Abstract
Summary: Innate sensors of viral infection detect viral products and initiate the signal cascades that lead to the antiviral response. Several proteins have been identified to play a role in this process, mostly members of the Toll‐like receptor and retinoic acid‐inducible gene I‐like receptor families. These receptors have been demonstrated to function in part by recognizing a diverse yet unique repertoire of nucleic acid substrates. Upon recognition of their ligands, these sensors activate distinct signaling pathways that lead to the secretion of type I interferon and inflammatory cytokines. It remains to be seen, however, if these sensors are redundant or whether each serves a unique function. In this work, we review the current knowledge of viral sensors, speculate on how they may function in vivo, and explore the potential reasons for their diversity.
Keywords: innate immunity, Toll‐like receptor, RIG‐I‐like receptor, pattern recognition receptors, viral immunity
Introduction
Viral pathogens have been discovered in all species from single‐cell bacteria to the most complex of mammals. To protect themselves from pathogenic effects of these invaders, organisms have been required to develop mechanisms to detect and prevent viral infection. In higher mammals, this requirement has evolved the adaptive immune system, which is able to generate highly specific antibodies and T cells that recognize specific viral proteins and peptides that either block infection or target infected cells for destruction. Initiation of the adaptive immune response, however, is a slow process that requires days to weeks for maximum effect. To provide protection during the initial hours and days of infection, we have maintained a system of pattern recognition receptors (PRRs) first seen in lower organisms that recognize broad motifs common to viral pathogens and thus serve as the initial sensors of viral infection. These sensors serve to initiate and maintain an antiviral response while the more specific adaptive response develops.
There are a wide variety of viral pathogens that are known to infect humans. Viruses can have genomes composed of double‐stranded RNA (dsRNA), single‐stranded RNA (ssRNA), or DNA and can replicate at different locations in the cell through different mechanisms that proceed through unique intermediates. Designing a system that can broadly detect all of these possibilities is challenging to say the least. To accomplish this, organisms use a variety of sensors from two main classes: Toll‐like receptors (TLRs) and retinoic acid‐inducible gene I (RIG‐I)‐like receptors (RLRs). These sensors protect different cellular compartments and signal through different adaptors to activate an antiviral response.
RLRs
RLRs are cytoplasmic proteins that recognize viral products that have gained access to the cytosol. There are currently three known members of this family: RIG‐I, melanoma differentiation‐associated gene 5 (MDA5), and laboratory of genetics and physiology‐2 (LGP2) (1). Both RIG‐I and MDA5 contain a DExD/H box helicase domain that binds dsRNA and two N‐terminal caspase recruitment (CARD) domains involved in signaling (2, 3, 4, 5). LGP2 contains the helicase domain but lacks the CARD domains and is thought to be a negative regulator (6, 7). Both RIG‐I and LGP2 also contain a C‐terminal repressor domain that blocks signaling in the absence of ligand binding (5). RIG‐I binds preferentially to ssRNAs that are phosphorylated at the 5′‐end (8, 9) and contain homopolyuridine or homopolyriboadenine motifs as well as short dsRNA (10, 11, 12). MDA5 recognizes long dsRNAs and does not require 5′‐phosphorylation (11, 12, 13, 14). The differences in ligand preferences of the two proteins result in specificity for the recognition of individual viruses, which is discussed later.
The pathways by which RLHs signal are shown in Fig. 1 . Both MDA5 and RIG‐I signal through CARD–CARD interactions with interferon‐β (IFN‐β) promoter stimulator 1 (IPS‐1) [also known as mitochondrial antiviral signaling (MAVS), virus‐induced signaling adapter (VISA), or CARD adapter‐inducing IFN‐β (Cardif)], which is located on the outer mitochondrial membrane (15, 16, 17, 18). Downstream of IPS‐1 (19), tumor necrosis factor (TNF) receptor‐associated factor 3 (TRAF3) activates TANK ‐binding kinase 1 (TBK1) and inhibitor of nuclear factor κB (NF‐κB) kinase ε (IKKε), which phosphorylate IFN regulatory factor 3 (IRF‐3) and IRF‐7 (20, 21). Activated IRF‐3 and IRF‐7 translocate into the nucleus and bind IFN‐stimulated response elements (ISREs), inducing the expression of type I IFNs (22). IPS‐1 also interacts with FAS ‐associated death domain (FADD)‐containing protein (23). FADD activates caspase‐8 and caspase‐10, and the activation of the caspase death effector domains activates NF‐κB, leading to the production of inflammatory cytokines (24). Thus, MDA5 and RIG‐I appear to activate both the IFN and inflammatory responses.
Figure 1.
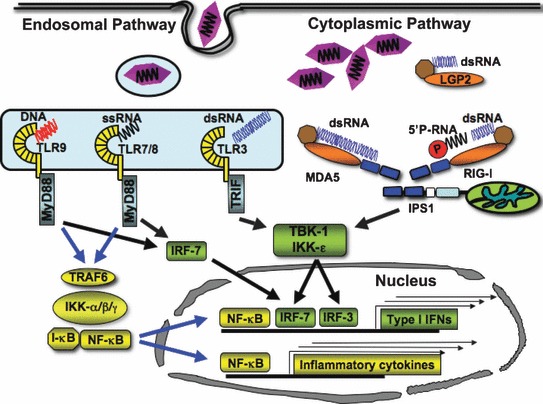
Cytoplasmic and endosomal sensors of viral nucleic acids. This figure illustrates the detection of viral products by retinoic acid‐inducible gene I (RIG‐I)‐like receptor (RLR) and Toll‐like receptor (TLR) family members. TLR3, TLR7/ 8, and TLR9 are located on endosomal compartments in which they sense their double‐stranded RNA (dsRNA), single‐stranded RNA (ssRNA), and CpG DNA ligands, respectively. TLR3 signals through the adapter protein Toll/interleukin‐1 (IL‐1) receptor (TIR) domain‐containing adapter‐inducing interferon‐β (TRIF), which activates tumor necrosis factor (TNF) receptor‐associated factor 3 (TRAF3) (not shown) and the TANK‐binding kinase 1 (TBK‐1) complex leading to interferon (IFN) regulatory factor 7 (IRF‐7) activation and IFN signaling. TRIF also signals through TRAF6, which leads to nuclear factor κB (NF‐κB) activation and inflammatory cytokine production. TLR7, TLR8, and TLR9 signal through the MyD88 adapter. MyD88 signals through a protein complex consisting of TRAF6 and IL‐1 receptor‐associated kinase 1/4 (IRAK1/4) (not shown), leading to the activation of type I IFN and NF‐κB signaling. RLR family members, melanoma differentiation‐associated gene 5 (MDA5), retinoic acid‐inducible gene I (RIG‐I), and laboratory of genetics and physiology‐2 (LGP2), are cytoplasmic proteins that detect viral products within the cytosol. MDA5 and RIG‐I signal through IFN‐β promoter stimulator 1 (IPS‐1), which is located on the mitochondrial membrane. IPS‐1 signals through TRAF3 and the TBK‐1/inhibitor of NF‐κB kinase ε complex to activate IRF‐3 and IRF‐7 and then type I IFN. IPS‐1 also signals through FAS‐associated death domain‐containing protein (FADD) leading to the activation of caspase‐8 and caspase‐10 (not shown), which causes NF‐κB activation and inflammatory cytokine production. LGP2 does not signal through IPS‐1 and is considered to be a negative regulator of RIG‐I.
TLRs
TLRs are transmembrane proteins that contain luminal leucine‐rich repeats (LRRs) that sense pathogen‐associated molecular patterns and cytoplasmic Toll/interleukin‐1 (IL‐1) receptor homology (TIR) domains that signal through downstream adapters (1). There are 10 members of the TLR family in humans and 13 in mice. TLRs involved in the detection of viral nucleic acids are located on the cell surface (TLR3) or in endosomal compartments (TLR3, TLR7, TLR8, and TLR9) (25). TLR3 recognizes dsRNA, which constitutes the genome of dsRNA viruses and is also an intermediate produced during replication of ssRNA viruses (26). TLR7 and TLR8 recognize ssRNA as well as imidazolequinilone compounds, which are known to have antiviral properties (25, 27, 28, 29, 30). TLR9 recognizes unmethylated CpG‐containing DNA, which is commonly found in the genomes of DNA viruses (31, 32).
TLR3 signals through the adapter protein TIR domain‐containing adapter‐inducing IFN‐β (TRIF) (33, 34) ( Fig. 1 ). TRIF interacts with TRAF3 and TRAF6 through TRAF‐binding motifs and with receptor‐interacting protein 1 (RIP1) and RIP3 through RIP homotypic interaction motifs (RHIM) (35, 36, 37). TRAF3 leads to the secretion of type I IFNs, while TRAF6 and RIP1 lead to NF‐κB activation and production of inflammatory cytokines (38). TLR7, TLR8, and TLR9 signal through the adapter protein myeloid differentiation primary response gene 88 (MyD88). MyD88 contains a TIR domain as well as a death domain that allows it to serve as an adapter for TLR signaling. MyD88 associates with a signaling complex consisting of TRAF6, Bruton’s tyrosine kinase (BTK), IL‐1 receptor‐associated kinase 4 (IRAK4), and IRAK1 (39). Signaling through this complex activates IRF7, NF‐κB, and mitogen‐activated protein kinase pathways (40, 41, 42). Thus, although RLRs and TLRs signal through different pathways, both appear to be able to activate the production of type I IFNs (i.e. IFN‐α and IFN‐β) and inflammatory cytokines.
Two additional TLR family members that signal through MyD88 have been implicated in the recognition of additional viral components. TLR2 is known to detect a variety of lipoproteins as well as yeast‐associated zymosan; however, it has also been demonstrated to have a role in the recognition of viral envelope proteins (43). Similarly, while TLR4 has traditionally been known as the sensor of LPS, it can also respond to virus‐derived envelope glycoproteins (44). In this review, we focus primarily on TLRs that recognize nucleic acids.
Additional sensors
The TLRs and RLRs have been shown to play a role primarily in RNA virus infection. Recently, the array of innate immune sensors of viral infection has been shown to include two additional cytosolic proteins that are involved in the recognition of DNA viruses. A DNA‐binding protein, named DNA‐dependent activator of IFN‐regulatory factors (DAI), Z‐DNA‐binding protein 1 (ZBP1), or DLM‐1, binds cytosolic DNA, inducing type I IFN and other genes involved in innate immunity (45, 46). Accordingly, RNA interference of mRNA for DAI in cells inhibits DNA‐mediated antiviral responses. Furthermore, Nacht domain‐, LRR‐, and PYD‐containing protein 3 (NALP3), a component of the cytosolic molecular complex termed the inflammasome, has been shown to recognize adenoviral DNA, inducing activation of caspase‐1 and maturation of pro‐IL‐1β in macrophages (47). Correspondingly, mice lacking NALP3 or its signaling adapter, apoptosis‐associated speck‐like protein containing a C‐terminal caspase (ASC), display reduced innate inflammatory responses to adenovirus particles. The discovery of these sensors has provided further insight into the innate response against DNA viruses.
Besides the RLR and TLR classes of sensors, other proteins are known to detect viral products and contribute to the immune response, especially RNase L and protein kinase R (PKR). RNase L has recently been reported to be involved in the RLR response to viral nucleic acids (48). It is proposed that 2′,5′‐linked oligoadenylate generated by viral infection activates RNase L to cleave self‐RNA into small RNA products, which are responsible for RLR signaling. However, it is not yet known how these small self‐RNAs are recognized by MDA5 and RIG‐I. PKR has been shown to dimerize upon binding of dsRNA. The activated PKR dimer phosphorylates eukaryotic initiation factor 2‐a (eIF2‐a), which results in the inhibition of translation, preventing viral replication (49). Like RLRs, RNase L and PKR are upregulated in response to type I IFN, demonstrating their important role in the preprogramed antiviral response.
Cytokine response to viruses
IFNs
The initiation of IFN production is an essential step in the antiviral response. Type I IFNs fight viruses both directly by inhibiting viral replication in cells and indirectly by stimulating the innate and adaptive immune responses. IFN‐α and IFN‐β bind to the IFN‐α receptor (IFNAR) in an autocrine or paracrine manner. Activation of this receptor leads to Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signal transduction pathways (50, 51). These genes increase the cellular resistance to viral infection and sensitize virally infected cells to apoptosis (52). Interestingly, several viral sensors are among those genes induced by IFN. They in turn enable the production of IFN, creating a positive feedback loop that creates a local cellular response. In addition, type I IFNs directly activate dendritic cells (DC) and natural killer (NK) cells and promote the survival and effector functions of T and B cells, providing a link between the innate response to infection and the adaptive immune response (53, 54, 55, 56).
TLRs signaling pathways also induce the recently identified type III IFNs. These include three proteins, named IFN‐λ1, IFN‐λ2, and IFN‐λ3, or IL‐29 (λ1) and IL‐28A/B (λ2/3). Although genetically distinct from type I IFNs, type III IFNs have similar biological antiviral functions (57, 58, 59). Whether RIG‐I and MDA5 transmit signals leading to the expression of type III IFN is yet unknown.
Inflammatory cytokines
In addition to IFN signaling, viral sensors are also known to initiate signaling for inflammatory cytokine and chemokine secretion. Both DCs and macrophages produce TNFα, IL‐6, monocyte chemotactic protein 1 (MCP‐1), and IL‐12 in response to viral infection. In addition, these same inflammatory cytokines are often detected in the serum of virally infected animals. Inflammatory cytokines activate the vascular endothelium as well as stimulate the recruitment of immune cells such as monocytes and neutrophils. While the resulting inflammatory response is important in the clearance of viral infection, a prolonged inflammatory state can also lead to adverse reactions including necrosis of local tissue and autoimmune diseases.
Understanding the diversity between viral sensors
It is not entirely clear whether viral sensors serve redundant or non‐redundant functions. One way in which viral sensors can be seen to have differential effects is by the recognition of different viruses. However, the sensors may also recognize different components of the same virus. Additionally, diversity could insure that different sensors activate the production of distinct cytokines. Finally, the differential expression of viral sensors in tissues and cell types is likely to contribute to their distinct roles in viral infection. In the following sections, we explore whether there is true redundancy or if there is specialization between the RLR and TLR families of sensors. This is also illustrated in Fig. 2 .
Figure 2.
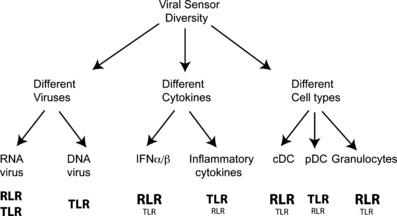
Hypothetical model of functional diversity among viral sensors. Three potential mechanisms by which viral sensors perform unique functions: (i) viral sensors detect different ligands resulting in the recognition of a variety of viral families and/or the recognition of different components of the same virus. (ii) Toll‐like receptor (TLR) and retinoic acid‐inducible gene I‐like receptor (RLR) signaling results in the production of different cytokine responses (i.e. type I interferon versus pro‐inflammatory cytokines). (iii) The expression of TLRs and RLRs in different cell types results in unique responses to viruses in different cells and tissues. Predominant sensors are indicated in bold, while secondary sensors are indicated below in smaller font.
Diversity by recognition of different viruses
RLRs
Among the RLRs, ligand preferences appear to determine which virus is recognized by which sensor. The current paradigm is that RIG‐I recognizes RNA‐containing 5′‐triphosphates, while MDA5 recognizes dsRNA. Therefore it is not surprising that RIG‐I has been shown to detect influenza A and B viruses, vesicular stomatitis virus (VSV), and some flaviviruses (Japanese encephalitis virus and hepatitis C virus) (13, 60, 61). Likewise, MDA5 detects picornaviruses such as encephalomyocarditis virus (EMCV), Mengo virus, and Theilers virus (13, 14) as well as caliciviruses (62). These viruses contain a 5′‐VPg cap instead of 5′‐triphosphate and make large amounts of dsRNA during replication. However, other results do not neatly fit this paradigm. RIG‐I and MDA5 play redundant roles in the recognition of West Nile virus (63), Dengue virus, (61) paramyxovirus, and reovirus (61), most of which contain 5′‐triphosphates. In addition, although Sendai virus has been shown to activate RIG‐I, it encodes for a protein, the V protein, that is a specific inhibitor of MDA5 (64). Furthermore, murine hepatitis virus, a member of the coronavirus family that does not contain VPg, has recently been shown to be recognized by MDA5 (65). One explanation is that, although RIG‐I preferentially recognizes 5′‐triphosphates and polyuridine‐rich regions, it can also recognize short dsRNA, while MDA5 recognizes long dsRNA (11). The ability of MDA5 and RIG‐I to specifically detect certain viruses while also detecting common pathogens illustrates the need for multiple sensors to recognize and control the wide variety of viral pathogens.
TLRs
Compared with that of the RLRs, the role of TLRs in antiviral responses is more intricate (66). TLR3 was originally shown to detect dsRNA (26). Accordingly, TLR3 has been implicated in the detection of several RNA viruses such as EMCV (67), respiratory syncytial virus (RSV) (68, 69), West Nile virus (70), and Punta Toro virus (PTV) (71). However, in one study TLR3 did not contribute to viral pathogenesis in VSV, lymphocytic choriomeningitis virus (LCMV), and reovirus infections (72). To make matters more confusing, TLR3 has been implicated in recognition of DNA viruses. TLR3‐deficient mice are more susceptible to murine cytomegalovirus (MCMV) infection than wildtype mice (73). Moreover, a recent human study has demonstrated that a dominant negative form of TLR3 causes susceptibility to neonatal herpes simplex‐1 encephalitis (HSE) (74). However, it is unclear why TLR3 plays such a major role in HSE, whereas it has no obvious role in other HSV‐1 infections, such as skin, eye, or mouth infections, or sepsis or in other DNA virus infections. Thus, TLR3 may recognize not only RNA viruses but also DNA viruses, most probably through RNA intermediates that are generated during viral replication.
TLR7 has been shown to contribute to the detection of RSV, Sendai virus, influenza, human immunodeficiency virus (HIV), VSV, and coxsackie virus B3 (CVB3) (75), while TLR8 has been implicated in the detection of influenza and paramyxovirus as well as HIV (28, 44, 76). TLR9 plays a role in the recognition of herpes simplex virus and cytomegalovirus infection (73, 77, 78, 79). TLR2 and TLR4 have been shown to play a role in the recognition of enveloped viruses. Both herpes viruses, which contain a DNA genome, and RSV, which has a ssRNA genome, have been reported to be recognized by these sensors (80, 81, 82). In summary, TLR7 and TLR8 recognize ssRNA viruses, while TLR9 recognize DNA viruses. TLR2 and TLR4 recognize enveloped viruses, while TLR3 plays a role in the recognition of both RNA and DNA viruses. Overall, TLR viral specificities exhibit a significant overlap with those of RLRs.
Diversity by distinct cytokine responses
Another potential explanation for the presence of different classes of sensors could be the induction of different cytokine responses. Although both RLRs and TLRs appear to be able to signal through both IFN and inflammatory cytokine pathways, there is evidence that the different classes have distinct functions in signal propagation leading to different immune responses.
Indeed, initial characterization of TLR3‐deficient mice in response to dsRNA analog polyinosinic–polycytidylic acid (polyI:C) revealed a defect in IL‐12 not type I IFN secretion in serum (26). Subsequently, it was shown that TLR3 induces a T‐helper 2 (Th2)‐type inflammatory response in airway epithelia cells infected with RSV infection (68, 69). Another study showed that TLR3 triggers an inflammatory response against West Nile virus infection that breaks down the blood–brain barrier, facilitating viral penetration and spreading in the brain and subsequent neuronal damage (70). Similarly, TLR3 plays an important role in liver pathology caused by PTV infection through the overproduction of inflammatory mediators (71). Altogether, these studies have suggested that TLR3 promotes a strong inflammatory response to RNA viruses, whereas it seems to have limited impact in type I IFN responses that control viral replication.
More recent studies have revealed differential signaling by RLRs and TLRs in individual cell types. One study found that influenza infection in bronchial epithelial cells led to TLR3‐dependent inflammatory cytokine induction and RIG‐I‐dependent IFN response (83). Another recent study has demonstrated that human keratinocytes contain functional TLR, RLR, and PKR signaling pathways and, with the use of small interfering RNA (siRNA) and small molecule inhibitors, has shown that TLR3 provides the main stimulus for NF‐κB signaling, while RLRs are the primary initiators of IRF3 and IFN signaling in this cell type (84).
The stimulation of different signaling pathways by TLRs and RLRs could have important implications. There are several viruses that are known to stimulate both TLR and RLR pathways, depending on whether infection is detected in endosomal or cytoplasmic compartments. It has been assumed that these sensors activate the same general cytokine responses; however, if the TLR and RLR pathways do indeed produce different cytokine products, then it is likely that they also produce different effects on the subsequent immune response. For example, it is known that the inflammatory cytokines can function not only to limit viral infection but also to cause excessive immunopathology. If we are able to distinguish which receptor is preferentially responsible for the production of each cytokine, then we may be able to target small molecules to the endosomal or cytoplasmic compartments to affect cytokine response. This could be a tremendous advantage to the fields of vaccine development as well as infectious diseases and tumor immunology, as antiviral sensors and their ability to induce cytokines are potential targets for therapy.
Diversity by differential distribution of sensors
The distribution of viral sensors in different cell and tissue types may be another mechanism to differentiate their actions. This is easily seen in comparison between conventional DCs (cDCs) and plasmacytoid DCs (pDC). cDCs are specialized for pathogen detection and antigen presentation. pDCs specialize in the secretion of type I IFNs in response to viruses (25, 85). In humans, cDCs express TLR1, TLR2, TLR3, TLR4, TLR5, TLR6, and TLR8, while pDCs preferentially express TLR7 and TLR9. cDCs are capable of expressing high levels of RIG‐I and MDA5, while pDCs also express these cytoplasmic sensors, but, paradoxycally, the sensors do not appear to function (86). Nevertheless, both cell types are able to respond to viruses. In human cDCs, this occurs in an MDA5‐ and TLR3‐dependent manner, leading to the production of IFN‐β, autocrine activation via IFNAR, and the production of IFN‐α. In pDCs, however, TLR3 is not present, MDA5 and RIG‐I may not be functional, and autocrine secretion and activation via IFN‐β does not occur, but high levels of IFN‐α are produced (87). This occurs because pDCs utilize TLR7 and TLR9 and express endogenously high levels of IRF7 (22), which primes them for IFN‐α production. In this situation, we can see that by varying the expression of TLRs and signaling components, different cell types have unique ways to detect pathogens. Extending this observation, a recent study demonstrated that human neutrophils respond to polyI:C through the RLR pathway rather than the TLR3 pathway (88).
A similar situation may occur in tissues. RIG‐I and MDA5 are IFN‐inducible genes that may be expressed in all cell types, while TLRs have a more restricted tropism. It has been demonstrated that MDA5 and RIG‐I are the predominant sensors for polyI:C and RNA virus infection in bone marrow‐derived DCs, macrophages, and fibroblasts. However, TLR3 has also been shown to play a role in different cell types. This may occur as a result of differential expression of the various sensors or their downstream adapter proteins. One specific example may occur in the brain. Several groups have shown that TLR3 is expressed in the brain, while MDA5 does not appear to be expressed. Most probably, this differential expression of the various viral sensors contributes to their importance in viral infection.
Conclusions
Both the TLR and RLR families of receptors contain multiple sensors that are important in viral infection. In this review, we examine the diversity of these sensors and the potential explanations for this diversity. The role of each sensor potentially may be distinguished by recognition of distinct viral pathogens, by stimulation of distinct cytokine signaling pathways, or by distribution of individual sensors in different cell and tissue types. Further studies are necessary to determine which of these possibilities most contributes, as well as the role of the individual sensors in viral infection in vivo.
Acknowledgements
This work was supported by JDRF grant no. 24‐2007‐420. S. M. is supported by training grant T32 AI007163.
References
- 1. Takeuchi O, Akira S. Recognition of viruses by innate immunity. Immunol Rev 2007;220:214–224. [DOI] [PubMed] [Google Scholar]
- 2. Yoneyama M, et al. The RNA helicase RIG‐I has an essential function in double‐stranded RNA‐induced innate antiviral responses. Nat Immunol 2004;5:730–737. [DOI] [PubMed] [Google Scholar]
- 3. Kang DC, et al. MDA‐5: an interferon‐inducible putative RNA helicase with double‐stranded RNA‐dependent ATPase activity and melanoma growth‐suppressive properties. Proc Natl Acad Sci USA 2002;99:637–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kovacsovics M, et al. Overexpression of Helicard, a CARD‐containing helicase cleaved during apoptosis, accelerates DNA degradation. Curr Biol 2002;12:838–843. [DOI] [PubMed] [Google Scholar]
- 5. Saito T, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG‐I and LGP2. Proc Natl Acad Sci USA 2007;104:582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rothenfusser S, et al. The RNA helicase Lgp2 inhibits TLR‐independent sensing of viral replication by retinoic acid‐inducible gene‐I. J Immunol 2005;175:5260–5268. [DOI] [PubMed] [Google Scholar]
- 7. Venkataraman T, et al. Loss of DExD/H box RNA helicase LGP2 manifests disparate antiviral responses. J Immunol 2007;178:6444–6455. [DOI] [PubMed] [Google Scholar]
- 8. Hornung V, et al. 5′‐Triphosphate RNA is the ligand for RIG‐I. Science 2006;314:994–997. [DOI] [PubMed] [Google Scholar]
- 9. Pichlmair A, et al. RIG‐I‐mediated antiviral responses to single‐stranded RNA bearing 5′‐phosphates. Science 2006;314:997–1001. [DOI] [PubMed] [Google Scholar]
- 10. Saito T, et al. Innate immunity induced by composition‐dependent RIG‐I recognition of hepatitis C virus RNA. Nature 2008;454:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kato H, et al. Length‐dependent recognition of double‐stranded ribonucleic acids by retinoic acid‐inducible gene‐I and melanoma differentiation‐associated gene 5. J Exp Med 2008;205:1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saito T, Gale M Jr. Differential recognition of double‐stranded RNA by RIG‐I‐like receptors in antiviral immunity. J Exp Med 2008;205:1523–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kato H, et al. Differential roles of MDA5 and RIG‐I helicases in the recognition of RNA viruses. Nature 2006;441:101–105. [DOI] [PubMed] [Google Scholar]
- 14. Gitlin L, et al. Essential role of MDA‐5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA 2006;103:8459–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawai T, et al. IPS‐1, an adaptor triggering RIG‐I‐ and MDA5‐mediated type I interferon induction. Nat Immunol 2005;6:981–988. [DOI] [PubMed] [Google Scholar]
- 16. Seth RB, et al. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF‐kappaB and IRF 3. Cell 2005;122:669–682. [DOI] [PubMed] [Google Scholar]
- 17. Kumar H, et al. Essential role of IPS‐1 in innate immune responses against RNA viruses. J Exp Med 2006;203:1795–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu LG, et al. VISA is an adapter protein required for virus‐triggered IFN‐beta signaling. Mol Cell 2005;19:727–740. [DOI] [PubMed] [Google Scholar]
- 19. Saha SK, et al. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J 2006;25:3257–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sharma S, et al. Triggering the interferon antiviral response through an IKK‐related pathway. Science 2003;300:1148–1151. [DOI] [PubMed] [Google Scholar]
- 21. Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 2003;4:491–496. [DOI] [PubMed] [Google Scholar]
- 22. Honda K, et al. IRF‐7 is the master regulator of type‐I interferon‐dependent immune responses. Nature 2005;434:772–777. [DOI] [PubMed] [Google Scholar]
- 23. Balachandran S, Thomas E, Barber GN. A FADD‐dependent innate immune mechanism in mammalian cells. Nature 2004;432:401–405. [DOI] [PubMed] [Google Scholar]
- 24. Takahashi K, et al. Roles of caspase‐8 and caspase‐10 in innate immune responses to double‐stranded RNA. J Immunol 2006;176:4520–4524. [DOI] [PubMed] [Google Scholar]
- 25. Iwasaki A, Medzhitov R. Toll‐like receptor control of the adaptive immune responses. Nat Immunol 2004;5:987–995. [DOI] [PubMed] [Google Scholar]
- 26. Alexopoulou L, et al. Recognition of double‐stranded RNA and activation of NF‐kappaB by Toll‐like receptor 3. Nature 2001;413:732–738. [DOI] [PubMed] [Google Scholar]
- 27. Diebold SS, et al. Innate antiviral responses by means of TLR7‐mediated recognition of single‐stranded RNA. Science 2004;303:1529–1531. [DOI] [PubMed] [Google Scholar]
- 28. Heil F, et al. Species‐specific recognition of single‐stranded RNA via toll‐like receptor 7 and 8. Science 2004;303:1526–1529. [DOI] [PubMed] [Google Scholar]
- 29. Lund JM, et al. Recognition of single‐stranded RNA viruses by Toll‐like receptor 7. Proc Natl Acad Sci USA 2004;101:5598–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beignon AS, et al. Endocytosis of HIV‐1 activates plasmacytoid dendritic cells via Toll‐like receptor‐viral RNA interactions. J Clin Invest 2005;115:3265–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Beutler B, et al. Genetic analysis of host resistance: Toll‐like receptor signaling and immunity at large. Annu Rev Immunol 2006;24:353–389. [DOI] [PubMed] [Google Scholar]
- 32. Bauer S, et al. Human TLR9 confers responsiveness to bacterial DNA via species‐specific CpG motif recognition. Proc Natl Acad Sci USA 2001;98:9237–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamamoto M, et al. Cutting edge: a novel Toll/IL‐1 receptor domain‐containing adapter that preferentially activates the IFN‐beta promoter in the Toll‐like receptor signaling. J Immunol 2002;169:6668–6672. [DOI] [PubMed] [Google Scholar]
- 34. Oshiumi H, et al. TICAM‐1, an adaptor molecule that participates in Toll‐like receptor 3‐mediated interferon‐beta induction. Nat Immunol 2003;4:161–167. [DOI] [PubMed] [Google Scholar]
- 35. Sato S, et al. Toll/IL‐1 receptor domain‐containing adaptor inducing IFN‐beta (TRIF) associates with TNF receptor‐associated factor 6 and TANK‐binding kinase 1, and activates two distinct transcription factors, NF‐kappa B and IFN‐regulatory factor‐3, in the Toll‐like receptor signaling. J Immunol 2003;171:4304–4310. [DOI] [PubMed] [Google Scholar]
- 36. Oganesyan G, et al. Critical role of TRAF3 in the Toll‐like receptor‐dependent and ‐independent antiviral response. Nature 2006;439:208–211. [DOI] [PubMed] [Google Scholar]
- 37. Meylan E, et al. RIP1 is an essential mediator of Toll‐like receptor 3‐induced NF‐kappa B activation. Nat Immunol 2004;5:503–507. [DOI] [PubMed] [Google Scholar]
- 38. Hacker H, et al. Specificity in Toll‐like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 2006;439:204–207. [DOI] [PubMed] [Google Scholar]
- 39. Suzuki N, Suzuki S, Yeh WC. IRAK‐4 as the central TIR signaling mediator in innate immunity. Trends Immunol 2002;23:503–506. [DOI] [PubMed] [Google Scholar]
- 40. Medzhitov R, et al. MyD88 is an adaptor protein in the hToll/IL‐1 receptor family signaling pathways. Mol Cell 1998;2:253–258. [DOI] [PubMed] [Google Scholar]
- 41. Burns K, et al. MyD88, an adapter protein involved in interleukin‐1 signaling. J Biol Chem 1998;273:12203–12209. [DOI] [PubMed] [Google Scholar]
- 42. Muzio M, et al. IRAK (Pelle) family member IRAK‐2 and MyD88 as proximal mediators of IL‐1 signaling. Science 1997;278:1612–1615. [DOI] [PubMed] [Google Scholar]
- 43. Morrison LA. The Toll of herpes simplex virus infection. Trends Microbiol 2004;12:353–356. [DOI] [PubMed] [Google Scholar]
- 44. Zhang SY, et al. Human Toll‐like receptor‐dependent induction of interferons in protective immunity to viruses. Immunol Rev 2007;220:225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takaoka A, et al. DAI (DLM‐1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 2007;448:501–505. [DOI] [PubMed] [Google Scholar]
- 46. Wang Z, et al. Regulation of innate immune responses by DAI (DLM‐1/ZBP1) and other DNA‐sensing molecules. Proc Natl Acad Sci USA 2008;105:5477–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 2008;452:103–107. [DOI] [PubMed] [Google Scholar]
- 48. Malathi K, et al. Small self‐RNA generated by RNase L amplifies antiviral innate immunity. Nature 2007;448:816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie 2007;89:799–811. [DOI] [PubMed] [Google Scholar]
- 50. Schindler C, Darnell JE Jr. Transcriptional responses to polypeptide ligands: the JAK–STAT pathway. Annu Rev Biochem 1995;64:621–651. [DOI] [PubMed] [Google Scholar]
- 51. De Veer MJ, et al. Functional classification of interferon‐stimulated genes identified using microarrays. J Leukoc Biol 2001;69:912–920. [PubMed] [Google Scholar]
- 52. Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity 2006;25:373–381. [DOI] [PubMed] [Google Scholar]
- 53. Tough DF. Type I interferon as a link between innate and adaptive immunity through dendritic cell stimulation. Leuk Lymphoma 2004;45:257–264. [DOI] [PubMed] [Google Scholar]
- 54. Theofilopoulos AN, et al. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol 2005;23:307–336. [DOI] [PubMed] [Google Scholar]
- 55. Marrack P, Kappler J, Mitchell T. Type I interferons keep activated T cells alive. J Exp Med 1999;189:521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Braun D, Caramalho I, Demengeot J. IFN‐alpha/beta enhances BCR‐dependent B cell responses. Int Immunol 2002;14:411–419. [DOI] [PubMed] [Google Scholar]
- 57. Ank N, West H, Paludan SR. IFN‐lambda: novel antiviral cytokines. J Interferon Cytokine Res 2006;26:373–379. [DOI] [PubMed] [Google Scholar]
- 58. Kotenko SV, et al. IFN‐lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol 2003;4:69–77. [DOI] [PubMed] [Google Scholar]
- 59. Sheppard P, et al. IL‐28, IL‐29 and their class II cytokine receptor IL‐28R. Nat Immunol 2003;4:63–68. [DOI] [PubMed] [Google Scholar]
- 60. Sumpter R Jr, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG‐I. J Virol 2005;79:2689–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Loo YM, et al. Distinct RIG‐I and MDA5 signaling by RNA viruses in innate immunity. J Virol 2008;82:335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McCartney SA, et al. MDA‐5 recognition of a murine norovirus. PLoS Pathog 2008;4:e1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fredericksen BL, et al. Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG‐I and MDA5 signaling through IPS‐1. J Virol 2008;82:609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Andrejeva J, et al. The V proteins of paramyxoviruses bind the IFN‐inducible RNA helicase, MDA‐5, and inhibit its activation of the IFN‐beta promoter. Proc Natl Acad Sci USA 2004;101:17264–17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Roth‐Cross JK, Bender SJ, Weiss SR. Murine coronavirus mouse hepatitis virus (MHV) is recognized by MDA5 and induces type I IFN in brain macrophages/microglia. J Virol 2008;82:9829–9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schroder M, Bowie AG. TLR3 in antiviral immunity: key player or bystander? Trends Immunol 2005;26:462–468. [DOI] [PubMed] [Google Scholar]
- 67. Hardarson HS, et al. Toll‐like receptor 3 is an essential component of the innate stress response in virus‐induced cardiac injury. Am J Physiol Heart Circ Physiol 2007;292:H251–H258. [DOI] [PubMed] [Google Scholar]
- 68. Rudd BD, et al. Deletion of TLR3 alters the pulmonary immune environment and mucus production during respiratory syncytial virus infection. J Immunol 2006;176:1937–1942. [DOI] [PubMed] [Google Scholar]
- 69. Rudd BD, et al. Differential role for TLR3 in respiratory syncytial virus‐induced chemokine expression. J Virol 2005;79:3350–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang T, et al. Toll‐like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med 2004;10:1366–1373. [DOI] [PubMed] [Google Scholar]
- 71. Gowen BB, et al. TLR3 deletion limits mortality and disease severity due to Phlebovirus infection. J Immunol 2006;177:6301–6307. [DOI] [PubMed] [Google Scholar]
- 72. Edelmann KH, et al. Does Toll‐like receptor 3 play a biological role in virus infections? Virology 2004;322:231–238. [DOI] [PubMed] [Google Scholar]
- 73. Tabeta K, et al. Toll‐like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc Natl Acad Sci USA 2004;101:3516–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang SY, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science 2007;317:1522–1527. [DOI] [PubMed] [Google Scholar]
- 75. Wang JP, et al. Cutting edge: antibody‐mediated TLR7‐dependent recognition of viral RNA. J Immunol 2007;178:3363–3367. [DOI] [PubMed] [Google Scholar]
- 76. Melchjorsen J, et al. Activation of innate defense against a paramyxovirus is mediated by RIG‐I and TLR7 and TLR8 in a cell‐type‐specific manner. J Virol 2005;79:12944–12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Krug A, et al. TLR9‐dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity 2004;21:107–119. [DOI] [PubMed] [Google Scholar]
- 78. Krug A, et al. Herpes simplex virus type 1 activates murine natural interferon‐producing cells through toll‐like receptor 9. Blood 2004;103:1433–1437. [DOI] [PubMed] [Google Scholar]
- 79. Lund J, et al. Toll‐like receptor 9‐mediated recognition of Herpes simplex virus‐2 by plasmacytoid dendritic cells. J Exp Med 2003;198:513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sato A, Linehan MM, Iwasaki A. Dual recognition of herpes simplex viruses by TLR2 and TLR9 in dendritic cells. Proc Natl Acad Sci USA 2006;103:17343–17348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kurt‐Jones EA, et al. Herpes simplex virus 1 interaction with Toll‐like receptor 2 contributes to lethal encephalitis. Proc Natl Acad Sci USA 2004;101:1315–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kurt‐Jones EA, et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat Immunol 2000;1:398–401. [DOI] [PubMed] [Google Scholar]
- 83. Le Goffic R, et al. Cutting edge: influenza A virus activates TLR3‐dependent inflammatory and RIG‐I‐dependent antiviral responses in human lung epithelial cells. J Immunol 2007;178:3368–3372. [DOI] [PubMed] [Google Scholar]
- 84. Kalali BN, et al. Double‐stranded RNA induces an antiviral defense status in epidermal keratinocytes through TLR3‐, PKR‐, and MDA5/RIG‐I‐mediated differential signaling. J Immunol 2008;181:2694–2704. [DOI] [PubMed] [Google Scholar]
- 85. Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat Immunol 2004;5:1219–1226. [DOI] [PubMed] [Google Scholar]
- 86. Coccia EM, et al. Viral infection and Toll‐like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte‐derived dendritic cells. Eur J Immunol 2004;34:796–805. [DOI] [PubMed] [Google Scholar]
- 87. Kumagai Y, et al. Alveolar macrophages are the primary interferon‐alpha producer in pulmonary infection with RNA viruses. Immunity 2007;27:240–252. [DOI] [PubMed] [Google Scholar]
- 88. Tamassia N, et al. Activation of an immunoregulatory and antiviral gene expression program in poly(I:C)‐transfected human neutrophils. J Immunol 2008;181:6563–6573. [DOI] [PubMed] [Google Scholar]


