Abstract
The interferons (IFNs) were originally described over 50 years ago, identified by their ability to confer viral resistance to cells. We now know that they are much more than just anti‐viral cytokines collectively having roles in both innate and adaptive immune responses, in tumor surveillance and defense, and modulation of immune cell function. Three types of IFN have now been described, simply referred to as type I, II and III. Distinguishable by the unique receptors that they rely on for signal transduction, the three types of IFN have specific and varied roles in the maintenance of human health and defense against pathogens. In mounting an IFN‐mediated immune response, the human body has developed the ability to regulate IFN‐mediated signal transduction. Like all cytokines, the ability of a cell to respond to IFN is completely dependent on the presence of its cognate receptor on the surface of the target cell. Thus, one of the major mechanisms used by the human body to regulate the strength and duration of the IFN response is through regulation of receptor levels, thereby altering the cytokine‐specific responsiveness of the target cell. This review will discuss the receptor system utilized by the type I IFNs and compare it with that of the type II and III IFNs, which also regulate immune responses through controlling receptor level on the cell surface.
Keywords: interferon signaling, receptors, regulation
Short abstract
The innate immune response recognises pathogens and sterile danger signals to generate effector cytokines, such as type I interferons (IFNs), with a high degree of sophistication. The May/June 2012 issue provides a series of reviews of the role of type I IFNs in regulating immune responses. Topics to be covered include the reason for the induction of IFNs, type I IFNs interactions with and regulation of target cells (e.g. T cells, B cells and Th10 cells) in the immune system, and the roles and mechanisms of IFNs in specific organs or diseases. The accompanying web focus presents links to related articles from across Nature Publishing Group to provide more background information about these proteins.
Discovered over 50 years ago, the interferons (IFNs) were first identified and are historically best known for their ability to elicit viral resistance to cells. 1 On the basis of this criteria the IFNs were initially classified into two types—the type I family composed of the acid‐stable forms IFNα and IFNβ, whereas the acid‐labile form, IFNγ, was classified as the lone type II IFN. 2 In recent years, a third type of IFN has been described, IFNλ. 3 Originally termed interleukin (IL)‐28a/b and IL‐29 these proteins have been re‐classified as IFNs based on the similar modes of induction and anti‐viral activities they share with the type I and type II IFNs. 3 However, although the type I and type III IFNs are induced during a viral infection and are, at least in part, involved in host defense against viruses, the type II IFN is primarily involved in the allergic response, in host defense against intracellular pathogens and in control of tumors.
The cytokines that make up the three types of IFNs share basic secondary structural elements with an overall five helical bundle architecture. The IFNs are all classified as class II alpha‐helical cytokines and thus in the same protein family as IL‐10, IL‐19, IL‐20, IL‐22, IL‐24, IL‐26. 4 But besides a conserved overall helical‐bundle fold, the IFNs otherwise share very limited homology, undoubtedly reflected by their use of distinct receptors for signal transduction. There are nine identified mammalian type I IFN subtypes including IFN‐α, of which there are 13 known subtypes, and single forms of IFN‐β, IFN‐ε, IFN‐κ, IFN‐ω, IFN‐δ, IFN‐τ, IFN‐v, IFN‐ζ. 4 These cytokines can share as much as 100% homology (between certain IFNα subtypes) to as little as ∼20% homology (in a triad between IFNα, IFNβ and IFNε subtypes). Although the most widely studied subtypes are IFNα and IFNβ, evolutionary conservation would suggest that each subtype has unique and perhaps tissue‐specific roles to play in human health and disease. However, not all subtypes are found in humans—IFN‐δ is found only in pigs whereas IFN‐τ is found only in ruminant animals. 4 The type I IFNs are acid stable, a feature which has assisted in the development of protocols for purification of these cytokines for therapeutic applications. 5 In contrast, there is only one type II IFN, IFNγ. 4 IFNγ is acid labile, a feature that distinguishes it from the type I IFNs. Also known as IL28A/IL28B and IL29, three type III IFNs have been identified. 3 Now termed IFNλs due to the common mode of viral induction they share with the type I IFNs, these cytokines have higher structural homology with IL‐10/IL‐22 rather than to the type I IFNs despite having higher amino‐acid identity with the IFNs. 6
Interferon receptor systems
The three IFN types are distinguished by the use of distinctive but related multi‐chain cell‐surface receptor complexes (see Figure 1, Table 1). All receptors involved in IFN signal transduction are classified as class II helical cytokine receptors (hCRs) sharing homologous structural folds and basic structural elements with other proteins including tissue factor, and the receptors for IL‐10, IL‐20 and IL‐22. 4 In the extracellular region, all members of this class of hCR have tandem domains consisting of ∼100 amino acids each housing a type III fibronectin (FBN‐III) domain with topology analogous to the immunoglobulin constant domain. With the exception of IFNAR1, which has a four‐domain architecture, all other IFN receptors consist of two FBN‐III domains. 7 Interestingly, although the receptors are unique to each IFN type, components of each signaling complex, namely IFNAR1, IFNAR2, IFNGR2 and IL10RB are encoded by genes clustered on human chromosome 21q22.1, suggesting common evolutionary conservation and thus a possible functional relationship between these systems.
Figure 1.
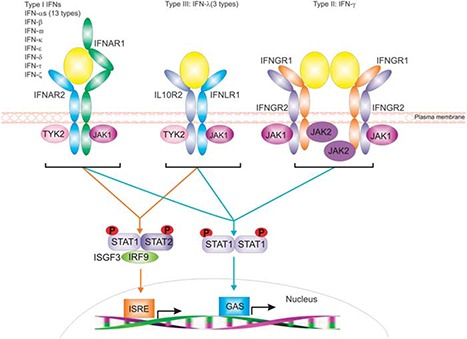
Representation of the distinct receptor systems employed by type I, type II and type III IFNs for signal transduction. Ligand engagement of these receptors initiates a signaling cascade that utilizes the receptor‐associated JAK kinases for receptor phosphorylation and subsequent STAT activation. 7 STATs activated following type I and type III IFN receptor engagement can drive expression of genes with either ISRE or GAS elements in their promoters, whereas signaling via the type II IFN receptor complex almost exclusively drives the expression of genes with GAS promoters elements. 27
Table 1.
The receptor systems and accessory signaling molecules used by the three IFN types for signal transduction
| IFN type | Interferons | Receptor | Signaling molecules |
|---|---|---|---|
| Type I | α (13 types), β, δ, ε, κ, τ, ω, ζ | IFNAR1, IFNAR2 | JAK1, Tyk2 STAT‐1, ‐2, ‐3, ‐4, ‐5 MAPK, PI3K, Akt, NFκB p53, PRMT1 |
| Type II | γ | IFNGR1, IFNGR2 | JAK1, JAK2 STAT‐1,‐2, ‐3, ‐5 MAPK, PI3K, Akt, NFκB |
| Type III | λ | IFNLR1, IL10RB | JAK1, Tyk2 STAT‐1, ‐2, ‐3, ‐4, ‐5 MAPK, PI3K, Akt |
Abbreviations: IFN, interferon; IL, interleukin.
Type I IFN
Despite their seemingly broad range of amino‐acid homologies, all type I IFNs signal through a common heterodimeric receptor composed of low‐ (IFNAR1) and high‐affinity (IFNAR2) receptor components 4 (see Figure 1). IFNAR2 exists as three isoforms transcribed from the same gene by exon skipping, alternative splicing and differential usage of polyadenylation sites. 8 The most well‐characterized form of IFNAR2, IFNAR2c, exists as a long transmembrane form with a full intracellular domain and is required for a complete type I IFN‐induced anti‐viral response via the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling cascade. 9 IFNAR2 also exists as a short transmembrane form lacking the intracellular domain (IFNAR2b) and a soluble form truncated before the transmembrane region (IFNAR2a), but possessing 11 additional carboxyl‐terminal hydrophobic amino acids not found in the extracellular domains of the other two forms. IFNAR2b reportedly acts as a dominant negative regulator of type I IFN activity at least in cell types where it is expressed. 10 Studies in our laboratory have shown that the soluble form of IFNAR2 (IFNAR2a) is found circulating in the blood and can exhibit both agonistic and antagonistic properties in certain circumstances. 11 Although the complete functionality of soluble IFNAR2 in type I IFN signal transduction requires further elucidation, soluble forms of receptors are clearly essential in other cytokine systems. 12
Structurally, while IFNAR2 is typical of all other class II hCRs, the low‐affinity receptor IFNAR1 is unique amongst the class II hCRs being comprised of four FBN‐III domains in the extracellular domain. 7 The functional consequences of a receptor with such an unusually elongated architecture, as compared with all other members of the same protein family, are yet to be fully demonstrated. One of the most compelling questions about the functionality of the type I IFN receptor system is how so many ligands can signal through the same heterodimeric receptor but drive a diverse array of biological signals. Although IFNα2 and IFNβ bind competitively to the IFNAR complex, a number of studies have shown that these cytokines engage the receptor components in ligand‐specific manners. 13 , 14 , 15 The consequences of the different modes of receptor engagement by these two ligands are reflected in the distinct gene sets they induce and the disparate biological responses they generate. 16 The affinity of the various ligands for IFNAR1, the stability of the ternary complex and the number of receptors presented on a cells surface have all been shown to participate in dictating ligand‐specific biological responses. 16 , 17 , 18 Recently, the crystal structures of both extracellular ternary IFNα and IFNω IFNAR signaling complexes were determined. 19 From these structures it is apparent that the ligands have conserved anchor points for receptor engagement but also make ligand‐specific receptor interactions that influence the biological outcome. 19 These structures confirm that it is the ligand‐specific interactions that influence ternary complex stability, and that the affinity of the ligands for IFNAR1 defines the resultant biological outcome of ligand engagement. 19 Although IFNAR1 in the ternary complex structures was truncated before the membrane proximal FBN‐III domain, the structures have given us a greater understanding of differential ligand engagement of a shared receptor but also suggest that every ligand engages the receptor complex in its own particular way.
To transduce signals via the JAK/STAT pathway, IFNAR1 and IFNAR2 are associated with tyrosine kinase 2 (Tyk2) and JAK1, respectively, for the kinase activity required for receptor phosphorylation and STAT recruitment to the receptor complex. 4 In the human system, Tyk2 has been shown to be required for stability of IFNAR1 on the cell surface. 20 A similar role for the IFNAR2‐associated JAK1 has not been demonstrated. Although IFNAR1 and IFNAR2 have not been found to be pre‐associated, they are both required for full type I IFN‐dependent STAT activation and the development of an effective anti‐viral state. 21
Type II IFN
Unlike the type I IFNs, which all appear to signal as monomeric cytokines, IFNγ signals as an anti‐parellel homodimer. 22 . The complex through which this cytokine signals is composed of four transmembrane‐spanning receptors; two chains of each of the high‐affinity (IFNGR1) and low‐affinity receptors (IFNGR2). 4 The IFNγ homodimer engages directly with the two IFNGR1 chains on opposing sides of the cytokine dimer. 22 IFNGR1 has been shown to be pre‐associated with IFNGR2 23 and although the ligand does not engage IFNGR2 directly, ligand‐induced conformational changes in both receptors have been reported. 23 Despite the fact that both IFNGR1 and IFNGR2 are not always present together on the surface of all cells (see Table 2), both receptor components are required for full activity of IFNγ. 24 For signal transduction via the JAK/STAT pathway, IFNGR1 binds to JAK1 whereas IFNGR2 binds to JAK2. 25 Although both kinases are necessary for signal transduction, only JAK1 has been demonstrated to be required for the formation of the full IFNγ signaling complex. 23
Table 2.
Cell type and tissue‐specific presentation of IFN receptors
| Cell/tissue | Interferon receptor | |||||
|---|---|---|---|---|---|---|
| Type I | Type II | Type III | ||||
| IFNAR1 | IFNAR2 | IFNGR1 | IFNGR2 | IFNLR1 | IL10R | |
| T cells | ||||||
| CD4+ | + 1 | + 1 | + 3 | + 3 | − 7 | + 8 |
| CD8+ | + 2 | + 2 | + 4 | − 4 | − 7 | + 8 |
| Th1 | + 5 , 6 | − 5 , 6 | ||||
| Th2 | + 5 , 6 | + 5 , 6 | ||||
| B cells | + 1 , 9 | + 1 , 9 | + 10 | + 10 | − 7 | + 11 |
| Astrocytes | +NS 12 | +NS 13 | − | + 13 | ||
| NK cells | + 14 | + 14 | + 15 | + 15 | − | ND |
| Epithelial cells | + 16 | + 16 | +/− 10 | +/− 10 | + 7 , 17 | + 18 |
| Endothelial cells | +NS 19 | + 20 | + 20 | − 17 | + 21 | |
| Plasmacytoid DCs | + 22 | + 22 | + 3 | + 3 | + 7 | + 23 |
| PBMCs | + 24 | + 24 | + 25 | + 25 | ND | + 26 |
| Tumor/cancer cells | + 27 , 28 , 29 | + 27 , 28 , 29 | + 15 | + 15 | ND | ND |
| Macrophages | + 30 | + 30 | + 10 | + 10 | − 7 | + 31 |
| Hepatocytes | + 32 | + 32 | +Low 10 | +Low 10 | + 33 | + 34 |
| Platelets | − 35 | − 35 | + 15 | + 15 | ND | + 36 |
| Fibroblasts | + 37 | ND | + 15 | + 15 | − 7 | + 38 |
| Phagocytes | ND | ND | + 15 | + 15 | ND | + |
| Eosinophils | + 39 | + 39 | + 40 | ND | + 41 | + 8 |
| Myeloid cells | + 42 | ND | ND | ND | ND | + 26 |
| Serum | ND | +(Soluble) 11 | ND | ND | ND | ND |
| Erythrocytes | ND | ND | − 15 | − 15 | ND | + 43 |
| Keratinocytes | + 44 | + 44 | + 45 | + 45 | + 7 | + 46 |
| Mouse oocytes | ND | ND | + 47 | + 47 | ND | ND |
| Megakaryocytes | + 35 | + 35 | ND | ND | ND | ND |
| Kidney | ND | ND | ND | ND | + 17 , 48 | + 49 |
| Brain | +/− 42 , a | Low a | Low a | +/− a | +/− 17 , a | Low 50 |
| CNS | + 42 | ND | + 51 | + 51 | ND | + 13 |
| Liver | + 52 | + 52 | Low 10 | Low 10 | − 17 | + 49 |
| Lung | + 53 | + 53 | ND | ND | + 17 , 48 | + 49 |
| Gastrointestinal tract | + 54 | + 54 | + 10 | + 10 | + 48 | + 49 |
| Preimplantation embryos | ND | ND | + 47 | + 47 | ND | ND |
Abbreviations: CNS, central nervous system; DCs, dendritic cells; IFN, interferon; IL, interleukin PBMCs, peripheral blood mononuclear cells.
‘+’ represents cell types experimentally demonstrated to present IFN receptors on the surface or for which this is inferred by responsiveness to the IFN type given. ‘−’ represents cell types that have been demonstrated to be negative. ‘ND’ represents cell types or tissues in which the presence of the relevant receptor components have not been demonstrated or are not reported as being found. ‘NS’ is used to demonstrate that the identity of the receptors were not specified.
References cited in the table are availalbe in Supplementary Information.
data for receptor distribution in the brain was also taken from the Allen Brain Atlas at http://www.brain-map.org.
Type III IFN
Similar to the type I IFNs, the type III IFNs signal as monomeric cytokines engaging one copy of each of their low‐affinity and high‐affinity receptors. However, unlike both the type I and type II IFNs, which employ their own dedicated receptors, the IFNλs utilize one unique receptor (IFNLR1) but also one required for signal transduction by IL‐10, IL‐22 and IL‐26 (IL10RB). 3 , 26 The receptor‐associated JAK kinases, JAK1 with IFNLR1 and Tyk2 with IL10RB are responsible for activation of the JAK/STAT pathway upon IFNλ engagement of this receptor complex. 7 As IL10RB is also common to the signaling complexes for IL‐10, IL‐22 and IL‐26, it remains to be seen whether there is any functional cross‐talk between the type III IFNs and these cytokines as a result of having a shared receptor.
Pathways of interferon signaling
All three types of IFN have some similarities and differences in the signal transduction pathways through which they exert their biological effects. All IFNs utilize the JAK/STAT pathway for signal transduction 7 (see Table 1). Much research has shown that specific combinations of STAT homo‐ and heterodimers activate target genes by binding to either interferon response factor 9 (IRF9), which couples the STAT/IRF complex to interferon‐stimulated response elements (ISREs) or directly to GAS elements found in the promoter regions of IFN target genes. 27 ISRE‐binding STAT combinations include STAT1/2 and STAT2/6 heterodimers and STAT2 homodimers; however, IFNγ has been shown to induce ISRE‐binding STAT1 homodimers. 28 GAS elements are generally recognized by STAT1, 3, 4, 5 homodimers and STAT1/2 or 1/3 heterodimers. 27 Despite co‐reliance on the JAK/STAT pathway, the STATs that each of the IFN types activate may be different. For anti‐viral activity, the type I IFNs activate primarily STAT‐1 and ‐2, whereas STAT‐3, ‐4, ‐5 and ‐6 can also be activated by IFNAR engagement in certain cell types (reviewed by Platanias 29 ). Although IFNγ signal transduction predominantly activates STAT1 homodimers, STAT3 homo‐ and STAT1–STAT3 heterodimers can also be generated. 27 Like the type I IFNs, the type III IFNs activate both STAT1 and STAT2 and can therefore drive the transcription of genes with either ISRE or GAS elements. 30 However, in some cell types the type III IFNs can also induce the activation of STAT‐3, ‐4 and ‐5. 31 As the type I and type III IFNs utilize the same JAKs and STATs for signal transduction, it is not surprising that these cytokines have similar biological functions in certain instances. Indeed, it has been demonstrated that IFNα and IFNλ induce a similar set of genes, albeit that the expression induced by IFNλ was weaker than that of IFNα in certain cell types. 32 It seems that the importance of type III IFN signaling lies in the narrow range of cells that have the ability to respond to these cytokines (see below).
Besides the JAK/STAT pathway, type I, II and III IFNs can also activate other signaling pathways, including the MAPK and PI3‐Kinase pathways 29 , 32 (see Table 1). The type I and type II IFNs have also been shown to activate and signal via the NFkB pathway; 33 , 34 however, although experimental evidence does not exist to support the activation of the NFkB pathway directly following IFNλ stimulation, bioinformatic analysis of the promoters of the three type III IFN genes suggests the presence of binding sites for NFkB. 29 The type I and II IFNs have also been shown to activate a CRKL‐dependent pathway important for activation of Rap1 and subsequent antagonism of the Ras pathway thus promoting tumor suppression and the growth inhibitory effects observed for IFNs 35 (reviewed by Platanias 29 ).
Effects of IFNs during immune responses
Generally, IFNs are produced by the body to fight infection or in an allergic response. 4 Following induction, ligand binding to the IFN receptor complexes initiates the transduction of signals that culminate in the transcriptional activation of gene sets, the nature of which is dependent on a number of factors including the stimulus and the IFN type/subtype. Although the defining activity of the IFNs is their ability to invoke an anti‐viral response in the host organism, these cytokines are collectively involved in many more immune responses. Of the IFNs, the type I IFNs have the broadest range of biological activities having both protective and counter‐protective effects in different immune situations. 36 Although type I IFNs are protective in viral infection, IFNβ signaling causes lethality during certain bacterial infections 36 but protection against certain protozoa and fungi. 37 , 38 , 39 Although the mechanism of IFNβ toxicity in sepsis is yet to be fully elucidated, it is apparent that IFNβ engagement of IFNAR1 during sepsis initiates the transmission of these lethal signals; Tyk2 also has a role. 40 , 41 Numerous studies have utilized IFNAR1‐null mice or cells, or anti‐IFNAR1‐neutralizing antibodies to demonstrate the importance of this receptor and type I IFN signal transduction for the maintenance of health and prevention of disease. Using these resources, IFNAR1 has been shown to have a role in tumor surveillance and cancer immunoediting, 42 and to alter the cell cycle, inhibit cell growth and suppress cancer. 43 This receptor also transmits signals involved in regulation of the immune system through the promotion of B‐cell survival, 44 , 45 induction of cross‐priming of T cells, 46 , 47 enhancement of humoral immunity, 48 and regulation of natural killer (NK) cell function 49 (reviewed in Hervas‐Stubbs et al. 50 ). Interestingly, the type I IFNs have also been shown to be components in seemingly conflicting activities of cell survival, 51 and in the promotion 52 and prevention of apoptosis. 44 These contrasting biological outcomes of IFN signaling may be due to differential receptor expression levels on the target cell and/or cell‐specific expression of components of the IFNAR receptor or an unknown cell‐specific regulatory mechanisms or elements that restrict IFN activities in certain cell types.
One organ in which IFNβ has significant importance is the CNS where, in the absence of IFNα, 53 IFNβ has been shown to be vital for viral immunity and protection from autoimmune disease such as experimental autoimmune encephalomyelitis. 54 In contrary to the lethality resulting from the IFNβ/IFNAR1 association seen in sepsis, this ligand‐receptor interaction is protective in the CNS. 54 In an elegant study by Prinz et al., 55 , 56 cell‐specific knockdown of IFNAR1 in CNS cells allowed the identification of myeloid cells as the effector target cell of IFNβ activity in a model of experimental autoimmune encephalomyelitis. In contrast, knockdown of IFNAR1 on B cells, T cells and neuroectodermal cells had no effect on disease progression. 55 This study suggests that despite the ubiquitous expression of the type I IFN receptor on all these cell types, the cell‐specific regulation of IFN signal transduction implies that there are as yet unknown mechanisms or elements that restrict IFN signaling in off‐target cells. It remains to be seen the exact nature of these cell‐specific regulatory mechanisms.
In contrast to the type I IFNs, IFNγ is induced in NK and NKT cells, CD8 T cells and Th1 CD4 effector T cells, has roles in immunity against viruses, intracellular bacteria and tumors and is generally anti‐inflammatory in allergy and asthma. 57 This cytokine thus has a very distinctive activity profile as compared with the type I IFNs. As mentioned, the type III IFNs utilize the same receptor‐associated JAKs and signal transduction pathways and thus induce very similar biological responses to the type I IFNs. 30 Furthermore, both IFN types are induced during a viral infection and are protective against viral pathogens. Where type III IFNs are unique from the type I IFNs is in the narrow range of cells able to respond to the former, as dictated by the presence of their receptor on the surface of these cells. 30 The type III IFNs are active against viral pathogens such as encephalomyocarditis virus and vesicular stomatitis virus (VSV) in several different cell types 3 , 26 , 58 and hepatitis B virus in hepatocytes. 59
Interferon receptor distribution
Apart from the fact that the different IFN types use distinct transmembrane receptors for signal transduction, it is clear that there is a great deal of redundancy in the use of JAKs, STATs and in the alternative signaling pathways employed by the cytokines. In particular, it would seem that the type I and type III IFNs may have some redundancy, as they are both induced by viral infection and utilize identical JAKs and STATs for an effective anti‐viral response. However, as the ability of cells to respond to cytokines is absolutely dependent on the presentation of the required receptor components on the cell surface, it is apparent that due to the strictly regulated distribution of IFN receptor components, the different types of IFN have either widespread or cell/tissue‐specific functions based on the presentation of their receptors on the surfaces of target cells. The receptors for the type I and II IFNs, and the IL10RB involved in IFNλ signaling are generally widely distributed and found on the surface of most cell types (see Table 2). Two major exceptions to this observation are in regards to the specific absence of IFNGR2 on the surface of Th1 cells 60 , 61 and the low expression level of membrane‐bound IFNAR2 in sections of the human brain (see Table 2; http://www.brain-map.org). As IFNγ specifically inhibits the activation of Th2 cells but not Th1 cells, regulation of the surface expression of IFNGR2 in this way restricts responsiveness of Th2 cells to this cytokine. 62 As we know that type I IFN (IFNβ) signaling is important in the brain, the low IFNAR2 expression suggests that either this receptor is not necessary for IFN signaling in this organ, that signaling via the complete IFNAR signaling complex is restricted to cells that have membrane‐bound IFNAR2 or that soluble IFNAR2 contributes to signaling in this organ. This observation could also suggest that canonical type I IFN signaling must be downregulated in the brain for protection of this critical region.
In contrast to type I and type II IFN receptor distribution, the cell surface expression of the high‐affinity receptor for the type III IFNs, IFNLR1 is more restricted thereby limiting cell‐specific responsiveness to these cytokines (see Table 2). Cells of epithelial origin, 30 , 58 particularly keratinocytes and cells from the kidney, lungs and the gastrointestinal tract have been shown to express significant levels of IFNLR1 on their cell surface. 58 , 63 Furthermore, dendritic cells 58 have also been shown to express IFNLR1. 58 Although it is clear that the cellular specificity of response to the type III IFNs lies with the restricted expression of IFNLR1, the receptor it shares with IL‐10, IL‐22 and IL‐26, IL‐10RB is widely distributed on the surface of many different cell types. 64
Factors that regulate interferon receptor presentation
Receptor engagement by the IFNs initiates signaling cascades that lead to the desired biological response. However, the response must be restrained in order to limit cellular responses and avoid the development of a ‘cytokine storm’ often associated with uncontrolled inflammation and lethality. Mechanisms underlying regulation of IFN signaling are multi‐factorial and can involve induction of negative regulators such as suppressor of cytokine signaling (SOCS) proteins, ligand‐induced receptor downregulation, ubiquitination and proteolytic receptor degradation. Both clathrin‐dependent and ‐independent mechanism of endocytosis have been demonstrated to be involved in the regulation of IFN receptor levels on the surface of target cells. Information in this section is summarized in Table 3.
Table 3.
Mechanisms of regulation of cell surface levels of the receptors for type I, type II and type III IFNs
| Interferon | Mechanism of regulation | Reference |
|---|---|---|
| Type I | Tyk2 association | 55 |
| SOCS1 | 56 | |
| Ubiquitination | 57 , 58 | |
| Endocytosis | 55 | |
| Lysosomal degradation | 59 | |
| LPS | 60 | |
| Bcr‐abl | 61 | |
| P38 | 62 | |
| VEGF | 63 | |
| Type II | TCR activation | 64 |
| Endocytosis | 65 | |
| Differential basal expression | 5 , 6 , 15 | |
| Bacterial/protozoan infection | 66 , 67 , 68 , 69 , 70 | |
| Other cytokines | 69 , 70 , 71 | |
| Type III | No information available |
Abbreviations: IFN, interferon; LPS, lipopolysaccharide; SOCS1, suppressor of cytokine signaling 1; TCR, T‐cell receptor; VEGF, vascular endothelial growth factor.
References cited in the table are availalbe in Supplementary Information.
Type I IFN
Regulation of type I IFN signaling can occur via basal, ligand‐dependent and ‐independent diminution of surface receptor levels, receptor ubiquitination promoting degradation, and may involve other and varied mechanisms. To complicate the matter further, the mechanisms of regulation vary between the two receptors and also upon the ligand stimulus. Basally, the decay of IFNAR1 has been shown to be more pronounced than that seen for IFNAR2 in certain cell types, reflecting differential regulation of the two receptors. 65 Also following the exogenous application of type I IFNs, IFNAR1 and IFNAR2 have both been shown to be differentially downregulated. 65 The extent and sustainment of the downregulation has been shown to be different for each receptor and has also been shown to vary with the ligand applied. 65 With respect to IFNAR1 in the human system, the Tyk2 constitutively associated with this receptor 66 has been shown to be involved in aiding stability of this receptor on the cell surface. The interaction between IFNAR1 and Tyk2 not only regulates surface expression levels of the receptor but has also been demonstrated to impede degradation of IFNAR1. 20 Recently, a linear endocytic motif has been identified within the intracellular domain of IFNAR1; it is hypothesized that Tyk2 may mask this motif thereby regulating receptor trafficking. 67 Studies in our laboratory have recently shown that SOCS1 negatively regulates type I IFN signaling 68 via an interaction with Tyk2 thereby controlling the activation status of the IFNAR1‐associated kinase. 69 This association is ligand‐dependent as type I IFN engagement of IFNAR induces expression of SOCS1 via the JAK/STAT pathway and regulates signaling in a negative feedback loop.
The intracellular domain of IFNAR1 contains a degron, a linear motif that directs initiation of receptor degradation via a ubiquitin‐dependent pathway. 70 A conserved serine residue (Ser535) within this motif is the target of kinases; phosphorylation of this serine leads to the recruitment of SCFβTrcp E3 ubiquitin ligase, ubiquitination and degradation of the receptor. 71 In a ligand‐dependent manner, Tyk2 is required for the phosphorylation of S535 on the intracellular domain of IFNAR1; however, at least another kinase, protein kinase D2 is also capable of phosphorylating this site 72 and therefore affecting the cell surface presentation of IFNAR1. Phosphorylation of Ser535 and subsequent ubiquitination and degradation of IFNAR1 is also reportedly mediated by casein kinase 1α (CK1α) in a ligand‐ and JAK kinase‐independent manner, 73 suggesting that there are multiple levels of regulation for this receptor. Interestingly, although IFNAR1 is ubiquitinated following IFN stimulation IFNAR2 is not, 65 suggesting differing mechanisms of regulation of these two receptors on the surface of cells. However, a ubiquitin‐specific protease that is known to be involved in the ISGylation and regulation of certain cellular substrates, UBP43, has been shown to inhibit type I IFN‐induced JAK/STAT signaling by blocking the interaction between IFNAR2 and JAK1. 74
Upon ligand engagement, the ternary IFNAR signaling complex is internalized rapidly by endocytosis. 75 , 76 Although heavily ubiquitinated IFNAR1 is routed for lysosomal degradation, the fate of IFNAR2 seems to depend on the cytokine stimulus. 65 Upon receptor engagement, internalized IFNAR2 has been shown to be either recycled to the cell surface (upon IFNα stimulation) or routed towards degradation (upon IFNβ stimulation). 65 Endocytosis of the ternary signaling complex has been reported to occur in a clathrin‐dependent manner resulting in lysosomal degradation of the IFNAR1 chain. 77 , 78 However, after ligand binding, IFNAR1 has been shown to be associated with lipid rafts 79 often associated with caveolae‐dependent endocytosis although the use of this pathway for the regulation of this receptor on the cell surface has not been directly shown. 75 It is unclear as to whether IFNAR2 can also be found in such lipid rafts or whether this is an IFNAR1‐specific association.
In the human system, a number of other IFNAR immunoregulatory pathways have been demonstrated that are both ligand‐dependent and ‐independent in nature. In mature dendritic cells, following lipopolysaccharide stimulation and subsequent IFNβ induction, IFNAR1 and IFNAR2 levels were shown to be downregulated from the surface of the cells. 80 This study also showed that although IFNAR2 levels returned to maximal after 24 h the levels of IFNAR1 remained low, supporting the suggestion of differential regulatory constraints on the two receptors. 80 Bcr‐abl signaling in chronic myeloid leukemia cells has been demonstrated to accelerate the degradation of IFNAR1 in a protein kinase D2‐dependent manner, 81 attenuating type I IFN signaling in these cells. Similarly, P38, a stress‐activated protein kinase, has been demonstrated to be an important regulator of IFNAR1 surface levels through participation in the phosphorylation of IFNAR1‐S532 and subsequent ubiquitination and degradation of this receptor. 82 Angiogenesis is stimulated by VEGF but inhibited by type I IFNs. 83 It has recently been demonstrated that VEGF can directly antagonize type I IFN signaling by promoting the protein kinase D2‐dependent downregulation of IFNAR1. 84
Many and varied mechanisms are utilized by the host to regulate type I IFN signal transduction; these mechanisms protect the host from the harmful effects of an uncontrolled immune response and also help to maintain homeostasis. In the type I IFN system, the literature suggests that the level of regulation that is applied to IFNAR1 is more stringent than for IFNAR2 implying necessity to do so. Taken from this, it would seem that IFNAR1 is the key to driving signaling via the type I IFNs and that in some instances type I IFN signaling can be strictly regulated through control of IFNAR1 surface levels.
Type II IFN
Similar to the type I IFN receptors, the IFNγ receptors IFNGR1 and IFNGR2 are regulated by distinct mechanisms. The negative regulator SOCS1, which suppresses type I IFN signaling also suppresses signaling via the type II IFN receptor complex specifically through a phosphotyrosine‐specific interaction with IFNGR1. 85 It remains to be seen whether SOCS1 directly affects the regulation of the IFNγ receptor components on the cell surface as it does for IFNAR1 in the type I IFN system described above. In a ligand‐independent manner, IFNGR1 has been shown to be downregulated following engagement of the T‐cell receptor (TCR) in naïve CD4+ T cells. 86 This receptor down‐modulation was found to be dependent on efficient nuclear translocation of nuclear factor of activated T‐cells induced by TCR signaling. 86
Upon ligand engagement of the receptor complex, IFNGR1 and IFNGR2 are internalized by endocytosis (reviewed by Claudinon et al. 75 ). In execution of this process, both caveolae‐ and clathrin‐dependent mechanisms have been reportedly associated with regulation of the type II IFN receptors on the surface of cells. 75 Alongside, IFNAR1, IFNGR1 and IFNGR2 have been shown to be associated with lipid rafts 79 and signaling by the type II IFN signaling complex can be inhibited by the disruption of lipid raft microdomains. 87 However, the association of IFNGR components with lipid rafts has been demonstrated to be both independent of ligand stimulation in certain cell types and dependent in others. 87 It remains to be seen how these seemingly contrasting mechanisms of regulation of IFNGR surface levels may confer cell‐specific constraints on signal transduction by the ternary IFNγ signaling complex.
The two IFNγ receptors are differentially regulated upon internalization via endocytic pathways. It seems that the endocytic pathway in which IFNGR1 is engaged routes the receptor through alternative intracellular pathways. Reports suggest that lipid raft‐associated IFNGR1 is routed to the nucleus to transduce signals via its associated STAT1, 87 whereas IFNGR1 endocytosed in a clathrin‐dependent manner routes IFNGR1 to be recycled to the cell surface (reviewed by Claudinon et al. 75 ). In contrast, after ligand binding the majority of IFNGR2 remains on the cell surface. 88 In human T lymphocytes, ligand‐dependent internalization IFNGR2 is regulated by intracellular trafficking between cytoplasmic stores and the cell surface, thereby limiting surface levels to regulate activity in these cells. 89 IFNGR1 but not IFNGR2 cell surface levels have also been shown to be downregulated in macrophages following treatment with mycobacteria and stimulation with TLR2 agonists. 90 The decrease in cell surface levels of IFNGR1 was shown to be dependent on clathrin and caveolae‐mediated endocytosis and proteasomal degradation. 90 IFNGR1 has also been shown to be downregulated following infection with Leishmania donovani 91 and Trypanosoma cruzi. 92 Interestingly, the type I IFNs, particularly IFNβ, induced following infection with Listeria monocytogenes have been shown to downregulate IFNGR1 surface levels thereby antagonizing the type II IFN response to infection. 93
Other IFN types can reportedly regulate the surface presentation of the type II IFN receptor components. Firstly, the type III IFN, IFNλ3 (IL‐29) has been shown to upregulate IFNGR1 levels on the surface of macrophages 94 demonstrating functional cross‐talk between the type II and III IFNs. As mentioned above, the type I IFNs have also been implicated in the regulation of the type II IFN receptors. 94 However, unlike the effect of IFNλ, the type I IFNs have been shown to downregulate IFNGR1 levels, thereby rendering the macrophages unresponsive to IFNγ treatment. 94 The application of exogenous IFNβ has also been shown to have the same affect on IFNGR1 levels in a mouse model of mycobacterial infection (reviewed by Rayamajhi et al. 93 ). As the type II IFNs have a protective role in the immune response to bacterial infections, the functional benefit that antagonism of IFNγ responsiveness has on the immune response of the host organism by the type I IFNs remains to be seen.
Type III IFN
Most probably due to the relatively limited number of research groups working on elucidating the functionality of the type III IFNs as compared with the type I or type II IFNs, there is at present minimal information on whether and how the type III receptors are regulated following IFNλ signal transduction. As far as can be ascertained from the current literature, there is no information available on whether they are regulated basally or upon ligand binding, whether they are internalized by endocytosis or any other mechanism. However, as expression of the receptors is restricted to cells of epithelial origin it is clear that basal expression of the receptors has a major part in the regulation of IFNλ signaling. Due to the role that Tyk2 has in facilitating SOCS1‐mediated regulation of IFNAR1 surface levels, and since Tyk2 is part of the type III IFN receptor complex through an interaction with IL10RB, it is possible that SOCS1 also has a role in the regulation of type III receptor surface expression. Experimental evidence to this effect has yet to be demonstrated.
Virally induced downregulation of IFN receptors
As a strategy for dampening the host immune response, certain viruses have developed mechanisms to downregulate receptors from both the type I and II IFNs. No information is currently available as to whether viruses can influence the presentation or regulate signaling via the receptors for the type III IFNs. The viruses that target type I IFN signal transduction through downregulation of receptor levels are Hepatitis C virus, Herpes simplex virus, VSV, Severe Acute respiratory syndrome (SARS) Coronavirus and West Nile virus (WNV). Via a ligand‐independent, P38‐activated pathway reliant on CK1α, HSV and VSV were shown to induce a phosphorylation‐dependent down regulation of IFNAR1 in the absence of classical STAT‐driven IFN signaling. 78 , 95 Similarly, a virally expressed accessory protein from the SARS coronavirus has been shown to induce stress within the infected host leading to activation of a ligand‐independent phosphorylation and degradation of IFNAR1. 96 WNV infection has recently been shown to dampen type I IFN responses by inducing a decrease in IFNAR1 levels and by inhibiting surface accumulation of this receptor in infected cells, a mechanism hypothesized to involve viral non‐structural proteins activating protein degradation pathways. 97 In the same study WNV was shown not to effect IFNAR2 surface expression. 97 Immunomodulation of IFNGR1 surface levels is also attributed to two proteins encoded by the human tumor‐inducing virus, Kaposi's sarcoma‐associated herpesvirus. 98 Both K3 and K5 proteins encoded by the virus were shown to downregulate surface levels of IFNGR1 and induce its degradation, thereby reducing responsiveness of the host cells to IFNγ. 98
Virally encoded IFN receptor mimetics
Apart from their effects on cell surface presentation of the IFN receptors, a number of viruses encode and secrete IFN receptor mimetics that directly bind the IFNs preventing an interaction with their receptors, thereby neutralizing cytokine activity. 99 , 100 , 101 , 102 Mimetics of the IFNγ receptor are encoded by vaccinia virus (VV), myxoma virus, ectromelia virus, cowpox virus, camelpox virus and yaba‐like disease virus (YLDV). 99 , 100 , 101 , 102 , 103 , 104 , 105 , 106 , 107 Similarly, VV and many other poxviruses also encode a soluble receptor mimetic that antagonizes type I IFN signaling. 103 , 104 , 105 , 107 , 108 Orthopoxviruses, such as YLDV encode orthologues of the type I IFN antagonist described above and have been shown to neutralize the activity of both type I and type III IFNs. 107
Conclusions and future directions
As with all cytokines and immunomodulatory molecules, the activity of IFNs must be tightly regulated to prevent deleterious effects while still mediating a targeted and efficacious immune response. IFN signal transduction is controlled at many levels, but initially through presentation of the high‐ and low‐affinity receptors expressed on target cells. The fact that viruses have evolved methods to downregulate IFN signaling, either by reducing IFN receptor expression or through production of IFN receptor mimetics, emphasizes the importance of the receptors as a key regulatory step in transducing IFN responses. As the IFNAR receptors are generally widely expressed, the type I IFNs have a broad range of target cells. This contrasts with the type II and type III IFNs, which show restricted cell‐specific activity due to the limited expression of components of their receptors. Regardless of the almost ubiquitous nature of type I IFN signaling, there are isolated examples of differential receptor expression for IFNAR2 with low levels of IFNAR2 message reported in the brain. To understand the implications of this to IFN signal transduction, there is a need for detailed studies of the relative levels of IFNAR1 and IFNAR2 protein expression in particular cell types/organs and during different cellular processes. Furthermore, even though the effects of most IFNs have been shown to be protective, IFNβ is the exception showing lethality in some models of bacterial infection but protection against viruses and autoimmune disease. Perhaps the process by which IFNβ transduces such contrasting functions involves differential use of the IFNARs in certain circumstances, thus leading to the induction of alternative gene sets. These examples of selective use or targeting of the IFNAR components may herald other as yet unidentified instances of differential IFNAR signaling during the regulation of immune responses by type I IFNs. This is an area that requires further study to fully elucidate the complete spectrum of regulatory constraints on IFN signal transduction.
ACKNOWLEDGEMENTS
This work is supported by funding from the National Health and Medical Research Council and the Victorian Government's Operational Infrastructure Support Program.
Supporting information
Supplementary Material
Footnotes
The Supplementary Information that accompanies this paper is available on the Immunology and Cell Biology website (http://www.nature.com/icb)
References
- 1. Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 1957; 147: 258–267. [PubMed] [Google Scholar]
- 2. Farrar MA, Schreiber RD. The molecular cell biology of interferon‐gamma and its receptor. Annu Rev Immunol 1993; 11: 571–611. [DOI] [PubMed] [Google Scholar]
- 3. Kotenko SV, Gallagher G, Baurin VV, Lewis‐Antes A, Shen M, Shah NK et al IFN‐lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 2003; 4: 69–77. [DOI] [PubMed] [Google Scholar]
- 4. Pestka S, Krause CD, Walter MR. Interferons, interferon‐like cytokines, and their receptors. Immunol Rev 2004; 202: 8–32. [DOI] [PubMed] [Google Scholar]
- 5. Pestka S. The interferons: 50 years after their discovery, there is much more to learn. J Biol Chem 2007; 282: 20047–20051. [DOI] [PubMed] [Google Scholar]
- 6. Gad HH, Dellgren C, Hamming OJ, Vends S, Paludan SR, Hartmann R. Interferon‐lambda is functionally an interferon but structurally related to the interleukin‐10 family. J Biol Chem 2009; 284: 20869–20875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kotenko SV, Langer JA. Full house: 12 receptors for 27 cytokines. Int Immunopharmacol 2004; 4: 593–608. [DOI] [PubMed] [Google Scholar]
- 8. Lutfalla G, Holland SJ, Cinato E, Monneron D, Reboul J, Rogers NC et al Mutant U5A cells are complemented by an interferon‐alpha beta receptor subunit generated by alternative processing of a new member of a cytokine receptor gene cluster. Embo J 1995; 14: 5100–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cohen B, Novick D, Barak S, Rubinstein M. Ligand‐induced association of the type I interferon receptor components. Mol Cell Biol 1995; 15: 4208–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gazziola C, Cordani N, Carta S, De Lorenzo E, Colombatti A, Perris R. The relative endogenous expression levels of the IFNAR2 isoforms influence the cytostatic and pro‐apoptotic effect of IFNalpha on pleomorphic sarcoma cells. Int J Oncol 2005; 26: 129–140. [PubMed] [Google Scholar]
- 11. Hardy MP, Owczarek CM, Trajanovska S, Liu X, Kola I, Hertzog PJ. The soluble murine type I interferon receptor Ifnar‐2 is present in serum, is independently regulated, and has both agonistic and antagonistic properties. Blood 2001; 97: 473–482. [DOI] [PubMed] [Google Scholar]
- 12. Jones SA, Richards PJ, Scheller J, Rose‐John S. IL‐6 transsignaling: the in vivo consequences. J Interferon Cytokine Res 2005; 25: 241–253. [DOI] [PubMed] [Google Scholar]
- 13. Lewerenz M, Mogensen KE, Uze G. Shared receptor components but distinct complexes for alpha and beta interferons. J Mol Biol 1998; 282: 585–599. [DOI] [PubMed] [Google Scholar]
- 14. Runkel L, Pfeffer L, Lewerenz M, Monneron D, Yang CH, Murti A et al Differences in activity between alpha and beta type I interferons explored by mutational analysis. J Biol Chem 1998; 273: 8003–8008. [DOI] [PubMed] [Google Scholar]
- 15. Russell‐Harde D, Wagner TC, Perez HD, Croze E. Formation of a uniquely stable type I interferon receptor complex by interferon beta is dependent upon particular interactions between interferon beta and its receptor and independent of tyrosine phosphorylation. Biochem Biophys Res Commun 1999; 255: 539–544. [DOI] [PubMed] [Google Scholar]
- 16. Jaitin DA, Roisman LC, Jaks E, Gavutis M, Piehler J, van der Heyden J et al Inquiring into the differential action of interferons (IFNs): an IFN‐alpha2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN‐beta. Mol Cell Biol 2006; 26: 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kalie E, Jaitin DA, Podoplelova Y, Piehler J, Schreiber G. The stability of the ternary interferon‐receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J Biol Chem 2008; 283: 32925–32936. [DOI] [PubMed] [Google Scholar]
- 18. Levin D, Harari D, Schreiber G. Stochastic receptor expression determines cell fate upon interferon treatment. Mol Cell Biol 2011; 31: 3252–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thomas C, Moraga I, Levin D, Krutzik PO, Podoplelova Y, Trejo A et al Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 2011; 146: 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ragimbeau J, Dondi E, Alcover A, Eid P, Uze G, Pellegrini S. The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. Embo J 2003; 22: 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hwang SY, Hertzog PJ, Holland KA, Sumarsono SH, Tymms MJ, Hamilton JA et al A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc Natl Acad Sci USA 1995; 92: 11284–11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walter MR, Windsor WT, Nagabhushan TL, Lundell DJ, Lunn CA, Zauodny PJ et al Crystal structure of a complex between interferon‐gamma and its soluble high‐affinity receptor. Nature 1995; 376: 230–235. [DOI] [PubMed] [Google Scholar]
- 23. Krause CD, Lavnikova N, Xie J, Mei E, Mirochnitchenko OV, Jia Y et al Preassembly and ligand‐induced restructuring of the chains of the IFN‐gamma receptor complex: the roles of Jak kinases, Stat1 and the receptor chains. Cell Res 2006; 16: 55–69. [DOI] [PubMed] [Google Scholar]
- 24. Pestka S, Kotenko SV, Muthukumaran G, Izotova LS, Cook JR, Garotta G. The interferon gamma (IFN‐gamma) receptor: a paradigm for the multichain cytokine receptor. Cytokine Growth Factor Rev 1997; 8: 189–206. [DOI] [PubMed] [Google Scholar]
- 25. Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol 1997; 15: 563–591. [DOI] [PubMed] [Google Scholar]
- 26. Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE et al IL‐28, IL‐29 and their class II cytokine receptor IL‐28R. Nat Immunol 2003; 4: 63–68. [DOI] [PubMed] [Google Scholar]
- 27. Wesoly J, Szweykowska‐Kulinska Z, Bluyssen HA. STAT activation and differential complex formation dictate selectivity of interferon responses. Acta Biochim Pol 2007; 54: 27–38. [PubMed] [Google Scholar]
- 28. Bluyssen HA, Muzaffar R, Vlieststra RJ, van der Made AC, Leung S, Stark GR et al Combinatorial association and abundance of components of interferon‐stimulated gene factor 3 dictate the selectivity of interferon responses. Proc Natl Acad Sci USA 1995; 92: 5645–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Platanias LC. Mechanisms of type‐I‐ and type‐II‐interferon‐mediated signalling. Nat Rev Immunol 2005; 5: 375–386. [DOI] [PubMed] [Google Scholar]
- 30. Donnelly RP, Kotenko SV. Interferon‐lambda: a new addition to an old family. J Interferon Cytokine Res 2010; 30: 555–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dumoutier L, Lejeune D, Hor S, Fickenscher H, Renauld JC. Cloning of a new type II cytokine receptor activating signal transducer and activator of transcription (STAT)1, STAT2 and STAT3. Biochem J 2003; 370: 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou Z, Hamming OJ, Ank N, Paludan SR, Nielsen AL, Hartmann R. Type III interferon (IFN) induces a type I IFN‐like response in a restricted subset of cells through signaling pathways involving both the Jak‐STAT pathway and the mitogen‐activated protein kinases. J Virol 2007; 81: 7749–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Du Z, Wei L, Murti A, Pfeffer SR, Fan M, Yang CH et al Non‐conventional signal transduction by type 1 interferons: the NF‐kappaB pathway. J Cell Biochem 2007; 102: 1087–1094. [DOI] [PubMed] [Google Scholar]
- 34. Gough DJ, Levy DE, Johnstone RW, Clarke CJ. IFNgamma signaling‐does it mean JAK‐STAT? Cytokine Growth Factor Rev 2008; 19: 383–394. [DOI] [PubMed] [Google Scholar]
- 35. Grumbach IM, Mayer IA, Uddin S, Lekmine F, Majchrzak B, Yamauchi H et al Engagement of the CrkL adaptor in interferon alpha signalling in BCR‐ABL‐expressing cells. Br J Haematol 2001; 112: 327–336. [DOI] [PubMed] [Google Scholar]
- 36. Decker T, Muller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol 2005; 5: 675–687. [DOI] [PubMed] [Google Scholar]
- 37. Orellana MA, Suzuki Y, Araujo F, Remington JS. Role of beta interferon in resistance to Toxoplasma gondii infection. Infect Immun 1991; 59: 3287–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morrell CN, Srivastava K, Swaim A, Lee MT, Chen J, Nagineni C et al Interferon‐{beta} suppresses the development of experimental cerebral malaria. Infect Immun 2011; 79: 1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meissner NN, Swain S, Tighe M, Harmsen A. Role of type I IFNs in pulmonary complications of Pneumocystis murina infection. J Immunol 2005; 174: 5462–5471. [DOI] [PubMed] [Google Scholar]
- 40. Kim JH, Kim SJ, Lee IS, Lee MS, Uematsu S, Akira S et al Bacterial endotoxin induces the release of high mobility group box 1 via the IFN‐beta signaling pathway. J Immunol 2009; 182: 2458–2466. [DOI] [PubMed] [Google Scholar]
- 41. Painz R, Walter I, Kolbe T, Rigler D, Vogl C, Steinborn R et al Organ‐specific and differential requirement of TYK2 and IFNAR1 for LPS‐induced iNOS expression in vivo . Immunobiology 2007; 212: 863–875. [DOI] [PubMed] [Google Scholar]
- 42. Dunn GP, Bruce AT, Sheehan KC, Shankaran V, Uppaluri R, Bui JD et al A critical function for type I interferons in cancer immunoediting. Nat Immunol 2005; 6: 722–729. [DOI] [PubMed] [Google Scholar]
- 43. Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem 1998; 67: 227–264. [DOI] [PubMed] [Google Scholar]
- 44. Badr G, Saad H, Waly H, Hassan K, Abdel‐Tawab H, Alhazza IM et al Type I interferon (IFN‐alpha/beta) rescues B‐lymphocytes from apoptosis via PI3Kdelta/Akt, Rho‐A, NFkappaB and Bcl‐2/Bcl(XL). Cell Immunol 2010; 263: 31–40. [DOI] [PubMed] [Google Scholar]
- 45. Kiefer K, Oropallo MA, Cancro MP, Marshak‐Rothstein A. Role of type I interferons in the activation of autoreactive B cells. Immunol Cell Biol 2012; 90: 498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Le Bon A, Etchart N, Rossmann C, Ashton M, Hou S, Gewert D et al Cross‐priming of CD8+ T cells stimulated by virus‐induced type I interferon. Nat Immunol 2003; 4: 1009–1015. [DOI] [PubMed] [Google Scholar]
- 47. Tough DF. Modulation of T cell function by type I interferon. Immunol Cell Biol 2012; 90: 492–497. [DOI] [PubMed] [Google Scholar]
- 48. Le Bon A, Schiavoni G, D'Agostino G, Gresser I, Belardelli F, Tough DF. Type i interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo . Immunity 2001; 14: 461–470. [DOI] [PubMed] [Google Scholar]
- 49. Trinchieri G, Santoli D. Anti‐viral activity induced by culturing lymphocytes with tumor‐derived or virus‐transformed cells. Enhancement of human natural killer cell activity by interferon and antagonistic inhibition of susceptibility of target cells to lysis. J Exp Med 1978; 147: 1314–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hervas‐Stubbs S, Perez‐Gracia JL, Rouzaut A, Sanmamed MF, Le Bon A, Melero I. Direct effects of type I interferons on cells of the immune system. Clin Cancer Res 2011; 17: 2619–2627. [DOI] [PubMed] [Google Scholar]
- 51. Yang CH, Murti A, Pfeffer SR, Kim JG, Donner DB, Pfeffer LM. Interferon alpha /beta promotes cell survival by activating nuclear factor kappa B through phosphatidylinositol 3‐kinase and Akt. J Biol Chem 2001; 276: 13756–13761. [DOI] [PubMed] [Google Scholar]
- 52. Chawla‐Sarkar M, Leaman DW, Borden EC. Preferential induction of apoptosis by interferon (IFN)‐beta compared with IFN‐alpha2: correlation with TRAIL/Apo2L induction in melanoma cell lines. Clin Cancer Res 2001; 7: 1821–1831. [PubMed] [Google Scholar]
- 53. Sandberg K, Eloranta ML, Campbell IL. Expression of alpha/beta interferons (IFN‐alpha/beta) and their relationship to IFN‐alpha/beta‐induced genes in lymphocytic choriomeningitis. J Virol 1994; 68: 7358–7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Paul S, Ricour C, Sommereyns C, Sorgeloos F, Michiels T. Type I interferon response in the central nervous system. Biochimie 2007; 89: 770–778. [DOI] [PubMed] [Google Scholar]
- 55. Prinz M, Schmidt H, Mildner A, Knobeloch KP, Hanisch UK, Raasch J et al Distinct and nonredundant in vivo functions of IFNAR on myeloid cells limit autoimmunity in the central nervous system. Immunity 2008; 28: 675–686. [DOI] [PubMed] [Google Scholar]
- 56. Kalinke U, Prinz M. Endogenous and therapeutically induced type I interferon responses differentially modulate Th1/Th17 mediated autoimmunity in the CNS. Immunol Cell Biol 2012; 90: 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schoenborn JR, Wilson CB. Regulation of interferon‐gamma during innate and adaptive immune responses. Adv Immunol 2007; 96: 41–101. [DOI] [PubMed] [Google Scholar]
- 58. Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB et al An important role for type III interferon (IFN‐lambda/IL‐28) in TLR‐induced antiviral activity. J Immunol 2008; 180: 2474–2485. [DOI] [PubMed] [Google Scholar]
- 59. Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol 2005; 79: 3851–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pernis A, Gupta S, Gollob KJ, Garfein E, Coffman RL, Schindler C et al Lack of interferon gamma receptor beta chain and the prevention of interferon gamma signaling in TH1 cells. Science 1995; 269: 245–247. [DOI] [PubMed] [Google Scholar]
- 61. Bach EA, Szabo SJ, Dighe AS, Ashkenazi A, Aguet M, Murphy KM et al Ligand‐induced autoregulation of IFN‐gamma receptor beta chain expression in T helper cell subsets. Science 1995; 270: 1215–1218. [DOI] [PubMed] [Google Scholar]
- 62. Gajewski TF, Fitch FW. Anti‐proliferative effect of IFN‐gamma in immune regulation. I. IFN‐gamma inhibits the proliferation of Th2 but not Th1 murine helper T lymphocyte clones. J Immunol 1988; 140: 4245–4252. [PubMed] [Google Scholar]
- 63. Sommereyns C, Paul S, Staeheli P, Michiels T. IFN‐lambda (IFN‐lambda) is expressed in a tissue‐dependent fashion and primarily acts on epithelial cells in vivo . PLoS Pathog 2008; 4: e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. The expanded family of class II cytokines that share the IL‐10 receptor‐2 (IL‐10R2) chain. J Leukoc Biol 2004; 76: 314–321. [DOI] [PubMed] [Google Scholar]
- 65. Marijanovic Z, Ragimbeau J, van der Heyden J, Uze G, Pellegrini S. Comparable potency of IFNalpha2 and IFNbeta on immediate JAK/STAT activation but differential down‐regulation of IFNAR2. Biochem J 2007; 407: 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Colamonici OR, Uyttendaele H, Domanski P, Yan H, Krolewski JJ. p135tyk2, an interferon‐alpha‐activated tyrosine kinase, is physically associated with an interferon‐alpha receptor. J Biol Chem 1994; 269: 3518–3522. [PubMed] [Google Scholar]
- 67. Kumar KG, Varghese B, Banerjee A, Baker DP, Constantinescu SN, Pellegrini S et al Basal ubiquitin‐independent internalization of interferon alpha receptor is prevented by Tyk2‐mediated masking of a linear endocytic motif. J Biol Chem 2008; 283: 18566–18572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fenner JE, Starr R, Cornish AL, Zhang JG, Metcalf D, Schreiber RD et al Suppressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity. Nat Immunol 2006; 7: 33–39. [DOI] [PubMed] [Google Scholar]
- 69. Piganis RA, de Weerd NA, Gould JA, Schindler CW, Mansell A, Nicholson SE et al Suppressor of cytokine signaling (SOCS)1 inhibits type I interferon (IFN) signaling via the IFNAR1 associated tyrosine kinase, Tyk2. J Biol Chem 2011; 286: 33811–33818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kumar KG, Krolewski JJ, Fuchs SY. Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination and degradation of the IFNAR1 subunit of type I interferon receptor. J Biol Chem 2004; 279: 46614–46620. [DOI] [PubMed] [Google Scholar]
- 71. Kumar KG, Barriere H, Carbone CJ, Liu J, Swaminathan G, Xu P et al Site‐specific ubiquitination exposes a linear motif to promote interferon‐alpha receptor endocytosis. J Cell Biol 2007; 179: 935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zheng H, Qian J, Baker DP, Fuchs SY. Tyrosine phosphorylation of protein kinase D2 mediates ligand‐inducible elimination of the Type 1 interferon receptor. J Biol Chem 2011; 286: 35733–35741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu J, Plotnikov A, Banerjee A, Suresh Kumar KG, Ragimbeau J, Marijanovic Z et al Ligand‐independent pathway that controls stability of interferon alpha receptor. Biochem Biophys Res Commun 2008; 367: 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY et al UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J 2006; 25: 2358–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Claudinon J, Monier MN, Lamaze C. Interfering with interferon receptor sorting and trafficking: impact on signaling. Biochimie 2007; 89: 735–743. [DOI] [PubMed] [Google Scholar]
- 76. Payelle‐Brogard B, Pellegrini S. Biochemical monitoring of the early endocytic traffic of the type I interferon receptor. J Interferon Cytokine Res 2010; 30: 89–98. [DOI] [PubMed] [Google Scholar]
- 77. Marchetti M, Monier MN, Fradagrada A, Mitchell K, Baychelier F, Eid P et al Stat‐mediated signaling induced by type I and type II interferons (IFNs) is differentially controlled through lipid microdomain association and clathrin‐dependent endocytosis of IFN receptors. Mol Biol Cell 2006; 17: 2896–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Marijanovic Z, Ragimbeau J, Kumar KG, Fuchs SY, Pellegrini S. TYK2 activity promotes ligand‐induced IFNAR1 proteolysis. Biochem J 2006; 397: 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Takaoka A, Mitani Y, Suemori H, Sato M, Yokochi T, Noguchi S et al Cross talk between interferon‐gamma and ‐alpha/beta signaling components in caveolar membrane domains. Science 2000; 288: 2357–2360. [DOI] [PubMed] [Google Scholar]
- 80. Severa M, Remoli ME, Giacomini E, Ragimbeau J, Lande R, Uze G et al Differential responsiveness to IFN‐alpha and IFN‐beta of human mature DC through modulation of IFNAR expression. J Leukoc Biol 2006; 79: 1286–1294. [DOI] [PubMed] [Google Scholar]
- 81. Bhattacharya S, Zheng H, Tzimas C, Carroll M, Baker DP, Fuchs SY. Bcr‐abl signals to desensitize chronic myeloid leukemia cells to IFNalpha via accelerating the degradation of its receptor. Blood 2011; 118: 4179–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bhattacharya S, Qian J, Tzimas C, Baker DP, Koumenis C, Diehl JA et al Role of p38 protein kinase in the ligand‐independent ubiquitination and down‐regulation of the IFNAR1 chain of type I interferon receptor. J Biol Chem 2011; 286: 22069–22076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Xiao HB, Zhou WY, Chen XF, Mei J, Lv ZW, Ding FB et al Interferon‐beta efficiently inhibited endothelial progenitor cell‐induced tumor angiogenesisi. Gene Ther 2011. (e‐pub ahead of print 10 November 2011; doi: 10.1038/gt.2011.171). [DOI] [PubMed] [Google Scholar]
- 84. Zheng H, Qian J, Varghese B, Baker DP, Fuchs S. Ligand‐stimulated downregulation of the alpha interferon receptor: role of protein kinase D2. Mol Cell Biol 2011; 31: 710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Qing Y, Costa‐Pereira AP, Watling D, Stark GR. Role of tyrosine 441 of interferon‐gamma receptor subunit 1 in SOCS‐1‐mediated attenuation of STAT1 activation. J Biol Chem 2005; 280: 1849–1853. [DOI] [PubMed] [Google Scholar]
- 86. Skrenta H, Yang Y, Pestka S, Fathman CG. Ligand‐independent down‐regulation of IFN‐gamma receptor 1 following TCR engagement. J Immunol 2000; 164: 3506–3511. [DOI] [PubMed] [Google Scholar]
- 87. Subramaniam PS, Johnson HM. Lipid microdomains are required sites for the selective endocytosis and nuclear translocation of IFN‐gamma, its receptor chain IFN‐gamma receptor‐1, and the phosphorylation and nuclear translocation of STAT1alpha. J Immunol 2002; 169: 1959–1969. [DOI] [PubMed] [Google Scholar]
- 88. Larkin J 3rd, Johnson HM, Subramaniam PS. Differential nuclear localization of the IFNGR‐1 and IFNGR‐2 subunits of the IFN‐gamma receptor complex following activation by IFN‐gamma. J Interferon Cytokine Res 2000; 20: 565–576. [DOI] [PubMed] [Google Scholar]
- 89. Rigamonti L, Ariotti S, Losana G, Gradini R, Russo MA, Jouanguy E et al Surface expression of the IFN‐gamma R2 chain is regulated by intracellular trafficking in human T lymphocytes. J Immunol 2000; 164: 201–207. [DOI] [PubMed] [Google Scholar]
- 90. Curry H, Alvarez GR, Zwilling BS, Lafuse WP. Toll‐like receptor 2 stimulation decreases IFN‐gamma receptor expression in mouse RAW264.7 macrophages. J Interferon Cytokine Res 2004; 24: 699–710. [DOI] [PubMed] [Google Scholar]
- 91. Ray M, Gam AA, Boykins RA, Kenney RT. Inhibition of interferon‐gamma signaling by Leishmania donovani. J Infect Dis 2000; 181: 1121–1128. [DOI] [PubMed] [Google Scholar]
- 92. Kierszenbaum F, Mejia Lopez H, Tanner MK, Sztein MB. Trypanosoma cruzi‐induced decrease in the level of interferon‐gamma receptor expression by resting and activated human blood lymphocytes. Parasite Immunol 1995; 17: 207–214. [DOI] [PubMed] [Google Scholar]
- 93. Rayamajhi M, Humann J, Kearney S, Hill KK, Lenz LL. Antagonistic crosstalk between type I and II interferons and increased host susceptibility to bacterial infections. Virulence 2010; 1: 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liu BS, Janssen HL, Boonstra A. IL‐29 and IFNalpha differ in their ability to modulate IL‐12 production by TLR‐activated human macrophages and exhibit differential regulation of the IFNgamma receptor expression. Blood 2011; 117: 2385–2395. [DOI] [PubMed] [Google Scholar]
- 95. Qian J, Zheng H, Huangfu WC, Liu J, Carbone CJ, Leu NA et al Pathogen recognition receptor signaling accelerates phosphorylation‐dependent degradation of IFNAR1. PLoS Pathog 2011; 7: e1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Minakshi R, Padhan K, Rani M, Khan N, Ahmad F, Jameel S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand‐independent downregulation of the type 1 interferon receptor. PLoS One 2009; 4: e8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Evans JD, Crown RA, Sohn JA, Seeger C. West Nile virus infection induces depletion of IFNAR1 protein levels. Viral Immunol 2011; 24: 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li Q, Means R, Lang S, Jung JU. Downregulation of gamma interferon receptor 1 by Kaposi's sarcoma‐associated herpesvirus K3 and K5. J Virol 2007; 81: 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Upton C, Mossman K, McFadden G. Encoding of a homolog of the IFN‐gamma receptor by myxoma virus. Science 1992; 258: 1369–1372. [DOI] [PubMed] [Google Scholar]
- 100. Alcami A, Smith GL. Vaccinia, cowpox, and camelpox viruses encode soluble gamma interferon receptors with novel broad species specificity. J Virol 1995; 69: 4633–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mossman K, Upton C, Buller RM, McFadden G. Species specificity of ectromelia virus and vaccinia virus interferon‐gamma binding proteins. Virology 1995; 208: 762–769. [DOI] [PubMed] [Google Scholar]
- 102. Mossman K, Upton C, McFadden G. The myxoma virus‐soluble interferon‐gamma receptor homolog, M‐T7, inhibits interferon‐gamma in a species‐specific manner. J Biol Chem 1995; 270: 3031–3038. [DOI] [PubMed] [Google Scholar]
- 103. Colamonici OR, Domanski P, Sweitzer SM, Larner A, Buller RM. Vaccinia virus B18R gene encodes a type I interferon‐binding protein that blocks interferon alpha transmembrane signaling. J Biol Chem 1995; 270: 15974–15978. [DOI] [PubMed] [Google Scholar]
- 104. Symons JA, Alcami A, Smith GL. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell 1995; 81: 551–560. [DOI] [PubMed] [Google Scholar]
- 105. Alcami A, Symons JA, Smith GL. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J Virol 2000; 74: 11230–11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Puehler F, Weining KC, Symons JA, Smith GL, Staeheli P. Vaccinia virus‐encoded cytokine receptor binds and neutralizes chicken interferon‐gamma. Virology 1998; 248: 231–240. [DOI] [PubMed] [Google Scholar]
- 107. Huang J, Smirnov SV, Lewis‐Antes A, Balan M, Li W, Tang S et al Inhibition of type I and type III interferons by a secreted glycoprotein from Yaba‐like disease virus. Proc Natl Acad Sci USA 2007; 104: 9822–9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Liptakova H, Kontsekova E, Alcami A, Smith GL, Kontsek P. Analysis of an interaction between the soluble vaccinia virus‐coded type I interferon (IFN)‐receptor and human IFN‐alpha1 and IFN‐alpha2. Virology 1997; 232: 86–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


