Abbreviations.
SARS, severe acute respiratory syndrome; SCoV, SARS coronavirus; MHV, mouse hepatitis virus; RT‐PCR, reverse transcriptase polymerase chain reaction; RNA, ribose nucleic acid.
Severe acute respiratory syndrome (SARS) is an emerging novel infectious disorder that was first diagnosed in Guangdong province in China in November 2002 and subsequently spread worldwide.1, 2, 3, 4, 5 Spread of SARS is via airborne droplets, and infection results in acute pulmonary inflammation and epithelial damage.6 During the initial outbreak, SARS was documented in approximately 8,000 persons globally, resulting in more than 700 deaths.
A growing body of evidence has proven that a novel coronavirus is the etiologic agent in SARS.7 Electron microscopic analysis of affected lung tissues revealed the distinctive structure of a coronavirus (Fig. 1). 8Genomic RNA of the SARS coronavirus (SCoV) has been isolated from sputum and tissue samples of SARS patients.1, 2, 3, 9 Koch's postulates were fulfilled with the demonstration that infection of primates with SCoV resulted in a similar clinical syndrome as observed in humans.9 Evidence suggests that SARS may have originated from an animal reservoir: Viruses similar to the SCoV have been found in Himalayan palm civets sold in live‐animal markets in Guangdong, China.10
Figure 1.
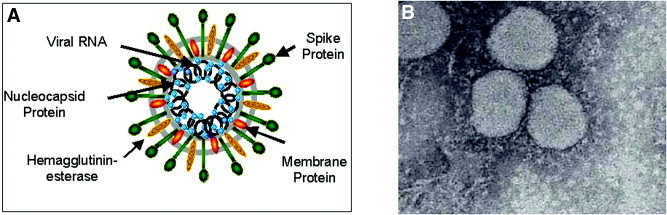
Structure of a coronavirus. (A) Schematic of the coronavirus MHV showing structural proteins and the genomic RNA. (B) Electron micrograph of the SCoV recovered from lung tissue from a patient who died of SARS.
The clinical severity of SARS can vary considerably, presumably because of the genetic diversity of host immune responses. Clinically, SARS is characterized by fever (100%), productive cough and shortness of breath (70%–100%), myalgia (70%), and diarrhea (25%–50%). In some cases, rapid progression of lung consolidation occurs, leading to respiratory failure and death.4, 5 Laboratory abnormalities include elevated lactate dehydrogenase (70%), lymphopenia (50%–70%), thrombocytopenia (50%), and hypocalcemia (60%). Liver enzyme abnormalities are common in SARS patients, although hepatic impairment has not been reported to be a prominent feature of this illness.
Coronaviruses are a diverse group of large, enveloped, positive‐stranded RNA viruses that cause a broad spectrum of diseases, including pneumonitis, hepatitis, nephritis, enteritis, and encephalitis in animals, and several of these viruses are significant veterinary pathogens.11 In humans, coronaviruses usually cause upper respiratory and enteric infections, and lower respiratory tract disease is rare. There are three groups of coronaviruses; each group is classified based on host range and genomic organization.11 Coronaviruses typically have narrow host ranges. Human coronaviruses belong to group 1 (HCoV‐229E) and group 2 (HCoV‐OC43). Although the SCoV does not strictly fall into any of the three groups of known coronaviruses, it bears closest resemblance to the group 2 coronaviruses, which also contain mouse hepatitis virus (MHV).
The type of disease incited by a given coronavirus is influenced by the age and genetic background of the host, the route of infection, and the biologic properties of the coronavirus serotype. The Coronaviridae share replicative and transcriptional features.7 The coronavirus genome is a single, nonsegmented RNA strand with an estimated molecular weight of 6 to 8 million Da.7 Coronaviruses contain an RNA‐dependent RNA polymerase that transcribes the positive template to synthesize both full‐length and subgenomic negative stranded RNAs that then serve as templates for mRNA synthesis. The polymerase is error prone, and RNA recombination occurs at a high frequency, with recombination sites observed throughout the genome, both in translated and untranslated regions, resulting in a very adaptable, mutable viral family. The three major structural proteins of coronaviruses are the nucleocapsid protein, the transmembrane protein, and the surface or spike glycoprotein.7 Coronaviruses of group 2 also have a hemagglutinin esterase protein. The spike glycoprotein binds to receptors on host cells and fuses the viral envelope with host cell membranes.7 Individual coronaviruses use different cellular receptors. Whereas the group 2 coronavirus (MHV) uses murine carcinoembryonic antigen‐related cell adhesion molecules, which are members of the immunoglobulin superfamily of receptors,12 a number of group 1 coronaviruses such as human coronavirus 229E require the zinc metalloprotease aminopeptidase nucleocapsid protein for entry into their target cells.13 Recently, the putative cell receptor for the SARS coronavirus has been identified as the metallopeptidase, angiotensin‐converting enzyme 2.14
In this issue of Hepatology, Chau et al.15 describe the clinical course and liver pathologic features in three SARS patients with liver impairment. All patients had moderate to marked elevation in serum alanine transaminase and underwent liver biopsy. Examination of these biopsy specimens suggested that liver impairment in SARS is the result of SCoV infection of the liver rather than other factors such as drug toxicity, superimposed bacterial sepsis, or a systemic inflammatory response. All three liver biopsies demonstrated hepatocyte apoptosis, and two of three had marked accumulation of cells in mitosis. Common pathologic features included ballooning of hepatocytes and mild to moderate lobular lymphocytic infiltration. Reverse transcriptase polymerase chain reaction (RT‐PCR) was positive for SARS coronavirus in liver tissue from all three patients, although virions could not be detected either by plaque assay on Vero cells or by electron microscopy.
The finding of a significant number of cells in the mitotic phase is interesting, and the authors raise the possibility of cell cycle disruption resulting from the SCoV, which has been reported in animal experimental models of MHV infection. In other studies, liver enzyme abnormalities have been reported commonly in SARS patients with an elevated alanine transaminase being the most common abnormality described. In an analysis of 138 cases of SARS, alanine transaminase was elevated in 23.4% of patients.4 Because the SCoV can be detected in many different types of tissues in infected patients,2 it is not surprising that direct infection of the liver with the virus can lead to liver enzyme abnormalities. However, this is the first report to associate hepatic SCoV infection and liver pathologic features.
Available data about MHV may have relevance to SARS and specifically to hepatic injury resulting from SARS. Although MHV replicates in all mice, certain strains develop a fulminant hepatitis and die, whereas others have little or no liver pathologic features and live. Within 24 hours of infection with MHV, livers from susceptible mice had focal necrotic lesions consisting of acidophilic degenerating hepatocytes and nuclear debris with a sparse inflammatory infiltrate.16 By 3 days after infection, these lesions became larger, more numerous, and were associated with an infiltrate of polymorphonuclear leukocytes (Fig. 2A). At 5 days, confluent necrosis was evident, and by day 7 to 10 days, the lesions were densely infiltrated with mononuclear cells and the adjoining liver parenchyma underwent hepatocyte regeneration as evidenced by many mitotic figures (Fig. 2B). Early in the course of the disease, virions could be detected by both plaque assay and electron microscopy, but by day 7, virions could not be detected by either assay, which may be relevant to the report by Chau et al.15 (Fig. 3).
Figure 2.
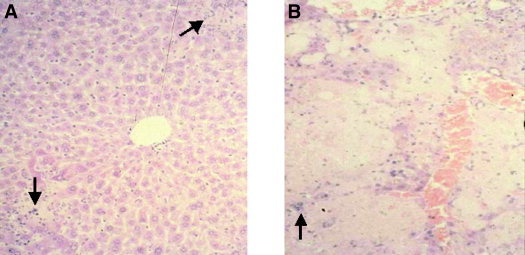
Liver histologic analysis in susceptible mice after MHV‐3 infection. (A) Areas of focal hepatic necrosis with polymorphonuclear infiltrates (arrow; original magnification, ×350). (B) Confluent hepatic necrosis with mononuclear cell infiltrates (arrow; original magnification, ×350).
Figure 3.
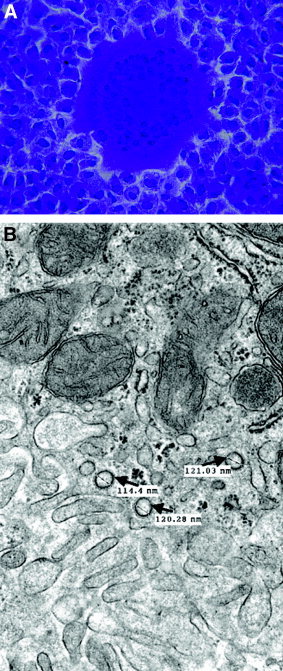
Assays for detection of coronavirus infection (MHV). (A) Plaque assay with fusion of cells (syncytia) on a monolayer of L2 cells stained with crystal violet. (B) Electron micrograph of liver at 3 days after infection showing typical MHV virions in a hepatocyte (arrow). The virions are round, mildly pleomorphic (100–120 nm), and show surface membrane thickening indicative of the corona peplomer structure.
Defining the basis for susceptibility to severe inflammatory outcomes after coronavirus infection has obvious implications for the study and treatment of clinical SARS. Although the exact mechanisms for resistance and susceptibility to MHV3 coronavirus infection have not be determined, studies have shown that in resistant mice, infection leads to a T helper type 1 (TH1)‐dominant response that, through the production of interferon, neutralizing antibodies, and cytotoxic T cells, results in viral clearance.17 In susceptible mice, viral infection results in a marked inflammatory response, with increased production of tumor necrosis factor, interleukin 1, and interferon‐γ associated with a T helper type 2 (TH2) cellular immune response and production of nonneutralizing antibodies.18 This response fails to control viral infection and results in endothelial cell activation, recruitment of inflammatory cells, vasoconstriction, intravascular thrombosis, and liver necrosis.19 Interestingly, fibrin thrombi and intimal swelling of pulmonary vessels have been described in autopsy samples of lung from SARS patients, and transcripts of a potent prothrombotic, fgl2/fibroleukin, have been detected. However, in the paper by Chau et al.,15 similar pathologic changes were not noted in the liver biopsies. This may reflect factors such as tropism of the SCoV agent for the lung, distribution and density of the receptor for SCoV in liver compared with lung tissue, as well as host genetic factors yet to be determined. The fact that a possible receptor for SCoV has recently been identified—angiotensin converting enzyme 2—will help in clarifying organ susceptibility to infection by SARS.
At present, no effective therapeutic strategy has been developed for patients with SARS. Treatments that were used commonly in the last outbreak of SARS included ribavirin and corticosteroids.4 Ribavirin was chosen because of its broad spectrum of activity against RNA viruses and steroids for their antiinflammatory properties. It is currently uncertain whether this treatment regimen had any positive impact on the outcomes. Furthermore, use of Ribavirin is associated with significant toxicity, including hemolysis resulting in discontinuation of its use. Kaletra is a combination of lopinavir and ritonavir, two human immunodeficiency virus protease inhibitors, that has been used in some patients with SARS. Again, efficacy of this medication against SARS is largely unknown. Immunoglobulin, interferons, and SCoV‐specific protease inhibitors and fusion inhibitors represent alternative therapeutic options for treating SARS patients. Interferon has in vitro activity against the SARS virus, and recent studies suggest this may be an effective agent, as has been shown for MHV infection in rodents.20, 21
Currently, it is unknown whether SARS will recur. A global infection control effort was effective in ending the first epidemic of SARS. However, if SARS behaves in a manner similar to other coronavirus, recurrences may be seasonal. In addition, animal reservoirs may serve as a source for future outbreaks. Our knowledge and understanding of SARS has progressed at a rapid rate, but much remains unknown about this new infectious threat.
References
- 1. Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003; 361: 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, et al. A novel coronaviruses associated with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1953–1966. [DOI] [PubMed] [Google Scholar]
- 3. Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003; 348: 1967–1976. [DOI] [PubMed] [Google Scholar]
- 4. Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, Ahuja A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med 2003; 348: 1986–1994. [DOI] [PubMed] [Google Scholar]
- 5. Poutanen SM, Low DE, Henry B, Finkelstein S, Rose D, Green K, Tellier R, et al. Identification of severe acute respiratory syndrome in Canada. New Engl J Med 2003: 348: 1993–2003. [DOI] [PubMed] [Google Scholar]
- 6. Donnelly CA, Ghani AC, Leung GM, Hedley AJ, Fraser C, Riley S, Abu‐Raddad LJ, et al. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet 2003; 361: 1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Holmes KV. SARS‐associated coronavirus. N Engl J Med 2003; 348: 1948–1951. [DOI] [PubMed] [Google Scholar]
- 8. Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, Chu CM, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet 2003; 361: 1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 2003; 362: 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003; 302: 276–278. [DOI] [PubMed] [Google Scholar]
- 11. Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003; 300: 1394–1399. [DOI] [PubMed] [Google Scholar]
- 12. Dveksler GS, Dieffenbach CW, Cardellichio CB, McCuaig K, Pensiero MN, Jiang GS, Beauchemin N, et al. Several members of the mouse carcinoembryonic antigen‐related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus‐A59. J Virol 1993; 67: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, Holmes KV. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 1992; 357: 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, et al. Angiotensin‐converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003; 426: 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chau TN, Lee KC, Yao H, Tsang TY, Chow TC, Yeung YC, Choi KW, et al. SARS‐associated viral hepatitis caused by a novel coronavirus: report of three cases. HEPATOLOGY 2004; 39: 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. MacPhee PJ, Dindzans VJ, Fung LS, Levy GA. Acute and chronic changes in the microcirculation of the liver in inbred strains of mice following infection with mouse hepatitis virus type 3. HEPATOLOGY 1985: 5: 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pope M, Chung S, Mosmann T, Leibowitz J, Gorczynski R, Levy G. Resistance of naïve mice to murine hepatitis virus strain 3 (MHV‐3) requires development of a Th1, but not Th2 response, whereas pre‐existing antibody protects against primary infection. J Immunol 1996; 156: 3342–3349. [PubMed] [Google Scholar]
- 18. Pope M, Rostein O, Cole E, Sinclair S, Parr R, Cruz B, Fingerote R, et al. Pattern of disease after murine hepatitis virus strain 3 (MHV‐3) infection correlates with macrophage activation and not viral replication. J Virol 1995; 69: 5252–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marsden P, Fung L, Mendicino M, Ghanekar A, Miller T, Chan C, Chan M, et al. The Fgl2/fibroleukin prothrombinase contributes to immunologically‐mediated thrombosis in experimental and viral hepatitis. J Clin Invest 2003; 112: 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cinatl J, Morgnestern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferons. Lancet 2003; 362: 293–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Loutfy MR, Blatt LM, Siminovitch KA, Ward S, Wolff B, Lho H, Pham DH, et al. Therapeutic potential of interferon alfacon‐1 plus corticosteroids in severe acute respiratory syndrome: a pilot study. JAMA 2003; 290: 3222–3228. [DOI] [PubMed] [Google Scholar]


