Abstract
Innovation in the life sciences depends on how much information is produced as well as how widely and easily it is shared. Policies governing the science commons – or alternative, more restricted informational spaces – determine how widely and quickly information is distributed. The purpose of this paper is to highlight why the science commons matters and to analyse its structure and function. The main lesson from our analysis is that both the characteristics of the physical resources (from genes to microbes, plants and animals) and the norms and beliefs of the different research communities – think of the Bermuda rules in the human genome case or the Belem declaration for bioprospecting – matter in the institutional choices made when organising the science commons. We also show that the science commons contributes to solving some of the collective action dilemmas that arise in the production of knowledge in Pasteur's Quadrant, when information is both scientifically important and practically applicable. We show the importance of two of these dilemmas for the life sciences, which we call respectively the diffusion–innovation dilemma (how readily innovation diffuses) and the exploration–exploitation dilemma (when application requires collective action).
Introduction
Which institutions have clamoured before the US Supreme Court to ensure that they can use patented inventions without having to get a licence or pay a royalty? Universities? No, they oppose the research exemption, and university technology managers have written a letter opposing proposed general research exemption in US law. Pharmaceutical companies fervently defend patents as the lifeblood of their industry, but in Merck v. Integra they argued for a broad exemption from patent infringement.
In the early history of the genome project, who decided to sequence genes and put the sequence information immediately into the public domain? The government? No, it opted to support such gene‐sequencing only in special instances and on a small scale. It was a large firm, Merck, that decided to fund this work and make it freely available. A consortium of companies teamed up with a few academic institutions to establish a collection of single nucleotide polymorphisms (SNPs). They filed patents with the full intention of abandoning them, in order to bolster the public domain. Why? To thwart the efforts of many other companies and universities to patent SNPs.
The purpose of this paper is to highlight the reasons why the science commons matter. Some of these reasons are obvious, but some are not so obvious and may even be counterintuitive. One way to understand these complex dynamics of innovation is through empirical studies, and some excellent work in this field does exactly that. The main approach we will take, however, is historical, focusing on genomics and microbiological resources as a field of study, but occasionally straying into related fields of biomedical research (such as bioinformatics or molecular and cellular biology) when they provide better examples to illustrate a point. We will also discuss examples from the broader field of biological resources in general, because of the many analogies between genomics, and studies of life sciences related to microbes and higher life forms.
There is some imprecision in the term “science commons”. The term “commons” has been used extensively in legal scholarship to designate goods to which there is open access (Benkler 1998; Lessig 1999). In the same vein, “Science Commons” is a specific organisation that has spun out of the Creative Commons movement. Science Commons moved from concept to action in the year 2005, with an office and executive director (John Wilbanks) to carry out its mission of “making it easier for scientists, universities, and industries to use literature, data, and other scientific intellectual property and to share their knowledge with others. Science Commons works within current copyright and patent law to promote legal and technical mechanisms that remove barriers to sharing”. 1 While we endorse their mission, they may not endorse our analysis; we have no direct connection to the organisation, and do not speak for it.
Some fuzziness around the edges of what constitutes a science commons concerns how it relates to “the public domain”. There are many terms marching under the banner of open science or public research. “Open access”, for example, can mean free access to view information, but not necessarily freedom to use it in all ways without restriction. To some, open science means no one can fence it in. Access to information, say through “viral” licensing or copyleft, may be conditional on agreeing not to restrict subsequent users. Information may also simply be put into the public domain, for example by depositing it at a freely available public database, for any and all subsequent uses, both proprietary and open. We focus on this last meaning, with information available to all at low or no cost. Sometimes there are restrictions on its use, but those restrictions must also involve minimal or zero costs. And we do not assume that once information is in the commons it is irreversibly fixed there. It can, in some cases, be used and removed with restrictions imposed; but then it is no longer part of the science commons.
In practice, open access can be organized through very different institutional means. In particular, the structure of the science commons in genomics and microbiology differs in important respects from the science commons of the “open source” communities or the Creative Commons project. That is why it is important to qualify what we mean by open access. Adapting the conventional categories from new institutional economics (see, for example, Schlager and Ostrom 1993) to the life sciences, we can distinguish three important categories of access and use rights. First, access can refer simply to the right to access a resource without being allowed to transform it or do any further research on it. This can be the case when a resource is used for educational purposes for instance. Second, accessing a resource can include the right to transform it and develop new lines of research. Third, in certain cases, permission is given to develop and commercialize follow‐on applications. Using these categories, we can distinguish between the following rights that define the components of the science commons in the life sciences:
-
•
access: the right to access a resource/information;
-
•
direct use: the right to change a resource/information;
-
•
follow‐on use: the right to change a resource/information and obtain ownership of the follow‐on applications;
-
•
management: the right to decide upon the way a resource/information (for instance, a database) is managed;
-
•
ownership: the right to exclude others from the use of a resource (exclusion right) and to sell the resource and all the related rights (alienation right).
From this institutional point of view, it is clear that the structure of the science commons differs widely when discussing cases such as GenBank, 2 MOSAICC 3 and GBIF 4 (see Table 1). For instance, as we will discuss below, for GenBank, “open access” does not mean that the user of the information automatically has the right to use it for commercial purposes or to develop follow‐on applications. If the sequences published on GenBank are the subject of patents, one has to get a licence to use them in research or product development. 5 For GBIF, the ownership of the resource and all the related rights are in the hands of the local data provider and hence access conditions vary according to the policies of the – mostly public – funding agencies. Access to the international culture collections network MOSAICC is open to all; however, when acquiring a resource, users have to sign a Material Transfer Agreement that should guarantee traceability of the resources and fair benefit‐sharing with their providers.
Table 1.
Comparison of the structure of the science commons in the life sciences to the Science Commons project
| Ownership and management | Access and direct use | Follow‐on use | |
|---|---|---|---|
| Science Commons | The author | Open access, conditions for direct use specified in the licence | Allowed if open access preserved |
| GenBank (International Nucleotide Sequence Database) | Public domain or patent | Open access, direct use allowed | Licence required if patented matter (in case of university research, settled practice of rational forbearance for suing) |
| GBIF (global portal to access databases of non‐human biological material) | Original database in the home countries | Open access, direct use allowed | Follow‐on applications specified in the original database |
| MOSAICC (international code of conduct adopted by an international network of culture collections) | The culture collections | Open access and direct use upon payment of a small handling fee | Allowed, with appropriate sharing of the benefits with the original provider of the material (if known) |
These different institutions have found different solutions to what the idea of a science commons means for the provision and use of knowledge. Hence, in evaluating historical examples in their particular context, the institutional structures and the type of collective action has to be specified for each case. What organisations played a major role in the establishment of this science commons? What were the norms of the communities that drove this evolution? And what were the characteristics that enabled collective action in successful cases of the science commons? To deal with these questions, we will first discuss the case of human genetics, because of the historical importance of this case and also because of the key role of genetics in general in the ongoing transformation of research in the life sciences. Next, we will broaden the discussion to other fields of the life sciences, such as plant genetics and general‐purpose biotech research tools, to show that in these fields also there is a growing need to systematically address data access and sharing of microbiological information and resources. The data we draw on come mainly from public documents and literature review of case studies in the field. However, informal contacts with officials from umbrella organisations such as the World Federation for Culture Collections (WFCC) and the International Council for Science (ICSU), as well as contacts with genome scientists and bio‐informatics researchers in our home universities, also played a key role in the process of sorting out the data.
Two final conceptual points need to be made before proceeding. First, there is extensive overlap between the different components of the science commons in the life sciences. In particular, research into the properties of whole organisms, ranging from microbes to animals, overlaps with molecular genetics and genomics, proteomics and the development of research tools for screening and genetic engineering. In practice, this has led to hybrid biological research facilities and networks, dedicated to collecting whole organisms but also key parts of organisms such as plasmids (circular DNA, used in biotechnology), cell lines and even entire organisms (in the case of microbes). Prominent examples include the international ex situ seed banks of the 15 Future Harvest Centers, 6 but the same evolution can be observed in the development of biobanks of human tissues (e.g., for cancer research) or vectors used in the highly touted field of genetic therapy. We will move back and forth between these different areas.
Second, there is extensive overlap between academic health research and the science commons in molecular biology. Academic science is important in many fields, not just the life sciences. In all lines of scientific and technical work, universities, not‐for‐profit research institutions, and government laboratories (referred to collectively here as academic research institutions) play key roles. Many people have been trained in academe, not just the people doing R&D, but also corporate managers and IT professionals, and have thus benefited from the exchange of ideas in academic science. Academe is also one place where the norms of Mertonian science have real traction, where the norms of openness, community, mutual criticism, and fair allocation of credit are supposed to be respected. In some circumstances, however, academic science is done in secret, or results are made available only at great cost or encumbered by restrictions on their use. Such science is not part of a science commons. Great science goes on in industry, including, or even particularly, in the life sciences, but no one expects the norms of openness to prevail in industrial R&D, even if in some circumstances they do. When they do, the results flowing from industrial R&D can become part of the science commons, and there are several instances of this in the case studies to follow.
The science commons thus is not the same as academic research. It remains true, nonetheless, that most of the science commons – at least in the life sciences – is based on academic research, and most academic research probably enlarges the science commons (although to our knowledge, no one has really verified this). One reason for interest in academia is that policies put in place over the past three decades have raised concerns about how big the science commons will be, and in particular, whether and to what degree government and not‐for‐profit funders and academic research institutions will maintain it.
Genomics: fights over public and private science in a fishbowl
In recent years genomics has been the ground for a vigorous, sometimes even vicious, fight over what should and should not be in the public domain, and under what conditions. Many of the fights have been over preserving the science commons. This has been a matter of explicit policy‐making in government, not‐for‐profit and academic institutions, and private firms since 1992 or 1993, when the commercial promise of genomics became apparent, and private funding for genomics in for‐profit companies began to accelerate.
The beginning of the Human Genome Project was marked by conflict between scientists who thought it was a poor use of resources and those who thought it a useful and efficient way to spend public research dollars. By broadening the project to include maps, tools, and organisms beyond just the human, most scientists came round to supporting it. As these controversies died down, an even more public conflict over sequencing the entire genome exploded, pitting a private company (Celera) against the public sector genome project. The battleground for both these conflicts was the science commons.
The story is often told as a race between J. Craig Venter, who in 1998 announced his intention to sequence the genome at a new start‐up company Celera, and the public Human Genome Project whose most conspicuous spokesmen were Francis Collins in the United States and Sir John Sulston in the United Kingdom. Collins was director of the National Human Genome Research Institute at NIH, and Sulston directed the Sanger Centre affiliated with the University of Cambridge and funded mainly by the Wellcome Trust and UK Medical Research Council. A consortium of laboratories funded by government agencies and not‐for‐profit organisations in North America, Europe and Japan constituted the “public genome project”. Sulston emerged as the rhetorical champion of that faction, 7 emphasising open science, rapid sharing of data and materials, and a passionate appeal to refrain from patenting bits of the human genome except when they could foreseeably induce investment in the development of end‐products such as therapeutic proteins. In 1996, the Wellcome Trust sponsored a Bermuda meeting of the major sequencing centres throughout the world. A set of “Bermuda Rules” emerged from the meeting, mandating daily public disclosure of DNA sequence data. The pledge to share data rapidly was linked to a plea not to patent DNA without significantly further characterising gene function and demonstrating utility. Characterising gene function was not the business of the publicly funded DNA sequencing centres, so the Bermuda Rules were in effect a “no patents” policy for the sequencing centres.
From 1998 until February 2001, when Science and Nature published rival articles with, respectively, the Celera and public genome project results (2001, 2001), there were two competing projects focused on sequencing the entire human genome. In addition, several other “genome projects” were running in parallel, in both the public and the private sectors. For five years before the extremely visible race between Celera and the “public genome programme” got underway for a complete reference sequence, two companies – Human Genome Sciences, Inc., and Incyte Genomics – were busily sequencing human genes. Many other companies were mapping and sequencing parts of the human genome, and thousands of laboratories were contributing sequencing and mapping information to databases and to scientific publications. By the time the initial genomic sequence publications came out, the ratio of private to public funding appeared to be roughly 2 private dollars for every 1 government or not‐for‐profit dollar (see Fig. 1). 8
Figure 1.
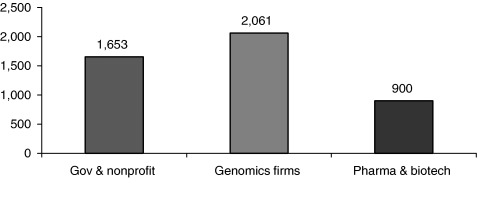
Research funding for genomics in the year 2000 (US $ million).
Source: Figure by the authors, references given in footnotes to accompanying text.
In 2001, the financial genome bubble burst. At the end of 2000, 74 publicly traded genomic firms were valued at $94 billion, of which the largest 15 accounted for approximately $50 billion. By the end of 2002, those 15 firms' market value had dropped to $10 billion, although their reported R&D expenditures climbed from $1 billion to $1.7 billion during the same 2‐year period (Kaufman et al. 2004).
All these numbers bear mention not to drone on about data, but to make three simple points. First, the private sector invested heavily in genomics, but these investments were made in expectation of financial returns. That was quite different from the public and not‐for‐profit funding of genomics which was mainly intended to produce public goods – knowledge and materials made widely available to advance knowledge and combat disease. Second, private R&D investment was a powerful adjunct to the public and not‐for‐profit funding. It followed public R&D in time, and it drew on the science commons without necessarily contributing to it. Its social benefit derived from developing goods and services that would otherwise not be produced. But third, and most to the point for policy purposes, it would be foolhardy to generalize from those happy circumstances where private R&D expands the science commons – to expect private R&D always to contribute to the science commons except in unusual circumstances, usually related to particular features of competition among firms in a particular industrial sector.
Applications in public health: when markets fail
To see why having a healthy science commons matters, we will first move away from genomics to make a general point about health research. Murphy and Topel (1999) estimated that the economic gains from increased life expectancy attributable to medical research were staggering – in the range of $2.8 trillion per year from 1970 to 1990 ($1.5 trillion of this from cardiovascular disease reduction). Many of the health benefits of discovering new information about health and disease come not from drugs, vaccines, or medical services, but from individuals acting on information. Cutler and Kadiyala (2001) attributed two‐thirds of the health gains in cardiovascular disease reduction to the effects of “public information”, such as stopping or reducing tobacco use, changing diet, getting more exercise, and monitoring blood pressure. The second largest determinant was technological change, such as the introduction of new drugs and services, followed by increasing cigarette taxes to reduce tobacco use (Cutler and Kadiyala 2001). The estimated return on investment in medical treatment was 4 to 1, but on public information it was 30 to 1.
Cutler and Kadiyala's result cannot be generalised, because smoking is a very large risk factor that is sui generis, and cardiovascular disease has proven far more amenable to many kinds of intervention than cancer and other chronic diseases. Cancer, diabetes, arthritis, and Alzheimer's disease, among others, appear far less tractable. Few if any risk factors will ever be found to rival tobacco use as predictors of poor health. But the finding that information can have an economic value irrespective of being translated into products and services in a paying market is nonetheless important. Even if public information will not be quite as powerful in reducing other chronic diseases as it has been for cardiovascular disease, the vector is likely to point in the same direction. We cannot say that public information will always prove more powerful than information channelled into new drugs, vaccines, biologics, devices, and medical services in the health care system. But if there are any more such instances – and the probability that there will be none seems vanishingly small – then the health science commons is essential, because it alone can supply the public information benefits. Proprietary science cannot produce public goods, both by definition and for instrumental reasons – to produce such benefits it would have to be shared at no cost. Both words in “public information” do a lot of work. We need new information that arises from science, but to capture many social benefits based on that knowledge, we also need it to be public.
Genomics provides several other examples of the value of public information. The 2002 report from the World Health organisation, Genomics and World Health gave the example of fosmidomycin (WHO 2002, p. 49). This drug is currently being tested in the treatment of malaria in Africa (Missinou et al. 2002). That use came to light as a consequence of sequencing the genome of the malaria parasite, and noticing a metabolic pathway not previously known to exist. The compound fosmidomycin was known to inhibit the pathway, and had been developed as a treatment for urinary tract infections. When the possibility of using the drug to treat malaria was revealed, it was pulled off the shelf and moved into clinical trials. This is a treatment that may never produce a profit for any company, but the social returns could be enormous if the drug works, because so many millions of people are infected with malaria. If not fosmidomycin, then perhaps other findings will lead to the prevention or treatment of malaria, enabled by the existence of the full genomic sequences for host, pathogen, and mosquito vector. 9 Making the information about these organisms available worldwide is essential to accruing the benefits of research. There is only a weak world market in drugs to treat malaria because it is largely an affliction in resource‐poor populations. The usual profit motives of the intellectual property system cannot create incentives where there is no prospect of profit to drive products through an expensive discovery and testing process. But networks of not‐for‐profit organisations, such as the Malaria Vaccine Initiative, the Global Fund, the WHO essential medicines programme, and other sources of “public” capital might be capable of discovering and developing new treatments despite the absence of commercial profit. Many of the scientists most motivated to study such diseases work in resource‐poor countries, but they have computers and access to public databases. They can use information that is posted in the science commons.
Another case study is SARS. Strains of the coronavirus that causes SARS were identified and sequenced within a month by at least three laboratories in Singapore, Canada, and the United States. That sequence information was shared widely, and a “chip” to detect the virus was available for research and possible clinical use just a few months later. Making progress with such speed required strong norms of open science, with obvious social benefits. 10
Public inputs to private science
Even if the public information impact of health research might be less important in the future than it has been in the past, would it diminish the role and importance of the science commons? In this section, the focus is not on social benefits foregone for lack of a robust commons. Rather the argument here shifts to the efficiency gains to private R&D that follow from private R&D being able to draw upon the commons.
Several lines of research corroborate the intuition that a pool of public information and materials must surely “raise all ships” to the benefit of each of them. The case is likely to be stronger in health research than in other lines of research, because of the well‐known deep mutualism between public and private R&D in health research.
The most direct line of evidence for this comes from the Carnegie‐Mellon Survey of industrial R&D managers. Cohen et al. (2002) conclude that “public research has a substantial impact on industrial R&D in a few industries, particularly pharmaceuticals”, and
the most important channels for accessing public research appear to be the public and personal channels (such as publications, conferences, and informal interactions), rather than, say, Licences or cooperative ventures. Finally, we find that large firms are more likely to use public research than small firms, with the exception that start‐up firms also make particular use of public research, especially in pharmaceuticals. (Cohen et al. 2002)
This certainly corroborates the stories of genomics start‐up companies, including upstart companies like Celera, depending heavily on their recent past in academic research, and their ongoing collaborations with (and sometimes markets in) this sector. And it highlights the role of large firms in preferring to draw inputs from a science commons, rather than having to collect atomised fragments of proprietary technologies and data. The history of genomics provides many examples of this, but two are particularly famous.
As the genome project took shape, the importance of maps of human and various “model organisms” was apparent. Which kinds of maps deserved substantial funding and concerted effort remained, however, a matter of ongoing dispute. One of the bones of contention was a “gene map” based on cDNA technology – that is, making DNA copies of the messenger RNA translated into protein within cells. The open question from 1987 through 1991 or so was whether the genome project would include cDNA sequencing, which would start sequencing efforts with DNA known to code for protein, and therefore certain to provide codes for most of the important building blocks of cells, while also providing targets for drug development.
It was the NIH decision not to fund cDNA sequencing that left the door open to Incyte and Human Genome Sciences to develop this field with private funding, because, in the absence of a big public effort, the low‐hanging fruit of the genome was there to be plucked, sequenced, and shipped off with claims to the patent office.
When Incyte and Human Genome Sciences began to go down this path, those who saw genes as increasingly important inputs to their R&D efforts – particularly large pharmaceutical companies – got concerned. Merck decided to take action (Williamson 1999). 11 It stepped forward to fund a public domain sequencing effort, starting with gene fragments and moving on to full‐length cDNAs (DNA copies of messenger RNA that encodes proteins). The sequencing was performed at Washington University in Saint Louis, home of one of the largest public genome sequencing facilities, and the data were to be moved quickly into the public domain.
Merck funded the work through a not‐for‐profit arm and had no privileged access to the data. Here was a large company funding data to flow into the science commons where it would be freely available to all. Why would it do this? Four reasons suggest themselves: (1) it poisoned the well for Incyte, Human Genome Sciences, and other start‐up firms, creating a public sector competitor to stop other companies securing exclusive property rights on genes, and thus limiting the number of genes that would have to be Licenced; (2) it built good will with scientists, vital collaborators in Merck's drug discovery efforts; (3) it was great public relations for the general public; and (4) it took advantage of not‐for‐profit funding. If Merck paid for it as corporate R&D, it could deduct the R&D as an expense, but would also have to justify the public domain science at the stockholders' expense. Through a not‐for‐profit arm, Merck funded great science, burnished Merck's image, and enhanced Merck's future freedom to operate freely, without having to show any returns on an “investment”.
The SNP Consortium story started five years later, but followed the same general lines. During the late 1990s, it became apparent that there were many single‐base‐pair differences among individuals in DNA sequences. These were dubbed single nucleotide polymorphisms, or SNPs. They could be used as DNA markers to trace inheritance, look for associations with diseases or traits, and study population differences: SNPs were valuable research tools. Many genomics firms, including Celera, began to signal that they were finding SNPs and filing patent applications. Given the uncertain rules over what patents the patent office would issue, it seemed possible that they would succeed in getting patents, meaning that anyone using a particular SNP would need to get a licence. This raised the prospect of needing to get licences on hundreds or even thousands of SNP sequences from some unknown (but potentially large) number of patent owners. This was just the kind of nightmare situation that Michael Heller and Rebecca Eisenberg had speculated might arise in their classic 1998 article on the “anticommons”– a situation when too many exclusive rights upstream needed to be assembled, thus thwarting the development of final products, such as drugs, vaccines, biologics, or instruments (Heller and Eisenberg 1998).
This threat awakened some companies and scientific institutions to forge an alliance to defeat patent rights in SNPs. 12 The SNP consortium was founded in 1999 with the aim of discovering SNPs, filing patent applications, mapping and characterising the SNPs, and then finally abandoning the patent applications. The expense and paperwork of this elaborate dance – filing then abandoning patents – was intended to ensure that SNPs were retained in the public domain, unfettered by patent rights. It was deemed necessary, as a defensive strategy, to ensure that consortium members had the standing of inventors should disputes over priority arise for related inventions (in patent parlance, interference proceedings, the administrative procedure to determine the real first inventor). Here a group of private firms of various sizes found common cause in preventing patents on research tools. They valued their freedom to operate highly and the threat of patenting sufficiently to pay for a complicated, expensive procedure to enlarge the public domain.
One interpretation of this story is that “the market” solved the problem. The wonder of capitalism worked its magic by creating public domain resources at private expense to forestall the undue private appropriation of rents from research tools. Does it generalise? Can we simply relax, and assume that the excesses of the patent system will be compensated by enlightened capitalists guarding their long‐term best interests and future freedom to operate? The Merck Gene Index and SNP Consortium cases show that sometimes the answer is yes. The nagging worry is that sometimes the answer may be no. Institutional diversity and eternal vigilance are probably the best insurance policies.
Science commons in microbiology
In our narrative of the structure and the function of the science commons, we discussed essentially the case of human genetics, because of the historical importance of this case and also because of the key role of genetics in general in the ongoing transformation of research in the life sciences. In this section, we will broaden the discussion to other fields of information sharing in non‐human microbiological data, contributing to important fields such as plant genetics and general‐purpose biotech research tools, to show that in these fields also there is a growing need to systematically address data access and sharing of microbiological information and resources.
Historically, it is the science commons in non‐human microbiological data and information that has been the main promoter of open access in the life sciences, through the organisation of public and non‐profit ex situ conservation facilities. In the last decade, these pre‐genomics ex situ facilities have progressively developed into multi‐service facilities called Biological Resource Centres, which organise the collection, organisation, curation, and exchange of biological resources and their associated data and information (BRCs, for a discussion of the concept, see OECD 2001). Today, the science commons in genetics extensively overlaps with this microbiological information commons as it is organised by the BRCs. In particular, life science research depends on the contribution of microbiology to the building of the general infrastructure of R&D in biotechnology and to the understanding, preservation, and sustainable use of biological diversity.
[The French Minister of Higher Education and Research (right) visits a biotechnological laboratory specialising in “DNA surgery” in Romainville, France, April 2006. AFP / Jacques Demarthon]
Our narrative of the microbiological science commons starts in the Yellowstone National Park (Ten Kate and Laird 2002, p. 237). In the summer of 1966, Dr Thomas Brock of Indiana University and one of his undergraduate students, Hudson Freeze, collected bacteria samples from the outflow channel of one of the hot springs at Yellowstone, at a temperature of about 69°C. The discovery of these bacteria was a scientific milestone, as it demonstrated that life could exist at much higher temperatures than had formerly been believed possible. But at that time they did not know that it would have far more important implications. The strain that was isolated from this discovery was deposited at the American Type Culture Collection (ATCC) in Washington DC, the largest culture collection in the world, holding over 70,000 strains from microbiological material from all over the world. They named the organism Thermus aquaticus (Taq) and published their work on its DNA in 1969.
A decade later, Kary Mullis, then a researcher at Cetus Corporation, in California, obtained a sample of this strain from ATCC, which distributed the samples in an open access regime upon the payment of a simple handling fee (at that time US $35). Mullis used the enzyme in his polymerase chain reaction, for which he won the Nobel Prize for Chemistry. The Taq enzyme greatly simplified this method, which makes millions of copies of DNA segments without using a living organism, and has become an extremely widely used method in molecular biology. This technique is still further developed and widely used in applications such as genomic fingerprinting, the diagnosis of infectious diseases, and the study of genetic mutations. After a long controversy over the patent rights, most rights on the PCR technique were finally acquired by the Swiss company Hoffmann‐La Roche in 1991.
There are numerous examples of cases where microbiological resources held in open access (in nature or in ex situ facilities) have been a key factor in new developments in the life sciences. The use of natural enzymes (to catalyse chemical reactions), as in the example of the enzyme derived from Thermus aquaticus, is widespread throughout the industry, which continues to rely on the provision of purified strains from natural specimen of bacteria kept in culture collections. Moreover, in health research, it is estimated that 25 per cent of all new drugs are based directly or indirectly on natural plants or organisms. 13 In agriculture and the food industry, wild varieties of plants or specimen of animals frequently travel around the world and are exchanged between different research and conservation facilities, before leading to new applications that are then marketed by private companies. For instance, the development of fish farming received an important impetus from the development of new varieties of tilapia, a tropical river fish that is found in several African countries (Greer and Harvey 2004, pp. 135–141). Several varieties of tilapia have been developed at the World Fish Centre in Malaysia (part of the Collaborative Group for International Agricultural Research (CGIAR) network) and are now used in fish farming in both developed and developing nations. The property rights on these breeds are still with the World Fish Centre, which does not restrict access for further research for non‐commercial purposes.
These cases illustrate how open access to microbiological material held in the public domain is fundamental both to our understanding of the basic processes of life and for R&D in biotechnology. However, in the context of the increasing influence of global intellectual property rights on the products of life science research, collectors now face the accusation of biopiracy (Sheldon and Balick 1995; Verma 2002). Especially when biological resources come from fragile ecosystems and in the case of traditional knowledge, fair compensation for their contribution to the production and conservation of resources and information is a legitimate claim by the local population. That is why a group of scientists involved in bioprospecting, who met in 1988 at Belem at an international meeting of the ethnobotanical society, adopted a series of rules for organising bioprospecting, requiring prior informed consent by the communities before biological resources and related information were accessed and used. The “Belem Declaration” adopted at that conference was co‐signed by leaders of indigenous communities that were invited to the conference (Posey and Dutfield 1997, pp. 146–149). This declaration has had a major influence on codes of conduct that were adopted later on, such as the MOSAICC code of conduct and the Bonn Guidelines of the Convention on Biological Diversity.
An important further step in the field of microbiology was the extension of these world‐wide networks for conservation and innovation in microbiological diversity to world‐wide information facilities that could be accessed through the Internet. In a manner resembling the human genome project, the organisation of a dialogue between the key actors led to the establishment of a global web portal, called the Global Biodiversity Information Facility (or GBIF). 14 This facility is physically based at the secretariat in Stockholm but can be accessed freely through the Internet and gives access to more than 1,000 nationally based databases providing up‐to‐date information about the microbiological world.
The science commons and economic efficiency: costs of data access
A final historical pastiche is relevant before closing out the arguments. If the history and geography had been different and database firms had turned their attention to genomics just a bit sooner, the story might have been quite different. As it was, the early algorithms for interpreting DNA sequences – such as the BLAST and Smith‐Waterman algorithms – were developed by individuals committed to open science. In more recent years, patents have begun to be issued on bioinformatic methods relevant to genomics. In some cases, these patents confer incentives to support “products” marketed by firms, with service and development teams to improve their quality. How this story will play out remains to be seen, but the ideas of “open genomics” are being tested in the real world alongside more proprietary models.
Databases themselves could become a focus of concern. The early years of the human genome project were marked by many decisions about the disposition of crucial databases. Human genetic disease and variation was lovingly catalogued by the team surrounding Victor McKusick at Johns Hopkins University, to produce the Online Mendelian Inheritance in Man (OMIM) database. Many other databases were established to retain data on human genetic maps of various types, and the same is true for other organisms. The DNA sequence data in the GenBank database were collected primarily by a trio of teams in the United States, Europe, and Japan, and these shared data among themselves. Creating and coordinating these databases, including the sequence databases, was a major struggle (Smith 1990), but the battle was waged with only limited concern for their commercial potential. The GenBank databases contain many errors (2005, 1999), and creating financial incentives sufficient to encourage careful curation and maintenance are one reason to support proprietary rights in databases. But that step should not be taken lightly.
We now have a decade‐long real‐world experiment to inform such decisions, with strong protection in Europe and only copyright protection for databases in the United States. How different might it have been had the genome project begun in Europe, just a decade later, when the European Community saw fit to create a new exclusive right in databases as an incentive for companies to create and maintain valuable data? The impact of this new form of intellectual property, database protection, has received particular attention from the scientific community. Scientists have become concerned that rights could hinder research. The landmark report on the topic was the 1997 Bits of Power report from the National Research Council in the US, which has led to a line of further work (National Research Council 1997). Much of the most advanced thinking has focused on weather, remote imaging, and other huge and complex data sets. There may be cause for concern, and not just for scientists, but for the innovation system as a whole. Exclusive property rights create friction and inefficiency. It may be that free access to data generated at government and not‐for‐profit expense is both more efficient and a more powerful prime for the economic engine than allowing every incremental advance to form the basis for rent‐seeking.
A brief political economy of the science commons in the life sciences
Where do we go from here? As our narrative has shown, the science commons is clearly important in furthering public interest objectives such as public health, food security and the conservation of biological diversity. However, we have also seen that some cases are more successful than others. There is no automatic move from the norms of the communities that favour the science commons to the provision of knowledge in the public interest. The latter still depends on the appropriate design of the institutions for collective actions that are required for broad diffusion and use of the knowledge base beyond the pursuit of individual interests.
When Robert Merton wrote about the sociology of science, the central task at hand was explaining how a set of social norms and practices yielded knowledge – what was different about science compared to, say, the humanities or the traditional professions (Merton 1973). John Ziman and others addressed what makes the methods of science produce “reliable knowledge” (Ziman 1978). Our concern here is about a related but distinct topic – how reliable knowledge can be turned into social benefit. The point of connection is science that is useful – science that falls squarely into “Pasteur's Quadrant” (Stokes 1997) where it both contributes to insights about how the world works and promises to make the world a better place through practical application (see Fig. 2). Producing knowledge in Pasteur's quadrant does not depend only on the norms and beliefs of the communities, but also on collective action for crossing borders between different communities and organising cooperation between public and private actors.
Figure 2.
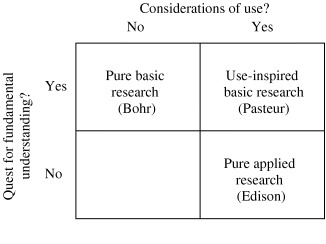
Science in Pasteur's Quadrant (the upper right box).
Source: Stokes (1997, p. 73).
Combining basic science and useful applications seems like trying to square the circle. However this is precisely what has characterised the technological revolution in the life sciences, where new insights in the field of genomics have contributed to our basic understanding of life, and have also contributed directly to applied research in diagnostics and drug design.
The specific dilemmas that arise from producing knowledge in Pasteur's quadrant have been analysed in more depth in the extensive literature on the knowledge economy (see Foray 2004). We have illustrated some of these dilemmas in Table 2. Two of them relate directly to the contribution of the science commons to the production and the use of information. The first is the diffusion–innovation dilemma, which deals with the problems that arise in the diffusion of research results and new applications. The second is the exploration–exploitation dilemma which highlights the need for collective action in exploring new lines of development that do not provide direct benefit to individual innovators.
Table 2.
Some incentive problems in Pasteur's quadrant: provision and use of basic scientific knowledge with potentially direct applications
| Incentive problems | |
|---|---|
| Provision of knowledge | Quality of data provided to global data portal Quality of resources managed in culture collections Exploration of new lines of development (exploration–exploitation dilemma) |
| Diffusion of knowledge | Under‐use: patent thickets, problem of anti–commons Diffusion: delay in diffusion of research results because of patent applications (diffusion–innovation dilemma) Under‐investment in follow‐on applications |
Source: authors.
The diffusion–innovation dilemma deals with striking the best balance between creating incentives for innovation for individual agents and organisations on the one hand and the broad diffusion of research results on the other. Well‐defined rules of ownership create greater incentives to innovate (Demsetz 1967; Schlager and Ostrom 1993), while diffuse ownership can lead to underinvestment and even depletion of valuable assets. 15 In particular, intellectual property rights such as patents on genetic information and biotechnological research tools have become key drivers for innovation in the life sciences. At the same time, they may undercut wide and early diffusion of research results, and so can have a long‐term negative effect on innovation. We can illustrate this dilemma with a case study on the history of plant breeding in the field of agriculture (Goeschl and Swanson 2002). Intellectual property in new seed varieties obtained through cross‐breeding has been an important driver of research. This has led to new crops such as rice, maize, and corn with great potential for increasing food security in developing countries. However, over the period from 1961 to 1999 for which data are available, the diffusion of innovation in the two crops with the highest protection, maize and sorghum, is lower than that in other crops. In this period, the gap between developing and developed countries in the productivity of maize and sorghum was 50 per cent higher, compared to the other crops, ranging up to 72 per cent; for maize and 62 per cent; for sorghum. The evidence further shows that enhanced property regimes work against the interests of the states that are furthest from the technological frontier of innovation. The poorest and least advanced fare worst. A similar story could be told about the drugs contributing to public health (such as anti‐AIDS drugs) or for basic research tools in biotechnology. The challenge in each of these cases is to find the right balance between investment in the diffusion of research results to the broadest group of stakeholders, and investment in innovation and the development of new market products.
The exploration–exploitation dilemma deals with the collective action required to create and apply general purpose knowledge. This knowledge is important because the development of new applications in the life sciences also depends on progress in the general knowledge base and the exploration of new lines of development with outcomes which are still uncertain and controversial, such as genetic therapy, bioremediation, or biofuels. Here the challenge is to strike a balance between exploitation of the research results, with a direct benefit for the public or private organisation investing in the research, and the exploration of new lines of development. The latter implies the organisation of collaborative learning and cooperation beyond the immediate benefits obtained by the individual participants. 16
Dealing with provision: examples of the exploration–exploitation dilemma
We can compare the relative performance on the exploration–exploitation dilemma of two important cases of innovation in the life sciences: GenBank for genomics and GBIF for culture collections. These are two cases of a science commons that have produced long‐term benefits for the science community as a whole, but with different institutional rules for organizing access and ownership.
In the case of GenBank, intellectual property on the gene sequences that are published depends on the data provider. 17 Some sequences are patented material, but most are in the public domain. However, for both patented and public‐domain sequences, important incentives exist to publish rapidly, and deposit data in GenBank. Having a sequence published on GenBank (or any alternative recognised international e‐repository) is a requirement for publishing scientific research on a new gene sequence in most journals. The provision of gene sequences to this international science commons is thus assured through its connection with publication, and thus with the collaborative effort of the basic scientific research community in the life sciences. 18 Making new genetic sequences of organisms rapidly publicly available is necessary both for fundamental research (as a common reference point for all research that refers to that genetic sequence) and for the rapid development of new applications (as in the development of a new drug for malaria discussed above).
In the case of culture collections, specimens of bacteria, fungi, microbes, and cell lines can be accessed upon the payment of a handling fee. The Global Biodiversity Information Facility (GBIF) makes information on these resources freely available through a global data portal on the Internet (James 2002). The situation for culture collections is thus in many respects similar to GenBank. Some of the incentives are also the same. The collection of strains by researchers is an important part of the discovery process in microbiology. Researchers deposit their strains in the national culture collections. To publish an article on a newly discovered strain, two samples have to be deposited in two different culture collections. Another important source of strains is the requirement to deposit strains in a culture collection when applying for patent protection. However, there are also some important differences between GenBank and GBIF. The information and quality management at GBIF is much more decentralised. GBIF only offers a common portal and data format allowing access to the several hundred nation‐based culture collections. Each of these collections manages its own database and has its own rules for intellectual property rights and conditions of use. 19
Further research is required to develop insights into the design of these collective action institutions and their contribution to the provision of general purpose knowledge. However, as these two cases clearly show, the science commons in life science research allow the development of institutional frameworks for successfully dealing with the exploration–exploitation dilemma.
Dealing with under‐use: examples of the diffusion–innovation dilemma
The science commons in the life sciences also provides interesting institutional solutions to the diffusion–innovation dilemma. As we have argued throughout the paper, the science commons plays an important role in making wide diffusion of research results possible, through both public and private actors. For instance, in the case of cDNA fragments, a private company funded the work to make cDNA sequences publicly accessible. This effort was important in making a basic research tool widely accessible and stimulating its rapid diffusion. In another consortium, the yeast sequencing programme in Europe, public and private partners collaborated in the first networked DNA sequencing enterprise (Cassier and Foray 1999). It was agreed that the fragments that were sequenced would remain private property during the development of the programme, but that once the complete genome was mapped all the sequences were made publicly accessible. This was done when the over‐6,000 genes of the whole genome had been sequenced, in an article published in Nature in 1997.
Another interesting way of dealing with the diffusion–innovation dilemma is illustrated by the case of the World Fish Centre and the International Rice Research Institute of the CGIAR network. In the World Fish Centre, 20 new breeds of tilapia, a tropical river fish originally found in Central and East Africa, were developed for use in aquaculture (Greer and Harvey 2004, pp. 135–141). New breeds were expected to become one of the most important species in aquaculture, and potentially to contribute to enhancing food security in developing countries. After the breeds were developed, discussions followed on how to transfer improvements to the local populations. In 1999, a corporate partner, GenoMar, was found. GenoMar received the exclusive licence to market a specific variety of a new breed, called “super‐tilapia”, in the Philippines, while the property rights on all the other varieties of new breeds remained with the World Fish Centre. Those varieties remain freely available for direct use in developing countries. Thus, in this case diffusion is organised by open access to the original varieties of new breeds, so that they can be used in aquaculture in developing countries, while the investment in the further development of specific commercial varieties is organised through and exclusive licence to the corporate partner.
A similar story could be told about other public‐private consortia. 21 The case of the International Rice Research Centre is interesting because it shows the multi‐layered character of the science commons. In this case, a resistant breed of rice was collected from a community in western Africa (Mali), and brought by an Indian researcher to the IRRC in the Philippines where it was crossbred to develop a commercial variety (Gupta 2004, pp. 75–102). Finally, the University of California (UC) Davis sequenced the genome of the new variety and acquired patent rights to it. In spite of this property right, the science commons was preserved through a variety of collective agreements. First, researchers at UC Davis created the genetic recognition fund to share the benefits of commercial exploitation with the farmers in the provider country. 22 Second, the IRRC retained the rights to do further research on the rice for non‐commercial purposes, and it was agreed that, if any new follow‐on application followed, no action would be taken to impede commercialisation of the new variety. 23
Conclusion
A science commons can supply the information needed to achieve social benefits that for‐profit markets in goods and services sometimes fail to provide. Even in markets well served by the profit motive, a science commons can in some circumstances improve efficiency – for example, when many disparate firms can draw on a common pool of knowledge and data, rather than having to construct the same information firm‐by‐firm (resulting in substantial duplication costs). The genomics cases we have highlighted suggest that theory of the science commons, or at least heuristic principles to maintain its health, may have some practical use in the real world of science and its application. We have illustrated a variety of social goals that can benefit from a robust scientific commons in genomics: advancing science, improving public health, improving food security, contributing to understanding and conserving biological diversity, and contributing to industrial R&D and commercialisation.
The main lesson that can be drawn from our narrative is the following: there is no single solution to organising a science commons in the life sciences in order to produce usable scientific knowledge. In particular, as we attempted to show, commercialisation of R&D in the life science is not the problem as such. In some cases, private sector initiatives contributed in important ways to the building of the science commons (cf. Table 3). The real challenge is how to provide incentives to invest into the production of general purpose knowledge on the one hand and into broad diffusion of the exploitable research results on the other. This is a challenge both for public and private sector research.
Table 3.
Different modes of involvement of private actors in the organisation of the the science commons (references in the footnotes to the case studies in the text)
| Examples of the life science commons discussed in the paper | Mode of involvement of the private sector | |
|---|---|---|
| Information sharing in genomics (human genetics) | GenBank/EMBL/DDBJ SNP Consortium cDNA sequencing funded by Merck | Publicly funded information clearing house, information provided by public, non‐profit and for profit entities Consortium of public and private partners Private company |
| Information sharing in non‐human genetics | Yeast sequencing programme International Rice Research Centre‐UC Davis exclusive licence World Fish Centre – GenoMar exclusive Licence | Consortium of public and private partners Public‐private partnership Public‐private partnership |
| Information sharing non‐human biological organisms | Global Biodiversity Information Facility (GBIF) | Publicly funded information clearing house, information provided by public, non‐profit, and for profit entities |
Source: authors.
Both the characteristics of the physical resources (from genes to microbes, plants and animals) and the norms and beliefs of the research communities determine the appropriate institutional choices for organising the science commons. Building capacity for developing new norms in these different communities, such as the norms set out in the Bermuda Rules in the human genome story, the prior informed consent rules adopted by the culture collections, or the corporate social responsibility in the case of Merck and the SNP Consortium are clearly important steps in this direction. The second lesson brings us back to the production of knowledge in Pasteur's Quadrant. Our hypothesis is that the science commons contributes to solving some of the dilemmas arising from the provision and use of knowledge in the general interest. Both our narrative and our brief review of the political economy of producing genomic knowledge show that this is in no way an automatic process. Diffusion of knowledge and the exploration of new lines of innovation depend on creating institutions for collective action. As we have seen in our evaluation, collective action has been highly successful in life sciences research in furthering the provision and use of knowledge. Sustaining this effort will, however, require continued institutional experimentation. And because of the institutional diversity, analysis of the conditions for success will be needed. How the collective action by involved stakeholders worked in these experiments will need to be studied, and a process of self‐evaluation and adjustment will surely play an important role in this.
Robert Cook‐Deegan has been the Director of the Institute for Genome Science and Policy's Centre for Genome Ethics, Law & Policy (Duke University) since July, 2002. He is the author of The Gene Wars: Science, Politics, and the Human Genome (New York: Norton, 1994; paperback 1996; tr. Korean 1995, Japanese 1996). Email: bob.cd@duke.edu
Tom Dedeurwaerdere is director of research at the Centre for Philosophy of Law and professor at the Faculty of Philosophy, both at the Université catholique de Louvain. Bibliographical information on his publications can be found on the website: http://www.cpdr.ucl.ac.be/perso/dedeurwaerdere Email: Dedeurwaerdere@cpdr.ucl.ac.be
Notes
*This paper built in part on a previous paper given by RC‐D at the University of Toronto to be published as “The Science Commons in Health Research: Structure, Function, and Value”, forthcoming in Journal of Techonology Transfer. RC‐D's contribution was supported in part by the US National Institute for Human Genome Research and Department of Energy through grant P50‐003391.
1. For further details see the Science Commons website: http://www.sciencecommons.org (accessed 2 April 2005). Executive Director, John Wilbanks, with headquarters based at the Massachusetts Institute of Technology.
2. GenBank is short for the International Nucleotide Sequence Database, publicly accessible through the DNA DataBase of Japan (http://www.ddbj.nig.ac.jp/Welcome.html), European Molecular Biology Laboratory Nucleotide Sequence Database (http://www.ebi.ac.uk/embl/index.html) and US National Centre for Biotechnology Information GenBank portals (http://www.ncbi.nlm.nih.gov). These are three mirror sites that exchange and update every night the new information on the sequences, respectively situated in Japan, the EU, and the USA. The information on DNA sequences is thus the same on the three sites, but each of them also offers specific services. Approximately 15 per cent of the user access is through the Japanese site, 15 per cent through the EU site, and 70 per cent through the US site.
3. Micro‐Organisms Sustainable use and Access regulation International Code of Conduct (http://www.belspo.be/bccm/mosaicc).
4. Global Biodiversity Information Facility, http://www.gbif.org (accessed 26 January 2006). Executive Director, Jim Edwards, with headquarters at the University of Copenhagen, Denmark.
5. It is unclear to what degree all research activities are subject to this restriction of the science commons. Most notably, in the case of university research, rational forbearance for suing seems to be a settled practice. However, absence of infringement action does not mean that university researchers do not change their research plans when using patented material with any plausible utility. For an in depth discussion and recent survey data, cf. 2006, 2005 and AAAS. (2006).
6. The 15 members of the Consultative Group on International Agricultural Research (CGIAR).
7. Sulston's model for the human genome project was the biology of the worm – a close‐knit community of scientists who studied nematodes, and had made immense scientific progress in a hub‐and‐spoke model of biology. Two central laboratories – one at the University of Cambridge and another at Washington University in Saint Louis – created large data sets centrally at large computing and instrumentation facilities dedicated to expensive genome‐wide mapping and sequencing projects on the worm genome. Those hubs shared data quickly and widely with the spokes – a network of smaller laboratories throughout the world. Sulston wrote The Common Thread with Georgina Ferry to tell the genome story from his point of view. His model was a public works project in genomics, with public funding producing a valuable scientific resource (Sulston and Ferry 2002).
8. In a snapshot taken of year 2000 genomics research funding, approximately 70 non‐profit and government funders provided an estimated $1.6–1.7 billion; 74 publicly traded firms dedicated wholly to or including genomics research as a major function reported over $2 billion in R&D expenditures; and assuming the reported 3–5 per cent of R&D in major pharmaceutical firms was for genomics (based on survey responses and rough informal estimates of pharma R&D managers), established firms were spending $800 million to $1 billion in genomics research (World Survey of Funding for Genomics Research, Stanford‐in‐Washington programme http://www.stanford.edu/class/siw198q/websites/genomics/entry.htm (accessed 2 April 2005)).
9. Sequences for both the most virulent pathogen, Plasmodium falciparum, and the most common vector, Anopheles gambiae, were published in 2002: Gardner, et al. (2002) and Holt et al. (2002).
10. This story of sharing sequence information is linked to a potential intellectual property story that could be complicated. At least three of the institutions that did the sequencing have applied for patents, and interference proceedings could be complex, as they are in different countries and on different strains that might need to be cross‐Licenced for many practical applications. A patent pool could emerge, or a monster interference proceeding to sort out the questions of inventorship. The legal costs could exceed the costs of deriving the sequence itself.
11. An excerpt from the press statement upon the first data release explains some details: “The Merck Gene Index is a broad collaborative effort, coordinated by Dr. Alan Williamson, Vice President, Research Strategy Worldwide, and Keith O. Elliston, Associate Director, Bioinformatics, of the Merck Research Laboratories. Dr. Greg Lennon's laboratory at the Lawrence Livermore National Laboratory (Livermore, California) has been supplying arrayed cDNA clones to Dr. Robert Waterston's laboratory (the Genome Sequencing Centre) at the Washington University School of Medicine (St. Louis, Missouri) for sequencing. The sequence data are being submitted to the Expressed Sequence Tag (EST) division of GenBank on a regular basis for immediate distribution. (GenBank, built and distributed by the National Centre for Biotechnology Information (NCBI) is a central repository of publicly‐available gene sequence information, widely known and heavily used by researchers in government, academe, and industry).” Press statement http://www.ncbi.nlm.nih.gov/Web/Whats_New/Announce/merck_feb10_95.html (accessed 3 April 2005).
12. See the website of the SNP Consortium (http://snp.cshl.org/about/ (accessed 3 April 2005)). See also Thorisson, and Stein (2003, pp. 124–7), and Holden (2002, p. 26).
13. “Natural product research is far from being the only source of novel active compounds; it is rather a complement to the chemical synthesis of new drugs. However a study made in 1989 in the US estimated that 25 per cent of drugs' active ingredients were extracted or derived from plants. Another study carried out in 1993 estimated that in the US 57per cent of the prescriptions contained at least one major active compound now or once derived after compounds derived from biodiversity” cf. Brahy (2005, 1996, 1989), J. Nat. Prod. (2003).
14. The idea of creating the GBIF developed from the discussions organised in the context of the OECD Megascience Forum, an intergovernmental forum where scientific ideas can be exchanged and consensus reached on the best way either to acquire new knowledge or to take advantage of a significant scientific development (James 2002, p. 5). The discussions that led to the GBIF took place in the Working Group on Biological Informatics between April 1996 and September 1998. These discussions allowed integrating the concerns of the established conservation community and the emerging bioinformatics community. As a result of the recommendations of this Working Group, an Interim Steering Committee was set up in 1999 under the auspices of the OECD ministers, which finally led to the establishment of the GBIF in autumn 2001.
15. As argued elsewhere in this special issue (Dedeurwaerdere, “The Institutional Economics of Sharing Biological Information”), well‐defined property rights do not necessarily mean private property. In the case of biological resources, innovation is often distributed amongst several actors, and forms of common property can be more efficient (Cassier and Foray 1999).
16. Such investment in exploration characterised, for instance, the early phases of the human genome project. The same is true for the GBIF consortium, where the establishment of the bioportal followed a phase of collaborative learning between bioinformaticians and microbiologists during exploratory research on the necessity and feasibility of common standards for data transmission.
17. Cf. for an introduction to this http://www.ncbi.nlm.nih.gov/Genbank/ (accessed 6 April 2006).
18. One important issue for collective action however is the quality management of the information in GenBank. Because of the pressure to publish rapidly, the information initially submitted is often incomplete and poorly verified. This is partially corrected for by review and use of the information by colleagues, but there is no systematic collaborative effort of quality management before submission. Depending on the portal of entry to the database, however, some routine error‐checking routines are done (e.g., checking for inclusion of sequences from common cloning vectors). Further refinements are possible for related databases, such as Gene and RefSeq, which contain more fully characterised gene sequences. But detailed annotation and reliability of data vary among GenBank sequences.
19. This decentralised management of culture collections has also led to important cooperation problems in the collection of strains. Indeed collection is a collaborative effort with the biodiversity rich countries that depends on clear agreements on benefit sharing and technology transfer. However, the competition between the culture collections has led to a “race to the bottom”, where collectors try to obtain the most strains with the fewest constraints.
20. Previously called the ICLARM, International Centre for Living Aquatic Resource Management (Greer and Harvey 2004, pp. 18–19), headquarters in Malaysia.
21. For instance the UK based CABI culture collection consortium developed a biological control agent from a fungi that can be used for killing insects on crops. After developing a spray based on this fungus, CABI was granted the exclusive property rights. CABI granted a licence to a corporate partner in South Africa to market one variety under the name “Green Muscle”. A less developed form of the spray was made available by CABI for free use through a general public licence (Ten Kate and Laird 2002, pp. 217–227).
22. However, this initiative was not very convincing. No commercial application resulted from the new resistant rice and the model of the genetic recognition fund did not receive broad support at UC Davis, which did not use it for other cases of patents on resources coming from developing countries.
23. These cases show that, as in the case of the Free Software Foundation (FSF) and Creative Commons, the attribution of property rights is an essential condition for promoting science commons. It was because the IRRI retained the rights to do further research on the rice that it could develop a pro‐commons policy. In the case of open software, the FSF even requires the authors of new software to have clear copyright assignment on their software in order that their GNU licence be enforceable (http://www.gnu.org (accessed 6 April 2006)).
References
- Benkler, Y. 1998. The commons as a neglected factor of information policy. Remarks at the Telecommunications Policy Research Conference (Sept. 1998). Available from: http://www.benkler.org/commons.pdf (Accessed July 2005).
- Brahy, N. 2005. “The contribution of databases and customary law to the protection of traditional knowledge”, Les Carnets du Centre de Philosophie du Droit, 117, 29. [Google Scholar]
- Cassier, M. , and Foray, D. 1999. The economics of high‐tech consortia. Case studies in bio‐medical research. Colline Working Paper Series, n° WP02.
- Cohen, W. M. , Nelson, R. R. , and Walsh, J. P. 2002. “Links and impacts: the influence of public research on industrial R&D”, Management Science, 48 (Jan), 1–23. [Google Scholar]
- Cutler, D. M. , and Kadiyala, S. 2001. The return to biomedical research: treatment and behavioral effects. Working Paper. http://post.economics.harvard.edu/faculty/dcutler/papers/cutler_kadiyala_for_topel.pdf (Accessed 3 April 2005).
- Demsetz, H. 1967. “Towards a theory of property rights”, American Economic Review, 62, 347–359. [Google Scholar]
- Foray, D. 2004. The economics of knowledge. Cambridge, MA: Massachusetts Institute of Technology. [Google Scholar]
- Gardner, M. J. , et al 2002. “Genome sequence of the human malaria parasite Plasmodium falciparum ”, Nature, 41, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks, W. R. , Audit, B. , De Angelis, D. , Tsoka, S. , and Ouzounis, Ch. A. 2005. “Percolation of annotation errors through hierarchically structured protein sequence databases”, Mathematical Biosciences, 193 (Feb), 223–234. [DOI] [PubMed] [Google Scholar]
- Goeschl, T. , and Swanson, T. 2002. “The diffusion of benefits from biotechnological developments. The impact of use restrictions on the distribution of benefits”. In : Swanson T., ed. The economics of managing biotechnologies. Dordrecht: Kluwer Academic Publishers, pp. 214–248. [Google Scholar]
- Greer, D. , and Harvey, B . 2004. Blue genes. Sharing and conserving the world's aquatic biodiversity. London: Earthscan. [Google Scholar]
- Grifo, F. T. , and Downes, D. R. 1996. “Agreement to collect biodiversity for pharmaceutical resource: major issues and proposed principles”. In : Brush S. and Stibansky D., eds Valuing local knowledge. Washington DC: Island Press. [Google Scholar]
- Gupta, A. K. 2004. “The role of intellectual property rights in the sharing of benefits arising from the use of biological resources and associated traditional knowledge”, World Intellectual Property organisation publications, 769 (E). [Google Scholar]
- Hansen, S. , Brewster, A. , Asher, J. , and Kisielewski, M. , 2006. The effects of patenting on the AAAS scientific community. Washington, DC: American Association for the Advancement of Science. Available online at: http://sippi.aaas.org/survey (Accessed 23 December 2006).
- Heller, M. A. , and Eisenberg, R. S. 1998. “Can patents deter innovation? The anticommons in biomedical research”, Science, 280 (1 May), 698–701. [DOI] [PubMed] [Google Scholar]
- Holden, A. L. 2002. “The SNP Consortium: summary of a private consortium effort to develop an applied map of the human genome”, Biotechniques, Jun Suppl, 22–24. [PubMed] [Google Scholar]
- Holt, R. A. , et al 2002. “The genome sequence of the malaria mosquito Anopheles gambiae ”, Science, 298, 129–149. [DOI] [PubMed] [Google Scholar]
- James, E. 2002. Establishing international scientific collaborations: lessons learned from the Global Biodiversity Information Facility. Report to the Sixth Meeting of the OECD Global Science Forum, January 2002.
- Kaufman, D. , Johnson, A. , and Cook‐Deegan, R. 2004. Stanford‐in‐Washington program, unpublished data (July).
- Lander, E. S. , et al 2001. “Initial sequencing and analysis of the human genome”, Nature, 409 (6822), 860–921. [DOI] [PubMed] [Google Scholar]
- Lessig, L. 1999. Code and the commons Keynote address at the Conference on Media Convergence, Fordham University Law School (9 February, 1999). Available from http://cyber.law.harvard.edu/works/lessig/fordham.pdf (Accessed July 2005).
- Merton, R. K. 1973. The sociology of science. Chicago: University of Chicago Press. [Google Scholar]
- Missinou, M. A. , Borrmann, S. , Schindler, A. , Issifou, S. , Adegnika, A. A. , Matsiegui, P. B. , Binder, R. , Lell, B. , Wiesner, J. , Baranek, T. , Jomaa, H. , and Kremsner, P. G. 2002. “Fosmidomycin for malaria”, The Lancet, 360 (14 Dec), 1941–1942. [DOI] [PubMed] [Google Scholar]
- Murphy, K. M. , and Topel, R. 1999. “The economic value of medical research”. Paper commissioned for Funding First and the Lasker Foundation. Available from: http://www.laskerfoundation.org/reports/pdf/economicvalue.pdf (Accessed 3 April 2005).
- National Research Council 1997. Bits of power: issues in global access to scientific data. Washington, DC: National Academy Press. [Google Scholar]
- OECD 2001. “Biological resource centres underpinning the future of life sciences and biotechnology”, OECD Science and Information Technology, Vol. 7, 68 pp. [Google Scholar]
- Pennissi, E. 1999. “Keeping genome databases clean and up to date”, Science, 286 (15 Oct), 447–450. [DOI] [PubMed] [Google Scholar]
- Posey, D. A. , and Dutfield, G. 1997. Le marché mondial de la propriété intellectuelle. WWF, CRDI. [Google Scholar]
- Principe, P. P. 1989. “The economic significance of plants and their constituents as drug”. In : Wagner H., Hikino H. and Farnsworth N. R., eds Economic and medicinal plant research. 3, pp. 1–17. London: Academic Press. [Google Scholar]
- Schlager, E. , and Ostrom, E. 1993. “Property rights regimes and coastal fisheries: an empirical analysis”. In : Anderson T. L. and Simmons R. T., eds The political economy of customs and culture: informal solutions to the commons problem. Lanham, MD: Rowman and Littlefield, pp. 13–41. [Google Scholar]
- Sheldon, J. W. , and Balick, M. J. 1995. “Ethnobotany and the search for balance between use and conservation”. In : Swanson T., ed. Intellectual property rights and biodiversity conservation: an interdisciplinary analysis of the values of medicinal plants. Cambridge: Cambridge University Press. [Google Scholar]
- Smith, T. F. 1990. “The history of the genetic sequence databases”, Genomics, 6, 701–707. [DOI] [PubMed] [Google Scholar]
- Stokes, D. E. 1997. Pasteur's quadrant: basic science and technological innovation. Washington, DC: Brookings Institution Press. [Google Scholar]
- Sulston, J. , and Ferry, G. 2002. The common thread: a story of science, politics, ethics, and the human genome. Washington, DC: National Academy Press. [Google Scholar]
- Ten Kate, K. , and Laird, S. A. 2002. The commercial use of biodiversity. London: Earthscan. [Google Scholar]
- Thorisson, G. A. , and Stein, L. D. 2003. “The SNP Consortium website: past, present and future”, Nucleic Acids Research, Jan 1, 31 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J. C. , et al 2001. “The sequence of the human genome”, Science, 291 (16 Feb), 1304–1351. [DOI] [PubMed] [Google Scholar]
- Verma, I. M. 2002. “Biopiracy: distrust widens the rich‐poor divide”, Molecular Therapy, 5 (2), 95. [DOI] [PubMed] [Google Scholar]
- Walsh, J. P. , Cho, Ch. , and Cohen, W.M 2005. Patents, material transfers and access to research inputs in biomedical research. Final Report to the National Academy of Sciences' Committee on Intellectual Property Rights in Genomic and Protein‐Related Inventions, 60 pp. Available from: http://tigger.uic.edu/~jwalsh/WalshChoCohenFinal050922.pdf (Accessed 22 September 2006).
- Williamson, A. R. 1999. “The Merck gene index project”, Drug Discovery Today, 4, 115–122. [DOI] [PubMed] [Google Scholar]
- World Health organisation , 2002. Genomics and world health. Geneva: World Health organisation. Advisory Committee on Health Research. [Google Scholar]
- Ziman, J. 1978. Reliable knowledge: an exploration of the grounds for belief in science. New York: Cambridge University Press. [Google Scholar]



