Abstract
The present study attempted to identify a target antigen for immunotherapy for cholangiocarcinoma. Forkhead box M1 (FOXM1) was selected as a candidate antigen based on the data of previous cDNA microarray analysis of clinical samples of cholangiocarcinoma. The level of FOXM1 mRNA was more than 4 times higher in cancer cells in comparison to adjacent normal epithelial cells, in all of 24 samples of cholangiocarcinoma tissues. An immunohistochemical analysis also detected FOXM1 protein in the cancer cells but not in the normal cells. Twenty‐three human FOXM1‐derived peptides predicted to bind to HLA‐A2 were analyzed to determine their ability to induce HLA‐A2‐restricted T cells in HLA‐A2 transgenic mice. FOXM1362‐370 (YLVPIQFPV), FOXM1373‐382 (SLVLQPSVKV), and FOXM1640‐649 (GLMDLSTTPL) peptides primed HLA‐A2‐restricted cytotoxic T lymphocytes (CTLs) in the HLA‐A2 transgenic mice. Human CTL lines reactive to these 3 peptides could also be established from HLA‐A2‐positive healthy donors and cancer patients. Natural processing of the 3 epitopes from FOXM1 protein was confirmed by specific killing of HLA‐A2‐positive FOXM1‐transfectants by peptide‐induced CTLs. FOXM1 is expressed in various types of cancers and it is also functionally involved in oncogenic transformation and the survival of cancer cells. Therefore, FOXM1 may be a suitable target for immunotherapy against various cancers including cholangiocarcinoma.
Keywords: FOXM1, cDNA microarray, CTL epitopes, tumor‐associated antigen, CTLs, HLA‐A2
Cholangiocarcinoma is a devastating malignancy, which is difficult to diagnose and treat. Unfortunately, only a few patients are considered suitable for surgery, and chemotherapy and radiotherapy have not yielded substantial improvements in the survival rate. The overall survival rate is poor, with less than 5% of patients surviving upto 5 years. There has been no significant change in this rate over the past 30 years.1 Recent studies have identified some therapeutic molecular targets in cholangiocarcinoma in preclinical studies, thus supporting the therapeutic potential of selected targeting strategies against cholangiocarcinoma.2 The results were encouraging, although they were obtained from in vitro studies and tumor cell xenograft models. Therefore, novel therapeutic strategies for cholangiocarcinoma are urgently needed.
In cancer immunotherapy, effective induction of cytotoxic T lymphocytes (CTLs) by tumor‐associated antigen (TAA) has shown promising results.3 The utilization of CTLs elicited by TAA is an ideal therapeutic approach, if they specifically attack tumor cells expressing the antigen and reveal little or no adverse effects on normal cells. The development of cDNA microarray technologies, coupled with genome information, has provided comprehensive profiles of the gene expression of malignant cells, which have been compared to those of normal cells.4, 5 TAAs should be expressed excessively and preferentially by the tumor cells but not by the normal tissues, and gene expression profiling with cDNA microarray technologies is an effective approach for the identification of new TAAs.6, 7, 8 Previous studies have identified several molecular targets useful for the diagnosis and immunotherapy by analyzing genome‐wide gene expression profiles of various cancerous tissues with cDNA microarrays.9, 10, 11, 12, 13, 14 The present study identified a candidate TAA, forkhead box M1 (FOXM1), based on the results of previous cDNA microarray analysis of the tissues from 25 patients with intrahepatic cholangiocarcinoma.15 Recently, an elevated expression of FOXM1 has been reported in a variety of human cancers,12, 15, 16, 17, 18, 19, 20 including lung cancer,21 which is the most common causes of cancer‐related deaths in Western countries.22 Among normal adult human organs, FOXM1 is expressed only in the testis, thymus, small intestine and colon.23
The identification of immunodominant epitopes of TAA has highlighted the utilization of these epitopes as promising therapeutic tools for cancer immunotherapy. However, the identification of CTL epitopes remains difficult, because many TAA‐derived peptides must be screened for their immunogenicity by using a large number of peripheral blood mononuclear cells (PBMCs) isolated from the patients with cancer. HLA transgenic mice (Tgm) provides versatile animal models for the preclinical evaluation of peptide‐based immunotherapy.24 Previous investigations have used HLA‐A2 Tgm for the identification of HLA‐A2‐restricted antigenic epitopes.11 The present study identified FOXM1‐derived HLA‐A2‐restricted epitopes by using HLA‐A2 Tgm. CTL lines reactive to these epitopes were also established from cancer patients and healthy donors. The data presented herein suggest that FOXM1 may therefore be a suitable target for immunotherapy of various cancers, including cholangiocarcinoma.
Abbreviations.
BM‐DC: bone marrow‐derived dendritic cell; CTL: cytotoxic T lymphocyte; DC: dendritic cell; ELISPOT: enzyme‐linked immunospot; FOXM1: forkhead box M1; HA: hemagglutinin; HLA: human leukocyte histocompatibility antigen; HRP: horseradish peroxidase; NSCLC: non‐small cell lung carcinoma; PBMC: peripheral blood mononuclear cell; TAA: tumor‐associated antigen; Tgm: transgenic mouse.
Material and Methods
cDNA microarray analysis
Intrahepatic cholangiocarcinoma tissue specimens were obtained from 25 patients who underwent hepatectomy at Kyoto University Hospital with written informed consents. All tumors were histologically diagnosed to be cholangiocarcinoma by pathologists. The profiling of gene expression by cDNA microarray analysis was carried out, as described previously.15
Mice, peptides and cell lines
HLA‐A2 transgenic mice described previously were kindly provided by Dr. F.A. Lemonnier (Department SIDA‐Retrovirus, Unite d' Immunite Cellulaire Antivirale, Institut Pasteur, France).11 Peptides (purity > 90%) were purchased from Anygen (Gwangju, South Korea). Three human cholangiocarcinoma (TFK, HuCCT‐1, MEC), two human hepatocellular carcinoma (HepG2 and SK‐Hep1), and the 23 human lung cancer cell lines including 7 adenocarcinomas (A427, A549, LC319, PC‐3, PC‐9, PC‐14 and NCI‐H1666), two bronchiolo‐alveolar carcinomas (NCI‐H1373, NCI‐H1781), 7 squamous cell carcinomas (EBC‐1, LU61, NCI‐H520, NCI‐H1703, NCI‐H2170, RERF‐LC‐AI and SK‐MES‐1), 2 adenosquamous carcinomas (NCI‐H226 and NCI‐H647), 1 large‐cell carcinoma (LX1), and 4 small‐cell lung cancers (DMS114, DMS273, SBC‐3 and SBC‐5) were used. Human SAEC cells were used as controls. A transporter associated with antigen processing (TAP)‐deficient and HLA‐A*0201‐positive cell line, T2, and a colon cancer cell line, Caco‐2, were obtained from the RIKEN Bioresource Center (Tsukuba, Japan). These cells were maintained in vitro in RPMI 1640 medium or DMEM supplemented with 10% heat inactivated fetal calf serum, except for Caco‐2 cells that were cultured in medium supplemented with 20% heat inactivated fetal calf serum. SAEC cells were grown in optimized medium (SAGM) purchased from Cambrex Bio Science (Walkersville, MD).
Blood and tissue samples
The clinical research using PBMCs from the donors was approved by the Institutional Review Board of Kumamoto University. Blood samples were obtained from healthy donors and from patients with either cholangiocarcinoma or lung cancers who provided their written informed consent. The expression of HLA‐A2 was examined by flowcytometry with an anti‐HLA‐A2 monoclonal antibody, BB7.2 (One Lambda, Canoga Park, CA), to select HLA‐A2‐positive donors. Cancerous tissues were excised from patients during surgery, after obtaining written informed consent.
RT‐PCR and Northern blot analysis
Total RNA was extracted from the cell lines and the tissues using the RNeasy kit (Qiagen, Dusseldorf, Germany) according to the manufacturers' instructions. Total RNA of human normal tissues was purchased from Clontech (Palo Alto, CA) and RT‐PCR was done as described previously.25 The primer sequences were: FOXM1, 5′‐CACCCCAGTGCCAACCGC TACTTG‐3′ and 5′‐AAAGAGGAGCTATCCCCTCCTCAG‐3′ (that can detect three splicing variants FOXM1a, FOXM1b and FOXM1c) or 5′‐CCCTGACAACATCAACTGGTC‐3′ and 5′‐GTCCACCTTCGCTTTTATTGAGT‐3′ (that cannot detect the variants); GAPDH, 5′‐CCCATCACCATCTTCCAGGAGC‐3′ and 5′‐CCAGTGAGCTTCCCGTTCAGC‐3′; ACTB, 5′‐GAGGTGATAGCATTGCTTTCG‐3′ and 5′‐CAAGTCAGTGTACAGGTAAGC‐3′. A Northern blot analysis was done as described previously.18 Gels with 20 μg of the total RNA of the human normal tissue (Clontech) per lane were blotted onto nylon membranes. The membranes were hybridized with 32P‐labeled FOXM1‐specific cDNA as a probe. The blots were then autoradiographed with intensifying screens at −80°C for 120 hr.
Immunohistochemical analysis
An immunohistochemical analysis was done as described previously.25 Sections of paraffin‐embedded cholangiocarcinoma and lung cancer specimens were stained with a mouse monoclonal anti‐human FOXM1 antibody (clone 3A9, Abnova, Taipei, Taiwan) by standard procedure. Staining signals were visualized using the DakoCytomation EnVision + System‐HRP (DakoCytomation, Carpinteria, CA).
Generation of FOXM1 transfectant
A cDNA encoding for FOXM1 tagged with Influenza virus Hemagglutinin (HA) was cloned into a pENTR vector (Invitrogen, Carlsbad, CA) and then transferred into a self‐inactivating lentivirus expression vector, CSII‐EF‐RfA,26 by LR recombination reaction (Invitrogen). Lentivirous‐mediated gene transfer was performed as described previously.27 The expression of the transgene‐derived FOXM1 protein was confirmed by Western blot analysis, using horseradish peroxidase (HRP)‐conjugated rat monoclonal anti‐HA antibody (clone 3F10, Roche, Mannheim, Germany). β‐actin was also detected by using mouse monoclonal anti‐β‐actin antibody (clone AC‐15, Sigma, St. Louis, MO) and HRP‐conjugated rabbit anti‐mouse IgG (Amersham Biosciences, Piscataway, NJ) antibody. The signals were developed and visualized using a chemiluminescence system (ECL, Amersham Biosciences).
Identification of HLA‐A2‐restricted CTL epitopes of FOXM1 using HLA‐A2 transgenic mice
The immunization of mice with peptide‐loaded bone marrow‐derived DCs (BM‐DCs) was done as previously described.12, 15 CD4− spleen cells were isolated from the immunized mice, and the frequency of cells producing IFN‐γ among 5 × 104 cells upon stimulation with syngeneic 1 × 104/well BM‐DCs, pulsed with and without each peptide, was assayed by an enzyme‐linked immunospot (ELISPOT) assay as previously descibed.28
HLA‐peptide‐binding assay
HLA‐A*0201‐positive and TAP‐deficient T2 cells, pre‐incubated overnight at 26°C, were cultured in the presence of 50 μM FOXM1 peptides and 5 μg/ml β2‐microglobulin (Sigma, St Louis, MO) in serum‐free medium at 26°C again for 1.5 hr and subsequently at 37°C for 18 hr. The level of expression of cell surface HLA‐A2 was analyzed by flow cytometry using anti‐HLA‐A2 monoclonal antibody, BB7.2. HLA‐A24‐restricted human Cytomegalovirus‐derived peptide (QYDPVAALF) as a negative control and SARS‐A2‐S‐7 peptide (NLNESLIDL), which is SARS coronavirus‐derived CTL epitope peptide with high binding affinity to HLA‐A2 (work in preparation), as a positive control were also used. The fluorescence index (FI) was calculated from the mean fluorescence intensity (MFI) of HLA‐A2 expressed on T2 cells determined by flow cytometry, using the formula; FI = (MFI [T2 cells with FOXM1 peptide]/MFI [T2 cells with CMV‐peptide]) − 1.
Induction of FOXM1‐reactive human CTLs
PBMCs were isolated from heparinized blood of HLA‐A2‐positive Japanese cancer patients or healthy donors by Ficoll‐Conray density gradient centrifugation. Peripheral monocyte‐derived dendritic cells (DCs) were generated as previously described.29 CD8+ T cells were isolated, using anti‐CD8 microbeads (Miltenyl Biotec, Bergisch Gladbach, Germany) from PBMCs of the same donors, and thereafter, peptide‐reactive CD8+ CTLs were generated as described.30 Six days after the last stimulation, antigen‐specific responses of the CTLs were analyzed by IFN‐γ ELISPOT assay as described above and also by 4 h‐51Cr release assay as described previously.31 Briefly, 51Cr labeled target cells (5 × 103) suspended in 100 μl of assay medium were seeded into wells of 96‐well plates. Various numbers of effector cells suspended in 100 μl of assay medium were added into the wells and were incubated for 4 hr, and the supernatant was collected from each well to measure the 51Cr radioactivity.
Statistical analysis
The statistical significance of the differential findings among the experimental groups was determined by Student's t‐test. Avalue of p < 0.05 was considered to be statistically significant.
Results
Markedly enhanced expression of FOXM1 in intrahepatic cholangiocarcinoma tissues
A laser microdissection‐based cDNA microarray analysis of 25 cases of intrahepatic cholangiocarcinoma has been reported previously.15 These data were used to search for genes showing a high relative expression ratio between cancer cells and normal epithelial cells of intrahepatic bile duct. This process identified 8 genes with greater that 4‐fold increased expression in cancer cells in more than 15 out of 25 tissue samples (Fig. 1 a). Among the 8 up‐regulated genes, the expression of FOXM1 exhibited the greatest increase in the expression in cancer cells in comparison to the normal cells (Fig. 1 a). Therefore, FOXM1 was evaluated as a candidate antigen for immunotherapy.
Figure 1.

cDNA microarray analysis revealed a markedly enhanced expression of FOXM1 mRNA in intrahepatic cholangiocarcinoma tissues. (a) A list of up‐regulated genes in intrahepatic cholangiocarcinoma tissue. These genes exhibited greater than 4‐fold increased expression in cancer cells in comparison to their normal counterparts in more than 15 out of 25 intrahepatic cholangiocarcinoma patients. The expression of FOXM1 mRNA in intrahepatic cholangiocarcinoma tissue specimens was markedly enhanced in 24 of 25 patients with intrahepatic cholangiocarcinoma. (b) The expression levels of FOXM1 in the normal tissues were far lower than those in non‐small cell lung carcinoma (NSCLC) or esophageal cancer tissues based on the cDNA microarray analysis.
The expression of FOXM1 was analyzed in 16 kinds of normal tissues including fetal kidney and liver and also non‐small cell lung cancer and esophageal cancer, using cDNA microarray (Fig. 1 b). FOXM1 was expressed in the testis, bone marrow, small intestine, thymus, and fetal liver among normal tissues. However, the levels of expression in these normal tissues were far lower than those in non‐small cell lung carcinoma and esophageal cancer tissues (Fig. 1 b).
cDNA microarray analysis of expression of FOXM1 in various cancer tissues
The previously reported data of cDNA microarray analysis of various kinds of cancerous tissues12, 13, 14, 15, 16, 17, 18, 19, 20, 21 were used to analyze the expression of FOXM1 in 21 different types of cancer tissues. Supporting Information Table 1 shows the expression of FOXM1 to be elevated in various cancer tissues. A particularly enhanced expression was recognized in bladder cancer, lung cancer (both small cell carcinoma and non‐small cell carcinoma) and pancreatic cancer.
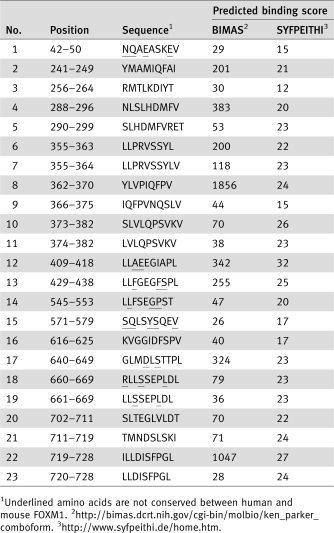
Table 1.
Human FOXM1‐derived peptides predicted to bind to HLA‐A2 (A*0201)
|
Expression of FOXM1 in various cancer cell lines and tissues
Further investigation of FOXM1 was conducted not only in cholangiocarcinoma but also in lung cancer, because lung cancer is the most common cause of cancer death in Western countries.22
The expression of the FOXM1 gene in cholangiocarcinoma, hepatocellular carcinoma and lung cancer cell lines (adenocarcinoma, bronchiolo‐alveolar carcinoma, squamous cell carcinoma, adenosquamous carcinoma, large cell carcinoma and small cell carcinoma), and lung cancer tissues was analyzed by RT‐PCR.
The FOXM1 gene contains 10 exons, 2 of which are alternatively expressed, giving rise to 3 differentially expressed mRNA isoforms.23 Cholangiocarcinoma cell lines, TFK, HuCCT‐1, and MEC, and hepatocellular carcinoma cell lines, HepG2 and SK‐Hep1 strongly expressed FOXM1b and FOXM1c (FOXM1b < FOXM1c) mRNA (Fig. 2).
Figure 2.
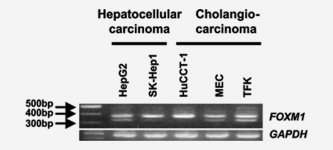
The expression of FOXM1 mRNA in cancer cell lines. RT‐PCR analysis was done to detect FOXM1 mRNA expression in cholangiocarcinoma and hepatocellular carcinoma cell lines. The same cDNA samples were tested for GAPDH for control.
FOXM1 was expressed in all the lung cancer cell lines examined (Supporting Information Fig. 1). In addition, all of the tissue samples of lung adenocarcinoma, squamous cell carcinoma, and small cell carcinoma also expressed FOXM1 mRNA, whereas normal lung epithelium did not (Supporting Information Fig. 1). FOXM1 was strongly expressed in all cancer cell lines examined, except for Caco‐2, which is a colon cancer cell line expressing a trace amount of FOXM1 mRNA (data not shown).
Among the normal tissue specimens, FOXM1 mRNA was expressed in the testis, fetal liver and thymus (FOXM1b > FOXM1c), stomach, small intestine and colon (FOXM1b < FOXM1c) (Fig. 3). A Northern blot analysis revealed relatively strong expression of FOXM1 mRNA in testis and weak expression in the fetal liver, thymus and gastrointestinal tracts (Fig. 3). These RT‐PCR and Northern blot analyses were consistent with the results of the cDNA microarray analysis (Fig. 1 b).
Figure 3.
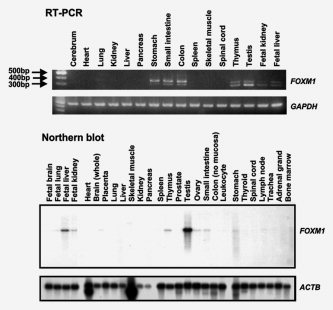
FOXM1 expression in various normal tissues analyzed by RT‐PCR and Northern blot analysis. The same Northern blot membrane was stripped and rehybridized with a β‐actin probe to prove the RNA integrity and to assess the loading of equal amounts of RNA.
Tissue samples of cholangiocarcinoma and lung cancer were analyzed immunohistochemically to determine the expression of FOXM1 protein in cancer cells (Fig. 4). An increased expression of FOXM1 protein was observed in the cancerous tissues, but not in the adjacent normal tissues.
Figure 4.
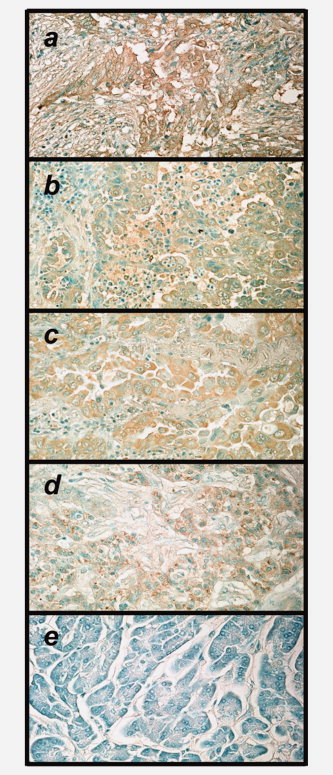
Immunohistochemical staining of FOXM1 protein in the intrahepatic cholangiocarcinoma (a), lung adenocarcinoma (b, c), cholangiocarcinoma (d), non‐cancerous lesion (pancreas) of d (e). The sections were stained with monoclonal anti‐FOXM1 antibody. Objective magnification was 200×.
Identification of HLA‐A2‐restricted FOXM1 epitopes by using HLA‐A2.1 Tgm
Human FOXM1‐derived peptides, carrying binding motifs for HLA‐A2 (HLA‐A*0201) were searched using the BIMAS software package (BioInformatics and Molecular Analysis Section, Center for Information Technology, NIH, Bethesda, MD, http://www-bimas.cit.nih.gov/) and SYFPEITHI (Biomedical Informatics, http://www.syfpeithi.de/). We selected 23 peptides predicted to bind to HLA‐A2 from the amino acid sequence conserved among human FOXM1a, FOXM1b and FOXM1c isoforms. The scores of the selected peptides calculated based on BIMAS and SYFPEITHI algorithms predicting HLA‐A2‐binding affinity are shown in Table 1.
Further investigations were conducted to identify HLA‐A2‐restricted epitopes from the selected 23 peptides, using HLA‐A2 Tgm. The 23 peptides were loaded onto BM‐DCs of HLA‐A2 Tgm and injected intraperitoneally twice into the mice with a 7‐day interval. Spleen cells were isolated from the mice 7 days after the second injection and cultured with BM‐DCs pulsed with the peptides. Subsequently, the cultured cells were subjected to an ELISPOT assay to analyze production of IFN‐γ upon stimulation with the same peptides. A series of repeated experiments revealed a reproducible response to FOXM1362‐370, FOXM1373‐382 and FOXM1640‐649 (designated as FOXM1‐8, FOXM1‐10 and FOXM1‐17, respectively; Table 1: Fig. 5). On the other hand, the peptide‐reactive CTL lines did not respond to the peptides pulsed onto non‐transgenic C57BL/6 mice‐derived BM‐DCs, thus verifying that the responses to these peptides were restricted by HLA‐A2.
Figure 5.
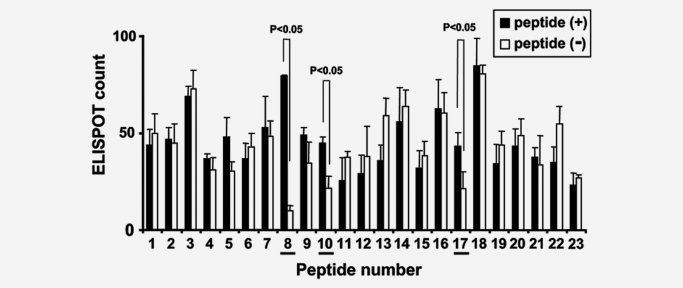
The identification of HLA‐A2‐restricted CTL epitopes of FOXM1 by using HLA‐A2 Tgm. HLA‐A2 Tgm were primed by injection of BM‐DCs (5 × 105) pulsed with the mixture of FOXM1‐derived peptides carrying HLA‐A2 (A*0201) binding motif into the peritoneal cavity of the Tgm twice with a 1 week interval. Seven days after the 2nd vaccination, spleens were isolated and CD4− spleen cells (5 × 106/well) were stimulated with syngeneic irradiated BM‐DCs (5 × 105/well) pulsed with each peptide for 6 days. Cultured CD4− spleen cells were subjected to ELISPOT assay to count IFN‐γ‐producing cells/5 × 104 CD4− spleen cells upon stimulation with 1 × 104 BM‐DCs with (black) or without (white) peptide loading. The columns and bars indicate mean of triplicate assays and SE, respectively. The data are each representative of 3 independent experiments. The underlines indicate the peptides that induced peptide‐specific responses (p < 0.05).
The amino acid sequences of FOXM1‐8 and FOXM1‐10 are conserved between human and mouse, whereas that of FOXM1‐17 is not conserved; human GLMDLSTTPL vs. mouse GLM ELNTTPL. To determine whether the immunization of the mice with the FOXM1 peptides caused autoimmunity, a histological analysis was done on various organs isolated from HLA‐A2 Tgm immunized with FOXM1‐8 or FOXM1‐10 peptides. We observe no pathological changes, such as lymphocyte infiltration or tissue destruction and repair in the brain, heart, lung, liver, small intestine, kidney, or testis of the immunized Tgm (data not shown). These results suggest that the CTL response elicited by immunization with FOXM1‐8 or FOXM1‐10 peptide may not cause damage to normal tissues.
Induction of FOXM1‐reactive human CTLs from HLA‐A2‐positive donors
To know whether or not the identified 3 FOXM1‐derived peptides actually bind to HLA‐A2, we analyzed the binding of the peptides to HLA‐A2 by using TAP‐deficient and HLA‐A2‐positive T2 cells. As shown in Figure 6, stabilization of cell surface HLA‐A2 by addition of FOXM1 peptides was observed, thus confirming the actual binding of the peptides to HLA‐A2.
Figure 6.
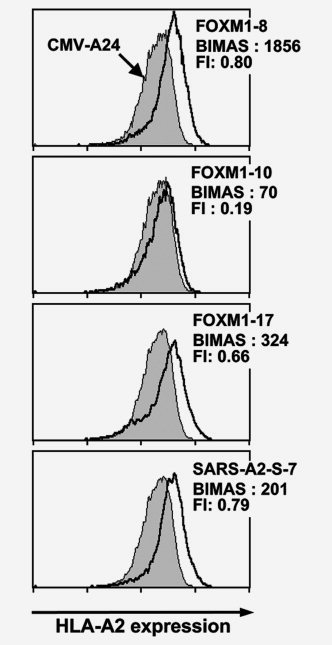
Binding of the FOXM1‐derived peptides to HLA‐A2. Three FOXM‐1 derived peptides were examined on the actual binding to HLA‐A2 by using a TAP‐deficient, HLA‐A*0201‐positive cell line T2. T2 cells, pre‐incubated overnight at 26°C, were cultured in the presence of 50 μM peptide and 5 μg/ml β2‐microglobulin in serum‐free medium at 26°C for 1.5 hr and subsequently at 37°C for 18 hr. After the culture, HLA‐A2 expression on the cell surface was measured by flow cytometry with anti‐HLA‐A2 monoclonal antibody BB7.2. A human cytomegalovirus‐derived peptide (QYDPVAALF) and a SARS‐A2‐S‐7 peptide (NLNESLIDL) were used as a negative and a positive control, respectively. The fluorescence index (FI) was calculated from the mean fluorescence intensity (MFI) of HLA‐A2 expressed on T2 cells determined by flow cytometry, using the formula FI = (MFI [T2 cells with FOXM1 peptide]/MFI [T2 cells with CMV peptide]) − 1. BIMAS score predicting HLA‐A2‐binding affinity of the peptides are also indicated.
Thereafter, we investigated human CTL response to FOXM1. We tried to establish FOXM1‐reactive CTL lines from PBMCs of HLA‐A2‐positive healthy individuals by stimulation with the FOXM1‐8, FOXM1‐10 or FOXM1‐17 peptides. CD8+ T cells sorted from the PBMCs were co‐cultured with autologous monocyte‐derived DCs pulsed with the peptides as described in the material and methods. CTL lines generated from HLA‐A2 positive healthy donors produced IFN‐γ specifically in response to re‐stimulation with T2 cells pulsed with the peptides (Fig. 7).
Figure 7.

CTL induction from PBMCs of HLA‐A2‐positive healthy donors. FOXM1 peptide‐reactive CTLs were generated from CD8+ T cells of HLA‐A2+ healthy donors. After 3 or 4 times of stimuli with autologous monocyte‐derived DCs pulsed with the FOXM1‐8, FOXM1‐10, or FOXM1‐17 peptide, the CTLs were subjected to IFN‐γ ELISPOT assay, using T2 cells pulsed with the same FOXM1 peptide as stimulators. The number of specific spots was determined by subtracting the mean number of spots in the absence of an antigen from that in the presence of the antigen: (ELISPOT number stimulated with T2 cells + peptide) − (ELISPOT number stimulated with T2 cells alone).
Further investigation examined whether the identified peptides were produced from FOXM1 protein by intracellular natural processing. FOXM1‐specific killing activity of CTLs induced with these peptides from HLA‐A2‐positive cancer patients were analyzed by 51Cr release assay. RT‐PCR analyses revealed that all of the HLA‐A2 positive cell lines that we examined expressed FOXM1 (Fig. 2, Supporting Information Fig. 1, and data not shown), except for Caco‐2 cells. Thus, we introduced Caco‐2 cells with an expression vector for FOXM1 by using lentivirus and used as target cells (Caco‐2‐FOXM1). Caco‐2 cells infected with lentiviruses without FOXM1‐gene were also used as negative control target cells (Caco‐2‐mock). The expression of transgene‐derived protein in the transfectants was confirmed as shown in Figure 8 a. The peptide‐induced CTL lines exhibited significantly higher cytotoxicity against Caco‐2‐FOXM1 cells than against Caco‐2‐mock cells. These results clearly indicate that these peptides were naturally processed from FOXM1 protein and presented in the context of HLA‐A2 to be recognized by peptide‐induced CTLs.
Figure 8.

Natural processing of FOXM1 peptides. (a) Total cell lysate from Caco‐2 cells infected with Lentiviruses without FOXM1‐gene (Caco‐2‐mock) or those introduced with HA‐tagged human FOXM1‐gene (Caco‐2‐FOXM1) was analyzed by Western blot analysis for transgene‐derived FOXM1expression, using anti‐HA antibody. As a loading control, the same membrane was blotted with an anti‐β‐actin antibody. (b) Cytotoxic activity of the CTL lines established from HLA‐A*0201‐positive healthy donors and patients with cancer by stimulation with the 3 FOXM1‐derived peptides against Caco‐2‐mock and Caco‐2‐FOXM1 (FOXM1+, HLA‐A*0201 +) was examined by 51Cr release assay. The percentage of specific lysis was calculated from the mean values of duplicate or triplicate assays.
Discussion
The identification of adequate target antigens which induce a potent and specific anti‐tumor immune response is important for the development of cancer immunotherapy. Although many TAAs in various cancers have been identified,32, 33 only a few antigens specifically expressed in cholangiocarcinoma have been reported34, 35 A previous cDNA microarray analysis identified multiple gene transcripts that are up‐ or down‐regulated in cholangiocarcinoma.15 The current study searched for TAA by using the cDNA microarray data and identified FOXM1 as a candidate for target antigen.
FOXM1 is over‐expressed in a number of aggressive human carcinomas.12, 15, 16, 17, 18, 19, 20, 21 An enhanced expression of FOXM1 was also observed in various cancer tissues in cDNA microarray analysis (Supporting Information Table 1). The expression of FOXM1 is significantly associated with poor overall survival of non‐small cell lung cancer and glioblastoma patients.36, 37 Therefore, FOXM1 may have the potential to serve as a good target of cancer immunotherapy for various types of cancers. To evaluate FOXM1 as a target for cancer immunotherapy, the present study examined the immunogenicity of FOXM1 in human immune system.
The FOXM1‐derived peptides predicted to bind to HLA‐A2 were analyzed for their capacity to induce CTLs. Stimulation of PBMCs obtained from healthy donors with FOXM1‐8 (YLVPIQFPV), FOXM1‐10 (SLVLQPSVKV) and FOXM1‐17 (GLMDLSTTPL) peptides resulted in the generation of peptide‐reactive CTL lines. Furthermore, CTLs reactive to the peptides generated from cancer patients exhibited specific cytotoxicity against HLA‐A*0201‐positive Caco‐2 cells introduced with an expression vector for FOXM1, but not against mock‐introduced Caco‐2 cells, thus verifying the natural processing of these peptides from FOXM1 protein in cancer cells. These results suggest that FOXM1 is a promising target for various types of cancers, and that the 3 peptides derived from FOXM1 are effective tools for the clinical application of peptide‐based immunotherapy for HLA‐A*0201‐positive cancer patients.
The human FOXM1 gene has a 10‐exon structure. Two exons, Va and VIIa, are alternatively spliced.38 Differential splicing of exons Va and VIIa in human FOXM1 yields 3 isoforms, FOXM1a, FOXM1b and FOXM1c. FOXM1a contains both alternative exons, FOXM1b contains none of the alternative exons and FOXM1c only retains exon Va.23 FOXM1b and FOXM1c exhibit transactivating activity because of the absence of VIIa, which is the inhibitory sequence. The presence of VIIa in FOXM1a renders it transcriptionally inactive.23 In the present study, FOXM1 was found to be expressed in various cancer cell samples, with a differential pattern of expression in normal testis and thymus. The FOXM1c isoform was more abundant in cultured cells, including those from the normal gastrointestinal tract and these results were consistent with the findings of other studies.39, 40
FOXM1, a transcription factor, plays an important role in regulating the expression of genes involved in cell growth, proliferation, differentiation, longevity and transformation. Expression profiling has identified a FOXM1‐regulated cluster of genes, including many G2/M‐specific genes (Cyclin B, CENP‐F, Nek‐2, etc).19, 41 The loss of FOXM1 expression in cancer cell lines results in mitotic spindle defects, delays cells in mitosis, and induces mitotic catastrophe.19, 41 These findings suggest that FOXM1 expression is essential for cancer cell growth and survival. Indeed, evident expression of FOXM1 mRNA was detected in most of the cell lines examined by RT‐PCR in the present study. If a molecule expressed by cancer cells plays an essential role in their survival, then cancer cells cannot escape from the immune cell attack directed to this molecule by losing expression of this molecule.42 In this regard, FOXM1 is thus considered to be a suitable target for anti‐cancer immunotherapy.
HLA Tgm is not only useful for the identification of HLA‐restricted antigenic epitopes but also for the evaluation of autoimmunity caused by immunization of peptides that are conserved between human and mouse FOXM1. The amino acid sequences of the FOXM1‐8 and FOXM1‐10 peptides are conserved between human and mouse. In the present study, vaccination with these peptides did not induce autoimmunity in HLA‐A2 Tgm. These results suggest that the use of these peptides for the HLA‐A*0201‐positive humans would be safe, although low levels of expression of FOXM1 was detected in some normal organs (Figs. 1 and 3).
A number of clinical trials have evaluated peptides derived from TAAs. However, tumor regression has not so far been observed as expected.43 The possible reasons for the lack of clinical efficacy could be an entry of advanced cancer patients in immunotherapy trials, and loss or down‐regulation of HLA class I molecules, co‐stimulatory molecules, and proteins associated with antigen presentation (such as TAP, low‐molecular‐weight protein (LMP) and β2‐microglobulin) in the tumor cells.44 Consequently, a significant portion of tumor cells could escape from CTLs specific to the TAA‐derived peptides presented by HLA class I molecules, even if CTLs could be successfully induced by TAA peptide vaccines capable of targeting the tumor cells themselves. The present study investigated the expression of tumor escape‐associated genes in 25 specimens of intrahepatic cholangiocarcinoma tissues using cDNA microarray. In this analysis, no down‐regulation of these genes associated with immune escape was observed (data not shown). These results suggested that the CTL‐mediated immunotherapy against the cholangiocarcinoma could be effective.
Recently, two inhibitors for FOXM1 transcriptional activity were identified and targeting FOXM1 with these inhibitors was an effective anticancer therapy. One is a peptide containing an amino acid sequence of p19ARF24‐49, which interacts with the FOXM1 C‐terminal transactivation domain; inhibition of FOXM1 transcriptional activity by the peptide caused selective apoptosis of hepatocellular carcinoma over‐expressing FOXM1 and a reduced proliferation and angiogenesis in hepatocellular carcinoma regions but not in the adjacent normal liver tissue in mice.45 The other is an antibiotic thiazole compound Siomycin A; this antibiotic inhibits anchorage‐independent growth and selectively induces apoptosis in FOXM1‐transformed cells.46 They could be potential anticancer drugs, although these results were based on the experiments with cell lines or transgenic mice.
Clinical trials of the epitope‐peptide vaccines derived from the TAAs identified by genome‐wide cDNA microarray analyses in different cancers are currently underway. The HLA‐A2‐restricted epitopes of FOXM1 identified in the present study may well be clinically tested in the near future.
Supporting information
Additional Supporting Information may be found in the online version of this article.
Supporting Table 1. Expression of FOXM1 in various cancers determined by genome‐wide cDNA microarray analysis.
Supplemental Fig. 1. Expression of FOXM1 mRNA in lung cancer cell lines, lung cancer tissues, and normal tissues. Lung cancer cell lines, lung cancer tissues, and normal airway epithelial cells were analyzed for the expression of FOXM1 by RT‐PCR. The same cDNA samples were tested for ACTB expression as a loading control: ADC, adenocarcinoma; BAC, bronchiolo‐alveolar carcinoma; SCC, squamous cell carcinoma; ASC, adenosquamous carcinoma; LCC, large cell carcinoma; SCLC, small cell carcinoma.
Acknowledgements
The authors thank the Cell Resource Center for Biomedical Research Institute of Development, Aging and Cancer, Tohoku University and RIKEN BioResource Center for providing the cell lines; Dr. F.A. Lemonnier (Department SIDA‐Retrovirus, Unite d' Immunite Cellulaire Antivirale, Institut Pasteur, France) for providing HLA‐A2 transgenic mice; Dr. Hiroyuki Miyoshi (RIKEN BioResource Center) for providing lentiviral vectors; Ms. Tatsuko Kubo (Department of Molecular Pathology, Kumamoto University) for technical assistance with immunohistochemical analyses.
References
- 1. Khan SA, Thomas HC, Davidson BR, Taylor‐Robinson SD. Cholangiocarcinoma. Lancet 2005; 366: 1303–14. [DOI] [PubMed] [Google Scholar]
- 2. Sirica AE. Cholangiocarcinoma: molecular targeting strategies for chemoprevention and therapy. Hepatology 2005; 41: 5–15. [DOI] [PubMed] [Google Scholar]
- 3. Stevanovic S. Identification of tumour‐associated T‐cell epitopes for vaccine development. Nat Rev Cancer 2002; 2: 514–20. [DOI] [PubMed] [Google Scholar]
- 4. Lin YM, Furukawa Y, Tsunoda T, Yue CT, Yang KC, Nakamura Y. Molecular diagnosis of colorectal tumors by expression profiles of 50 genes expressed differentially in adenomas and carcinomas. Oncogene 2002; 21: 4120–8. [DOI] [PubMed] [Google Scholar]
- 5. Hasegawa S, Furukawa Y, Li M, Satoh S, Kato T, Watanabe T, Katagiri T, Tsunoda T, Yamaoka Y, Nakamura Y. Genome‐wide analysis of gene expression in intestinal‐type gastric cancers using a complementary DNA microarray representing 23,040 genes. Cancer Res 2002; 62: 7012–7. [PubMed] [Google Scholar]
- 6. Mathiassen S, Lauemoller SL, Ruhwald M, Claesson MH, Buus S. Tumor‐associated antigens identified by mRNA expression profiling induce protective anti‐tumor immunity. Eur J Immunol 2001; 31: 1239–46. [DOI] [PubMed] [Google Scholar]
- 7. Schmidt SM, Schag K, Muller MR, Weinschenk T, Appel S, Schoor O, Weck MM, Grunebach F, Kanz L, Stevanovic S, Rammensee HG, Brossart P. Induction of adipophilin‐specific cytotoxic T lymphocytes using a novel HLA‐A2‐binding peptide that mediates tumor cell lysis. Cancer Res 2004; 64: 1164–70. [DOI] [PubMed] [Google Scholar]
- 8. Uchida N, Tsunoda T, Wada S, Furukawa Y, Nakamura Y, Tahara H. Ring finger protein 43 as a new target for cancer immunotherapy. Clin Cancer Res 2004; 10: 8577–86. [DOI] [PubMed] [Google Scholar]
- 9. Nakatsura T, Yoshitake Y, Senju S, Monji M, Komori H, Motomura Y, Hosaka S, Beppu T, Ishiko T, Kamohara H, Ashihara H, Katagiri T, et al. Glypican‐3, overexpressed specifically in human hepatocellular carcinoma, is a novel tumor marker. Biochem Biophys Res Commun 2003; 306: 16–25. [DOI] [PubMed] [Google Scholar]
- 10. Nakatsura T, Kageshita T, Ito S, Wakamatsu K, Monji M, Ikuta Y, Senju S, Ono T, Nishimura Y. Identification of glypican‐3 as a novel tumor marker for melanoma. Clin Cancer Res 2004; 10: 6612–21. [DOI] [PubMed] [Google Scholar]
- 11. Komori H, Nakatsura T, Senju S, Yoshitake Y, Motomura Y, Ikuta Y, Fukuma D, Yokomine K, Harao M, Beppu T, Matsui M, Torigoe T, et al. Identification of HLA‐A2‐ or HLA‐A24‐restricted CTL epitopes possibly useful for glypican‐3‐specific immunotherapy of hepatocellular carcinoma. Clin Cancer Res 2006; 12: 2689–97. [DOI] [PubMed] [Google Scholar]
- 12. Yoshitake Y, Nakatsura T, Monji M, Senju S, Matsuyoshi H, Tsukamoto H, Hosaka S, Komori H, Fukuma D, Ikuta Y, Katagiri T, Furukawa Y, et al. Proliferation potential‐related protein, an ideal esophageal cancer antigen for immunotherapy, identified using complementary DNA microarray analysis. Clin Cancer Res 2004; 10: 6437–48. [DOI] [PubMed] [Google Scholar]
- 13. Imai K, Hirata S, Irie A, Senju S, Ikuta Y, Yokomine K, Harao M, Inoue M, Tsunoda T, Nakatsuru S, Nakagawa H, Nakamura Y, et al. Identification of a novel tumor‐associated antigen, cadherin 3/P‐cadherin, as a possible target for immunotherapy of pancreatic, gastric, and colorectal cancers. Clin Cancer Res 2008; 14: 6487–95. [DOI] [PubMed] [Google Scholar]
- 14. Harao M, Hirata S, Irie A, Senju S, Nakatsura T, Komori H, Ikuta Y, Yokomine K, Imai K, Inoue M, Harada K, Mori T, et al. HLA‐A2‐restricted CTL epitopes of a novel lung cancer‐associated cancer testis antigen, cell division cycle associated 1, can induce tumor‐reactive CTL. Int J Cancer 2008; 123: 2616–25. [DOI] [PubMed] [Google Scholar]
- 15. Obama K, Ura K, Li M, Katagiri T, Tsunoda T, Nomura A, Satoh S, Nakamura Y, Furukawa Y. Genome‐wide analysis of gene expression in human intrahepatic cholangiocarcinoma. Hepatology 2005; 41: 1339–48. [DOI] [PubMed] [Google Scholar]
- 16. Okabe H, Satoh S, Kato T, Kitahara O, Yanagawa R, Yamaoka Y, Tsunoda T, Furukawa Y, Nakamura Y. Genome‐wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res 2001; 61: 2129–37. [PubMed] [Google Scholar]
- 17. van den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS, Sommer C, Reifenberger G, Hanash SM. Characterization of gene expression profiles associated with glioma progression using oligonucleotide‐based microarray analysis and real‐time reverse transcription‐polymerase chain reaction. Am J Pathol 2003; 163: 1033–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nakamura T, Furukawa Y, Nakagawa H, Tsunoda T, Ohigashi H, Murata K, Ishikawa O, Ohgaki K, Kashimura N, Miyamoto M, Hirano S, Kondo S, et al. Genome‐wide cDNA microarray analysis of gene expression profiles in pancreatic cancers using populations of tumor cells and normal ductal epithelial cells selected for purity by laser microdissection. Oncogene 2004; 23: 2385–400. [DOI] [PubMed] [Google Scholar]
- 19. Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res 2005; 65: 5181–9. [DOI] [PubMed] [Google Scholar]
- 20. Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, Lyubimov A, Costa RH. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res 2006; 66: 1712–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, Major ML, Gusarova GA, Yoder HM, Costa RH, Kalinichenko VV. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res 2006; 66: 2153–61. [DOI] [PubMed] [Google Scholar]
- 22. Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol 2001; 2: 533–43. [DOI] [PubMed] [Google Scholar]
- 23. Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG, Roebuck KA, Costa RH. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol 1997; 17: 1626–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Firat H, Garcia‐Pons F, Tourdot S, Pascolo S, Scardino A, Garcia Z, Michel ML, Jack RW, Jung G, Kosmatopoulos K, Mateo L, Suhrbier A, et al. H‐2 class I knockout, HLA‐A2.1‐transgenic mice: a versatile animal model for preclinical evaluation of antitumor immunotherapeutic strategies. Eur J Immunol 1999; 29: 3112–21. [DOI] [PubMed] [Google Scholar]
- 25. Nakatsura T, Senju S, Yamada K, Jotsuka T, Ogawa M, Nishimura Y. Gene cloning of immunogenic antigens overexpressed in pancreatic cancer. Biochem Biophys Res Commun 2001; 281: 936–44. [DOI] [PubMed] [Google Scholar]
- 26. Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self‐inactivating lentivirus vector. J Virol 1998; 72: 8150–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Irie A, Harada K, Tsukamoto H, Kim JR, Araki N, Nishimura Y. Protein kinase D2 contributes to either IL‐2 promoter regulation or induction of cell death upon TCR stimulation depending on its activity in Jurkat cells. Int Immunol 2006; 18: 1737–47. [DOI] [PubMed] [Google Scholar]
- 28. Yokomine K, Nakatsura T, Minohara M, Kira J, Kubo T, Sasaki Y, Nishimura Y. Immunization with heat shock protein 105‐pulsed dendritic cells leads to tumor rejection in mice. Biochem Biophys Res Commun 2006; 343: 269–78. [DOI] [PubMed] [Google Scholar]
- 29. Nakahara S, Tsunoda T, Baba T, Asabe S, Tahara H. Dendritic cells stimulated with a bacterial product, OK‐432, efficiently induce cytotoxic T lymphocytes specific to tumor rejection peptide. Cancer Res 2003; 63: 4112–8. [PubMed] [Google Scholar]
- 30. Watanabe T, Suda T, Tsunoda T, Uchida N, Ura K, Kato T, Hasegawa S, Satoh S, Ohgi S, Tahara H, Furukawa Y, Nakamura Y. Identification of immunoglobulin superfamily 11 (IGSF11) as a novel target for cancer immunotherapy of gastrointestinal and hepatocellular carcinomas. Cancer Sci 2005; 96: 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Monji M, Nakatsura T, Senju S, Yoshitake Y, Sawatsubashi M, Shinohara M, Kageshita T, Ono T, Inokuchi A, Nishimura Y. Identification of a novel human cancer/testis antigen, KM‐HN‐1, recognized by cellular and humoral immune responses. Clin Cancer Res 2004; 10: 6047–57. [DOI] [PubMed] [Google Scholar]
- 32. Renkvist N, Castelli C, Robbins PF, Parmiani G. A listing of human tumor antigens recognized by T cells. Cancer Immunol Immunother 2001; 50: 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev 2002; 188: 22–32. [DOI] [PubMed] [Google Scholar]
- 34. Swierczynski SL, Maitra A, Abraham SC, Iacobuzio‐Donahue CA, Ashfaq R, Cameron JL, Schulick RD, Yeo CJ, Rahman A, Hinkle DA, Hruban RH, Argani P. Analysis of novel tumor markers in pancreatic and biliary carcinomas using tissue microarrays. Hum Pathol 2004; 35: 357–66. [DOI] [PubMed] [Google Scholar]
- 35. Watanabe H, Enjoji M, Nakashima M, Noguchi K, Kinukawa N, Sugimoto R, Kotoh K, Nakamuta M, Nawata H, Watanabe T. Clinical significance of serum RCAS1 levels detected by monoclonal antibody 22‐1‐1 in patients with cholangiocellular carcinoma. J Hepatol 2003; 39: 559–63. [DOI] [PubMed] [Google Scholar]
- 36. Takahashi K, Furukawa C, Takano A, Ishikawa N, Kato T, Hayama S, Suzuki C, Yasui W, Inai K, Sone S, Ito T, Nishimura H, et al. The neuromedin U‐growth hormone secretagogue receptor 1b/neurotensin receptor 1 oncogenic signaling pathway as a therapeutic target for lung cancer. Cancer Res 2006; 66: 9408–19. [DOI] [PubMed] [Google Scholar]
- 37. Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, Aldape KD, Xie TX, Pelloski CE, Xie K, Sawaya R, Huang S. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res 2006; 66: 3593–602. [DOI] [PubMed] [Google Scholar]
- 38. Korver W, Roose J, Clevers H. The winged‐helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res 1997; 25: 1715–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res 2002; 62: 4773–80. [PubMed] [Google Scholar]
- 40. Ma RY, Tong TH, Cheung AM, Tsang AC, Leung WY, Yao KM. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. J Cell Sci 2005; 118: 795–806. [DOI] [PubMed] [Google Scholar]
- 41. Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol 2005; 7: 126–36. [DOI] [PubMed] [Google Scholar]
- 42. Anderson MH, Becker JC, Thor Straten P. Regulator of apoptosis: suitable targets for immune therapy of cancer. Nat Rev Drug Discov 2005; 4: 399–409. [DOI] [PubMed] [Google Scholar]
- 43. Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med 2004; 10: 909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gabrilovich D. Mechanisms and functional significance of tumour‐induced dendritic‐cell defects. Nat Rev Immunol 2004; 4: 941–52. [DOI] [PubMed] [Google Scholar]
- 45. Gusarova GA, Wang IC, Major ML, Kalinichenko VV, Ackerson T, Petrovic V, Costa RH. A cell‐penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest 2007; 117: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Radhakrishnan SK, Bhat UG, Hughes DE, Wang IC, Costa RH, Gartel AL. Identification of a chemical inhibitor of the oncogenic transcription factor forkhead box M1. Cancer Res 2006; 66: 9731–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article.
Supporting Table 1. Expression of FOXM1 in various cancers determined by genome‐wide cDNA microarray analysis.
Supplemental Fig. 1. Expression of FOXM1 mRNA in lung cancer cell lines, lung cancer tissues, and normal tissues. Lung cancer cell lines, lung cancer tissues, and normal airway epithelial cells were analyzed for the expression of FOXM1 by RT‐PCR. The same cDNA samples were tested for ACTB expression as a loading control: ADC, adenocarcinoma; BAC, bronchiolo‐alveolar carcinoma; SCC, squamous cell carcinoma; ASC, adenosquamous carcinoma; LCC, large cell carcinoma; SCLC, small cell carcinoma.


