiratory disease in calves. Vet. Rec. 31, 203. [DOI] [PubMed] [Google Scholar]
- Vanopdenbosch, E. , Wellemans G., Oudewater J., and Petroff K., 1992: Prevalence of torovirus infections in Belgian cattle and their role in respiratory, digestive and reproductive disorders. Vlaams Diergeneesk. Tijdschr. 61, 187–191. [Google Scholar]
- Woode, G. N. , Reed D. E., Runnels P. L., Herrig M. A., and Hill H. T., 1982: Studies with an unclassified virus isolated from diarrheic calves. Vet. Microbiol. 7, 221–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woode, G. N. , Saif L. J., Quesada M., Winand N. J., Pohlenz J. F., and Kelso Gourley N., 1985: Comparative studies on three isolates of Breda virus of calves. Am. J. Vet. Res. 46, 1003–1010. [PubMed] [Google Scholar]


