Abstract
Purpose of Review
In this review, we discuss the current implications of the changing genomic epidemiology of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), etiological agent of the Coronavirus Disease 2019 (COVID-19) and its potential relationship with the change of clinical manifestations in patients with confirmed infection.
Recent Findings
Over the course of the current pandemic, the virus has been found more diverse in new countries. Simultaneously, also new clinical manifestations are observed, particularly more prominent gastrointestinal and neurological findings.
Summary
SARS-CoV-2/COVID-19 is changing not only its epidemiology, but also its genomic diversity and clinical manifestations, both aspects coupled, needs to be considered in the study of this ongoing pandemic.
Keywords: Genomic Epidemiology, Clinical Manifestations, SARS-CoV-2, COVID-19, Chile, Latin America
Epidemics are evolving scenarios by nature, and the Coronavirus Disease 2019 (COVID-19) pandemic is not an exception to the rule [1–3]. Since its beginning, in Wuhan, China, on December 2019, the worldwide spread of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has shown multiple expected and unexpected epidemiological and clinical outcomes [4–10]. Furthermore, the rapid expansion and large geographical extension of its transmission over a relatively short period are now affecting more than 180 countries, ramping up to more than 1,270,000 cases and over 69,000 reported deaths, [11–14] mainly in the highest populated regions of the world, including all largest cities in Latin America [15–17]. Then, we would like to present a summary of the pattern of clinical manifestations of COVID-19 in Chile, comparing them with what is seen in other regions, and provide a hypothesis on its relationship with the genetic diversity of isolates.
In the beginning and throughout the first 2 months of the COVID-19 pandemic, the principal reported clinical manifestations of the disease in most countries included fever, cough, dyspnea, myalgia and fatigue, sputum production, sore throat, diarrhea, and, less frequently, headache (8.0%) [4–10]. However, more recently, several countries, including Germany, the UK, and Italy [18], have reported an increasing number of neurological manifestations such as anosmia (the loss of the ability to detect one or more smells, temporary or permanent), hyposmia (decreased sensitivity to some or all smells), ageusia (loss of taste functions of the tongue), dysgeusia (or parageusia, is the distortion of the sense of taste), and hypogeusia (the decrease in taste sensitivity) in patients with confirmed SARS-CoV-2 infection [19].
Up to April 5, 2020, there were 26,259 COVID-19 confirmed cases reported in Latin America, 23,622 in South America, and 2637 in Central America [20]. In Chile, where the first confirmed case of COVID-19 was identified on March 3, 2020, in the city of Talca, 315 km from the capital Santiago, a different pattern of clinical findings among rRT-PCR confirmed cases is being described. After the reporting of the index case and throughout March 23, 2020, 10,768 cases have been investigated, and 922 (4.7%) were positive [21]. Of these, 597 (64.8%) presented with headache as a cardinal symptom, while only 8.5% and 49.0% presented fever and cough, respectively, among other symptoms (Table 1) [21]. The presence of headache in the majority of these patients may suggest the potential neurotropism and neurovirulence of SARS-CoV-2 like seen for other naturally neurotropic human coronaviruses such as SARS-CoV and HCoV-OC43 and -229E [22–25]. If so, recognition of headache within the clinical scenario of SARS-CoV-2 and the detailed description of its nuances and attributes within the timeline of infection is of utmost importance and an aspect deserving further investigations. Up to date, suspected case definition in Chile is still focused on fever, acute respiratory syndrome, and epidemiological nexus for testing purposes. Then, considering the findings in Europe and now in Chile, the case definition probably should be updated, considering neurological manifestations such as headache, anosmia, and dysgeusia [18].
Table 1.
Main clinical findings reported in the first 922 notified cases of COVID-19 in Chile, up to March 23, 2020
| Symptoms | n | % |
|---|---|---|
| Headache | 597 | 64.8 |
| Dyspnea | 498 | 54.0 |
| Cough | 452 | 49.0 |
| Thorax pain | 407 | 44.1 |
| Sore throat | 394 | 42.7 |
| Fever | 78 | 8.5 |
| Abdominal pain | 41 | 4.4 |
| Myalgia | 32 | 3.5 |
During the COVID-19 pandemic, significant efforts have been made globally in order to characterize the virus, by multiple genome-based techniques, including the next-generation sequencing revolution, in order to have a real approach to the genomic epidemiology of SARS-CoV-2 [26]. More than two thousand genomes of SARS-CoV-2 have been sequenced from different countries and continents, allowing unprecedented levels of resolution and insight in the evolution and epidemic diffusion of the virus. These genomes are available at the public database of the Global Initiative on Sharing All Influenza Data (GISAID) [27]. GISAID was originally developed for genomic data sharing on influenza [28] but now has a comprehensive, dynamic, and constantly updated SARS-CoV-2 database, in addition to other repositories or databases such as the GenBank.
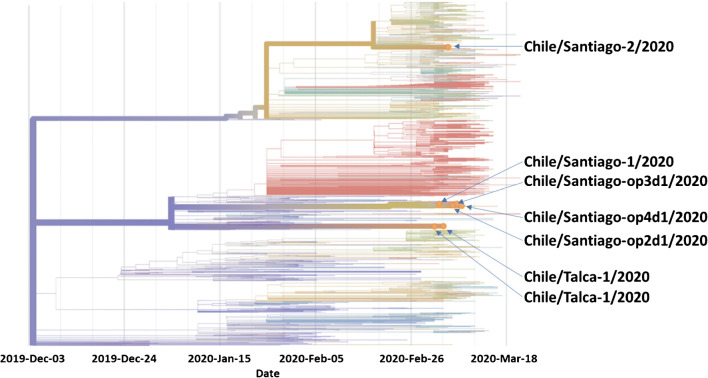
To date, Chile through its Institute of Public Health has sequenced and incorporated a total of seven SARS-CoV-2 genomes from the cities of Talca and Santiago (Fig. 1) [27, 29] with a general divergence ranging from 5.983 to 7.984 (Table 2). The Chile/Santiago-2/2020 isolate, obtained from a 40-year-old returning traveler from Italy [27, 29], revealed, in addition, a single nucleotide mutation (C3393T) with an AA mutation, at the ORF1a gene, A1043V [27, 29]. Overall, these genomes show multiple introduction events to Chile.
Fig. 1.
Phylogeny of SARS-CoV-2 reconstructed by the GISAD collaboration (https://nextstrain.org/ncov) from 1455 genomes including seven strains sequenced from Chile
Table 2.
Nucleotide and AA mutations, and divergence reported for these isolates
| Isolate | Nucleotide mutations | AA mutations | Divergence | Clade |
|---|---|---|---|---|
| Chile/Talca-1/2020 | 0 | 0 | 5.983 | B |
| Chile/Talca-2/2020 | 0 | 0 | 5.983 | B |
| Chile/Santiago-1/2020 | 0 | 0 | 6.981 | B |
| Chile/Santiago-2/2020 | C3393T | ORF1a: A1043V | 7.984 | A2a |
| Chile/Santiago-op2d1/2020 | 0 | 0 | 6.981 | B |
| Chile/Santiago-op3d1/2020 | 0 | 0 | 6.981 | B |
| Chile/Santiago-op4d1/2020 | 0 | 0 | 6.981 | B |
Then, the question is whether phylogenetic heterogeneity among different strains of SARS-CoV-2 could be associated with the clinical spectrum of manifestations of COVID-19. We know that previously, other epidemic emerging conditions, such as Zika, diverged into clinical differences and impact by its two lineages, African (ZIKVAF) and Asian (ZIKVAS) genotypes. In the type-I interferon receptor-deficient mouse model, ZIKVAF causes severe disease, whereas a much milder infection was seen after challenge with ZIKVAS [30]. Studies showed that there are strain-specific virulence differences between the African and Asian lineages in ZIKV mouse models [31]. Comparing the clinical findings between infected animals with different lineages of the viruses may help to understand the impact of genomic evolution and divergence and to surveillance by analyzing their sequences periodically. Currently, GISAID has analyzed in detail over 1300 genomes of SARS-CoV-2 and classified the sequences in at least three relevant clades, S (containing 439 genomes), G (404), V (108), and other clades (424) [27]. S clade contains most of America’s genomes. However, six of the Chilean isolates belong to the clade B and the Chile/Santiago-2/2020 to the clade A2a (Fig. 1), which maybe is related to the different clinical pattern observed in Chilean patients confirmed with SARS-CoV-2 infection. Currently, only SARS-CoV-2 sequences from isolates of Brazil, Peru, and Chile are available from South America, in addition to Mexico and Panama for the rest of Latin America.
In addition, to the clades classification, population genetic analyses of the SARS-CoV-2 genomes suggested that these viruses evolved into two major types (designated L and S), which are well defined by two different SNPs that show nearly complete linkage across the viral strains sequenced to date. Apparently, the L type was more prevalent in the early stages of the outbreak in Wuhan, China, and its frequency decreased after early January 2020. The S type, which is evolutionarily older and less aggressive, might have increased in relative frequency due to relatively weaker selective pressure [32]. In a recent study from Chile on the first four cases of SARS-CoV-2/COVID-19 in Chile, authors suggest that at least two different viral variants entered into the country, from Europe and Asia [33].
This hypothesis should encourage more studies on the genomic epidemiology of SARS-CoV-2 in Latin America. As this pandemic and other similar events have taught us in the past, a virus that spillover from animals to humans and reach adequate transmissibility can quickly spread with little pressure changes on its genome. Therefore, it is of paramount importance to also look into hosts’ genomes for clues to explain these genomic divergences and its translation into the differing symptomatic presentation [34]. Furthermore, it will allow countries, especially in Latin America, to adopt new measures for epidemiological surveillance, testing, and clinical protocols. For Latin American countries, preparedness and response should be consistent with their states’ capacities [15, 16].
Footnotes
This article is part of the Topical Collection on COVID-19 in the Tropics: Impact and Solutions
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Azamfirei R. The 2019 novel coronavirus: a crown jewel of pandemics? J Crit Care Med (Targu Mures) 2020;6(1):3–4. doi: 10.2478/jccm-2020-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan S, Siddique R, Ali A, Xue M, Nabi G. Novel coronavirus, poor quarantine, and the risk of pandemic. J Hosp Infect. 2020. 10.1016/j.jhin.2020.02.002. [DOI] [PMC free article] [PubMed]
- 3.Watts CH, Vallance P, Whitty CJM. Coronavirus: global solutions to prevent a pandemic. Nature. 2020;578(7795):363. doi: 10.1038/d41586-020-00457-y. [DOI] [PubMed] [Google Scholar]
- 4.Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020. 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed]
- 5.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020. 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed]
- 6.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China The Lancet. 2020;395(10223):497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020. 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed]
- 8.Phan LT, Nguyen TV, Luong QC, Nguyen TV, Nguyen HT, Le HQ, et al. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020. 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed]
- 9.Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020. 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed]
- 10.Rodriguez-Morales AJ, Cardona-Ospina JA, Gutiérrez-Ocampo E, Villamizar-Peña R, Holguin-Rivera Y, Escalera-Antezana JP, et al. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis. 2020;101623. 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed]
- 11.World Health Organization. Statement on the meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). https://www.who.int/news-room/detail/23-01-2020-statement-on-the-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov). 2020.
- 12.World Health Organization. Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). https://www.who.int/news-room/detail/30-01-2020-statement-on-the-second-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-outbreak-of-novel-coronavirus-(2019-ncov). 2020.
- 13.World Health Organization. Pneumonia of unknown cause – China. https://www.who.int/csr/don/05-january-2020-pneumonia-of-unkown-cause-china/en/. 2020.
- 14.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020. 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed]
- 15.Rodriguez-Morales AJ, Sánchez-Duque JA, Hernández-Botero S, Pérez-Díaz CE, Villamil-Gómez WE, Méndez CA, et al. Preparación y control de la enfermedad por coronavirus 2019 (COVID-19) en América Latina. Acta Medica Peruana. 2020;37(1):3–7. doi: 10.35663/amp.2020.371.909. [DOI] [Google Scholar]
- 16.Rodriguez-Morales AJ, Gallego V, Escalera-Antezana JP, Mendez CA, Zambrano LI, Franco-Paredes C, et al. COVID-19 in Latin America: the implications of the first confirmed case in Brazil. Travel Med Infect Dis. 2020;101613. 10.1016/j.tmaid.2020.101613. [DOI] [PMC free article] [PubMed]
- 17.Zambrano LI, Fuentes-Barahona IC, Bejarano-Torres DA, Bustillo C, Gonzales G, Vallecillo-Chinchilla G, et al. A pregnant woman with COVID-19 in Central America. Travel Med Infect Dis. 2020;101639. 10.1016/j.tmaid.2020.101639. [DOI] [PMC free article] [PubMed]
- 18.Giacomelli A, Pezzati L, Conti F, Bernacchia D, Siano M, Oreni L, et al. Self-reported olfactory and taste disorders in SARS-CoV-2 patients: a cross-sectional study. Clin Infect Dis. 2020. 10.1093/cid/ciaa330. [DOI] [PMC free article] [PubMed]
- 19.Hopkins C, Kumar N. Loss of sense of smell as marker of COVID-19 infection. https://www.entuk.org/sites/default/files/files/Loss%20of%20sense%20of%20smell%20as%20marker%20of%20COVID.pdf. 2020. [DOI] [PMC free article] [PubMed]
- 20.PAHO. Cumulative suspected and confirmed COVID-19 cases reported by countries and territories in the Americas, as of 29 March 2020. https://www.paho.org/en/documents/cumulative-suspected-and-confirmed-covid-19-cases-reported-countries-and-territories-3. 2020.
- 21.Ministerio de Salud de Chile. Informe de situación COVID-19. N° 16 - Chile al 23 de marzo de 2020. Departamento de Epidemiología. Ministerio de Salud de Chile. https://www.minsal.cl/wp-content/uploads/2020/03/Informe_10_COVID_19_Chile.pdf. 2020.
- 22.Yeh EA, Collins A, Cohen ME, Duffner PK, Faden H. Detection of coronavirus in the central nervous system of a child with acute disseminated encephalomyelitis. Pediatrics. 2004;113(1 Pt 1):e73–e76. doi: 10.1542/peds.113.1.e73. [DOI] [PubMed] [Google Scholar]
- 23.Morfopoulou S, Brown JR, Davies EG, Anderson G, Virasami A, Qasim W, Chong WK, Hubank M, Plagnol V, Desforges M, Jacques TS, Talbot PJ, Breuer J. Human coronavirus OC43 associated with fatal encephalitis. N Engl J Med. 2016;375(5):497–498. doi: 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- 24.Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, Zou W, Zhan J, Wang S, Xie Z, Zhuang H, Wu B, Zhong H, Shao H, Fang W, Gao D, Pei F, Li X, He Z, Xu D, Shi X, Anderson VM, Leong AS. Multiple organ infection and the pathogenesis of SARS. J Exp Med. 2005;202(3):415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arbour N, Day R, Newcombe J, Talbot PJ. Neuroinvasion by human respiratory coronaviruses. J Virol. 2000;74(19):8913–8921. doi: 10.1128/jvi.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaiarsa S, De Marco L, Comandatore F, Marone P, Bandi C, Sassera D. Bacterial genomic epidemiology, from local outbreak characterization to species-history reconstruction. Pathog Glob Health. 2015;109(7):319–327. doi: 10.1080/20477724.2015.1103503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.GISAID. Genomic epidemiology of novel coronavirus. https://nextstrain.org/ncov. 2020.
- 28.Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22(13). 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed]
- 29.Castillo AE, Parra B, Tapia P, Acevedo A, Lagos J, Andrade W et al. hCoV-19/Chile/Talca-1/2020 - EPI_ISL_414577. https://platform.gisaid.org/epi3/frontend#2df63c2020.
- 30.Dowall SD, Graham VA, Hewson R. Lineage-dependent differences of Zika virus infection in a susceptible mouse model are associated with different profiles of cytokines, chemokines, growth factors and acute phase proteins. Cytokine. 2020;125:154864. doi: 10.1016/j.cyto.2019.154864. [DOI] [PubMed] [Google Scholar]
- 31.Dowall SD, Graham VA, Rayner E, Hunter L, Atkinson B, Pearson G, Dennis M, Hewson R. Lineage-dependent differences in the disease progression of Zika virus infection in type-I interferon receptor knockout (A129) mice. PLoS Negl Trop Dis. 2017;11(7):e0005704. doi: 10.1371/journal.pntd.0005704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang X, Wu C, Li X, Song Y, Yao X, Wu X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 2020. 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed]
- 33.Castillo AE, Parra B, Tapia P, Acevedo A, Lagos J, Andrade W, et al. Phylogenetic analysis of the first four SARS-CoV-2 cases in Chile. J Med Virol. 2020. 10.1002/jmv.25797. [DOI] [PMC free article] [PubMed]
- 34.Ollarves-Carrero MF, Rodriguez-Morales AG, Bonilla-Aldana DK, Rodriguez-Morales AJ. Anosmia in a Healthcare Worker with COVID-19 in Madrid, Spain. Trav Med Infect Dis. 2020. [DOI] [PMC free article] [PubMed]