Abstract
Importance
An ongoing outbreak of COVID-19 has exhibited significant threats around the world. We found a significant decrease of T lymphocyte subsets and an increase of inflammatory cytokines of hospitalized patients with COVID-19 in clinical practice.
Methods
We conducted a retrospective, single-center observational study of in-hospital adult patients with confirmed COVID-19 in Hubei Provincial Hospital of traditional Chinese and Western medicine (Wuhan, China) by Mar 1, 2020. Demographic, clinical, laboratory information, especially T lymphocyte subsets and inflammatory cytokines were reported. For patients who died or discharge from hospital, the associations of T lymphocyte subsets on admission were evaluated by univariate logistic regression with odds ratios (ORs) and 95% confidence intervals (CIs), warning values to predict in-hospital death were assessed by Receiver Operator Characteristic (ROC) curves.
Results
A total of 187 patients were enrolled in our study from Dec 26, 2019 to Mar 1, 2020, of whom 145 were survivors (discharge = 117) or non-survivors (in-hospital death ==28). All patients exhibited a significant drop of T lymphocyte subsets counts with remarkably increasing concentrations of SAA, CRP, IL-6, and IL-10 compared to normal values. The median concentrations of SAA and CRP in critically-ill patients were nearly 4- and 10-fold than those of mild-ill patients, respectively. As the severity of COVID-19 getting worse, the counts of T lymphocyte drop lower.28 patients died in hospital, the median lymphocyte, CD3+ T-cell, CD4+ T-cell, CD8+ T-cell and B-cell were significantly lower than other patients. Lower counts (/uL) of T lymphocyte subsets lymphocyte (<500), CD3+T-cell (<200), CD4+ T-cell (<100), CD8+ T-cell (<100) and B-cell (<50) were associated with higher risks of in-hospital death of CIVID-19. The warning values to predict in-hospital death of lymphocyte, CD3+ T-cell, CD4+ T-cell, CD8+ T-cell, and B-cell were 559, 235, 104, 85 and 82, respectively.
Conclusion
We find a significant decrease of T lymphocyte subset is positively correlated with in-hospital death and severity of illness. The decreased levels of T lymphocyte subsets reported in our study were similar with SARS but not common among other virus infection, which may be possible biomarkers for early diagnosis of COVID-19. Our findings may shed light on early warning of high risks of mortality and help early intervention and treatment of COVID-19.
Keywords: COVID-19, T cell subsets, Retrospective study, Immunity
Introduction
An ongoing outbreak of novel coronavirus disease COVID-19 has exhibited significant threats to international health and economy. COVID-19 caused by the virus SARS-Cov-2.1 The most likely ecological reservoirs for SARS-Cov-2 are bats,2 but it is believed that the virus transmitted from animal to human by some intermediated animal host. In China, the first case of COVID-19 was reported in Wuhan, Hubei province at Dec,2019.3 The first series of patients in Wuhan with conformed COVID-19 worked or lived around the local Huanan seafood wholesale market,4 selling wild animals as well. Shortly after that, infected patients exploded without contact history of the market.5 The number of COVID-19 globally were 76,769 till Feb 21, 2020, including 75,569 patients in China.6 Unfortunately, no target treatment for this virus has been proved until now. We cannot totally know of this virus and its related diseases COVID-19. Therefore, this severe infectious disease garnered great research interest. Previous clinical studies revealed the epidemiological and clinical characteristics of infected patients of special population7 or series cases.8, 9, 10, 11 They focused on general clinical characteristics and laboratory results of patients with confirmed COVID-19. Meanwhile, several studies reported the damaged immunity system caused by SARS-Cov-2 and a significant decrease of lymphocyte in blood routine examination. But no research focused on the T lymphocyte subsets in COVID-19. In a general way, immune system is the first to be affected with virus infection. It has been confirmed that coronavirus infectious diseases such as severe acute respiratory (SARS) and Middle East respiratory syndromes (MERS) play important roles in immunity system injury. The T-cell and B-cell immunity may predict the disease severe level of coronavirus infection as well as the clinical outcomes.12 T lymphocyte plays a vital role in cell-mediated and humoral immunity.13 T lymphocyte includes CD3+ T cells, CD4+ T cells (helper T cells) and CD8+ T cells (cytotoxic T cells, or killer T cells), natural killer (NK) cells and B cells, which can only recognize protein-based receptor-bound antigens. From Dec 26, 2019 to now, we included over hundreds of patients with conformed COVID-19. We are one of the hospitals which primary diagnosed and treated COVID-19 patients and reported this new infectious disease to Chinese Center for Disease Control and Prevention (CDC) at first. In clinical practice, we find that most patients showed significant changes of T lymphocyte subset and several inflammatory cytokines. Therefore, we are interested in the relationship between T lymphocyte subset and in-hospital death and severity of illness among hospitalized patients with COVID-19. In this study, we focus on the suppressed T lymphocyte subsets and increased inflammatory cytokines. Furthermore, we measured the association of T lymphocyte subsets and in-hospital death as well as the severity of COVID-19.
Methods
Study design and participants
For this retrospective, single-center observational study, we included patients from Dec 26, 2019 to Mar 1,2020 at Hubei Provincial Hospital of traditional Chinese and Western medicine, Wuhan, China. Our hospital located closely to the Huanan seafood wholesale market. Our hospital is the first institution reported the new infectious disease to Chinese Center for Disease Control and Prevention (CDC). A continuous series of patients who were diagnosed as confirmed COVID-19.
This study was reviewed and approved by the medical Ethical Committee of Hubei Provincial Hospital of traditional Chinese and Western medicine (NO.2020013). Written informed consent was obtained from every enrolled patient.
Data collection
We reviewed clinical records, laboratory findings, T lymphocyte subsets, chest CT scans, severity of illness, in-hospital death for continuing 187 patients with confirmed COVID-19. All information was obtained and curated with a customized data collection form. Two clinical investigators (XU and FAN) independently reviewed the data collection forms to verify data accuracy. We extract information of 187 patients of confirmed COVID-19, including demographic data, medical history, severity of illness, clinical symptoms, signs laboratory parameters on different time points, supporting therapy (anti-virus, antibiotics, immune stimulants and respiratory support), survival states of patients. The admission time of included patients was between Dec. 26, 2019 and Mar 1, 2020.
Laboratory procedures
Maternal throat swab samples were collected and tested for SARS-CoV-2 of our hospital laboratory department and CDC in Wuhan, Hubei, following WHO guidelines for qRT-PCR. Positive confirmatory cases of COVID-19 infection were defined as those with a positive test result.
Routine blood examinations were complete blood count, coagulation profile, serum biochemical tests (including renal and liver function, creatine kinase and electrolytes), T lymphocyte subsets CD3+ T-cell, CD4+ T-cell (CD3+CD4+), CD8+ T-cell (CD3+CD8+), B-cell (CD3−CD19+) and NK-cell (CD3−(CD16+CD56)+), myocardial enzymes, interleukin-6 (IL-6), IL-1β, IL-10, serum amyloid A (SAA) and c-reactive protein (CRP). T lymphocyte subsets were measured by the flow cytometry. Chest computed tomographic (CT) scans were done for all inpatients. Frequency of examinations were determined by the physician in charge.
Evaluation criteria of severity of illness
The severity levels of COVID-2019 are measured as follows:
-
1.
mild illness: without symptoms and signs of severe and critically infection
-
2.severe illness (according to one of these criteria):
-
a.breathing difficulties, respiratory rate≥30 bpm;
-
b.SpO2 ≤93% at rest;
-
c.PaO2/FiO2 ≤ 300 mmHg.
-
a.
-
3.critically illness: (according to one of these criteria):
-
a.respiratory failure, and need mechanical ventilation assistant;
-
b.shock;
-
c.multi-organs failure, need transport to intensive care unit (ICU).
-
a.
Statistical analysis
The continuous data was presented as median with interquartile rage (IQR), and categorical data was present as frequencies with percentages. The severity of illness was categorized with mild, severe and critically ill. Clinical data were classified and analyzed according to in-hospital death and severity of illness. We adopted Mann-Whitney U test, χ2 test or Fisher exact test to compare differences between groups. To measure the possible association of T lymphocyte subsets with in-hospital death, we reported odd ratios (ORs) and 95% confidence intervals (CIs) which were generated by univariate logistic regression. Considering the total numbers of in-hospital death (n = 28) in our study, we did not adopt multivariable analysis for reasons of potential overfitting. The possible warning values of T lymphocyte subsets counts were assessed by Receiver Operator Characteristic (ROC) curves. The area under the ROC curve (AUC), specificity and sensitivity were all reported.
All reported statistics of the data were two sided with a significant difference of 5%, and CIs were reported as two sided 95% CIs. Statistical analysis of the data performed in R v3.6.1 software (http://www.R-project.org).
Results
The clinical characteristics of patients
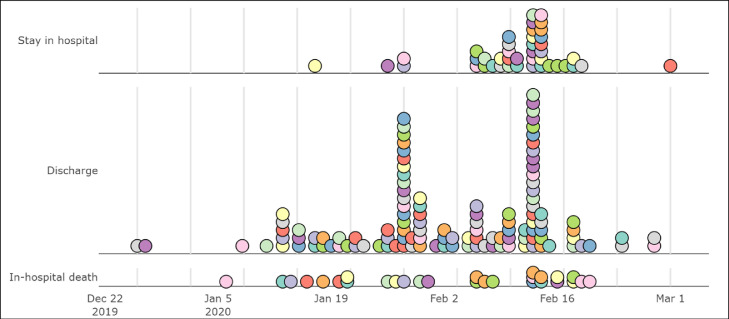
In this study, a total of 187 patients were enrolled, including 103 (55%) males and 84 (45%) females, median age 62-year old (IQR [48.5,71]). The admission time of patients was from Dec 26,2019 to Mar 1,2020 (Fig. 1 ). By the end of Mar 5, 2020, 117 patients were discharged from hospital, 28 patients died in hospital, and 45 patients still accepted treatment in hospital. Only 9 patients were clustered and had exposure history of Huanan seafood market and 48 patients had a contact history with confirmed COVID-19.
Fig. 1.
The admission time of patients with COVID-19 grouped by hospital states.
According to the categorical criteria mentioned in methods, 80 patients (42.8%) were mild ill including 30 males (37.5%), 45 (24.1%) patients were severe ill (31 males, 68.9%). 62 (33.1%) patients (42 males, 67.7%) were critically ill, among which 28 patients died in hospital. More Patients who died in hospital comorbid with hypertension (n = 24, proportion, 38.7%) and CCD (n = 13, proportion, 21%) than alive patients. Both in severe-ill and critically-ill including in-hospital death group, we found older age (median age≥60) than other groups (P<0.001). In severe-ill and critically-ill patients, male gender (higher than 60%) were more than mild-ill patients (P<0.001).
On admission, the common symptoms of patients were weak (159 patients, 85%), fever (153 patients, 81.8%), poor appetite (151 patients, 80.7%), cough (145patients, 77.5%) and shortness of breath (137 patients, 73.3%). Other common symptoms included diarrhea (43 patients, 23%), and muscle pain (34 patients, 18.2%). The comorbidity of patients included hypertension (50 patients, 26.7%), cardio-cerebrovascular disease (CCD) (19 patients, 10.2%), metabolic diseases (21 patients, 11.2%) and pulmonary diseases (3 patients, 1.6%). CCD included coronary heart disease, myocardial/cerebral infarction, and stroke. Metabolic diseases included type 2 diabetes, gout, and hyperlipemia.
All patients underwent chest CT on admission. For mild ill of patients, 9 (11.2%) patients showed one lateral lesion, 71 (88.8%) patients showed bilateral lesion, 70 (87.5%) patients showed multi-segment lesion; For severe patients, 45 (100%) patients showed multi-segment lesion; For critically ill patients, 62 (100%) patients showed bilateral or multi-segment lesion.
Treatment and laboratory parameters on admission
All patients were treated in separation. All patients received antivirus treatment, including Arbidol, Ribavirin, Oseltamivir, Kaletra and Interferon. 154 (82.4%) patients were given antibiotic treatment, including 51 (64.6%) mild-ill patients, 41 (91.1%) severe-ill patients and 62 (100%) critically ill patients. 2 (3.2%) critically ill patients received anti-fungi treatment. 94 (53.6%) patients received immune globin treatment. 113 (60.4%) patients received hormone treatment. The median length of hormone treatment was 4 days. All patients received oxygen support, including nasal catheter and mask oxygen therapy (n = 133), high-flow nasal cannula oxygen therapy (n = 36), non-invasive (n = 36) and invasive (n = 13) mechanical ventilation.
On admission, all patients exhibited a significant decrease under lower limit of lymphocyte counts in blood routine examination (Lymp) (median=0.88, IQR [0.55,1.27], G/L). Conversely, inflammatory cytokines including SAA (median=213.24,IQR [62.85, 301.00], mg/L), CRP (median=32.69, IQR [7.83,82.44], mg/L), IL-6 (median=14.70,IQR [4.50,28.60], pg/mL), IL-10 (median=6.33, IQR [4.90,12.70], pg/mL) were significantly higher than normal values. (Table 1–1 ). Other parameters including Aspartate aminotransferase (AST), Alanine aminotransferase (ALT), blood urea nitrogen (BUN), Creatine kinase and urea acid were range to normal values. (Table1-1)
Table 1–1.
Baseline characteristics of included patients (overall and categorized by survival states).
| Overall (n = 187) | Discharge (n = 117) | Died (n = 28) | Stay in hospital (n = 42) | P | |
|---|---|---|---|---|---|
| Gender (M%) | 103 (55.1) | 59 (50.4) | 17 (60.7) | 27 (64.3) | 0.244 |
| Age | 62.00 [48.50, 71.00] | 56.00 [43.00, 66.00] | 73.00 [68.00, 77.25] | 68.50 [58.25, 76.00] | <0.001 |
| Disease history | |||||
| Health (%) | 92 (49.2) | 70 (59.8) | 7 (25.0) | 15 (35.7) | 0.001 |
| HP (%) | 50 (26.7) | 21 (17.9) | 10 (35.7) | 19 (45.2) | 0.001 |
| CCD (%) | 19 (10.2) | 8 (6.8) | 7 (25.0) | 4 (9.5) | 0.017 |
| Kidney (%) | 6 (3.2) | 2 (1.7) | 2 (7.1) | 2 (4.8) | 0.277 |
| Lung (%) | 3 (1.6) | 2 (1.7) | 0 (0.0) | 1 (2.4) | 0.732 |
| Metabolic (%) | 21 (11.2) | 13 (11.1) | 4 (14.3) | 4 (9.5) | 0.824 |
| Severity (%) | <0.001 | ||||
| mild | 80 (42.8) | 68 (58.1) | 0 (0.0) | 12 (28.6) | |
| severe | 45 (24.1) | 37 (31.6) | 0 (0.0) | 8 (19.0) | |
| critically ill | 62 (33.2) | 12 (10.3) | 28 (100.0) | 22 (52.4) | |
| Length in hospital | 17.00 [12.00, 21.00] | 18.00 [14.00, 21.00] | 18.00 [12.00, 25.00] | 16.00 [11.00, 20.00] | 0.009 |
| Clinical symptoms | |||||
| Fever (%) | 153 (82.3) | 96 (82.1) | 23 (82.1) | 34 (82.9) | 1 |
| Cough (%) | 145 (78.0) | 89 (76.1) | 23 (82.1) | 33 (80.5) | 0.788 |
| Sign (%) | 137 (73.7) | 72 (61.5) | 27 (96.4) | 38 (92.7) | <0.001 |
| Chest pain (%) | 13 (7.0) | 8 (6.8) | 2 (7.1) | 3 (7.3) | 1 |
| Muscle soreness (%) | 34 (18.3) | 28 (23.9) | 3 (10.7) | 3 (7.3) | 0.032 |
| Headache (%) | 8 (4.3) | 5 (4.3) | 1 (3.6) | 2 (4.9) | 1 |
| Dizziness (%) | 17 (9.2) | 10 (8.5) | 5 (17.9) | 2 (5.0) | 0.207 |
| Sore throat (%) | 9 (4.8) | 6 (5.1) | 2 (7.1) | 1 (2.4) | 0.618 |
| Diarrhea (%) | 43 (23.1) | 28 (23.9) | 6 (21.4) | 9 (22.0) | 1 |
| Nausea (%) | 23 (12.4) | 11 (9.4) | 7 (25.0) | 5 (12.2) | 0.091 |
| Poor appetite (%) | 151 (81.2) | 91 (77.8) | 27 (96.4) | 33 (80.5) | 0.054 |
| Weakness (%) | 159 (85.5) | 94 (80.3) | 28 (100.0) | 37 (90.2) | 0.011 |
| Complicated infection | |||||
| Bacterial infection (%) | 23 (12.4) | 9 (7.7) | 11 (39.3) | 3 (7.3) | <0.001 |
| Fungi infection (%) | 2 (1.1) | 1 (0.9) | 1 (3.6) | 0 (0.0) | 0.327 |
| Treatment | |||||
| Oxygen support | |||||
| nasal catheter | 110 (59.1) | 77 (65.8) | 10 (35.7) | 23 (56.1) | 0.013 |
| mask oxygen | 23 (12.4) | 8 (6.8) | 8 (28.6) | 7 (17.1) | 0.004 |
| High-flow nasal cannula oxygen therapy | 5 (2.7) | 1 (0.9) | 2 (7.1) | 2 (4.9) | 0.077 |
| non-invasive mechanical ventilation | 36 (19.4) | 9 (7.7) | 13 (46.4) | 14 (34.1) | <0.001 |
| invasive mechanical ventilation | 13 (7.0) | 0 (0.0) | 9 (32.1) | 4 (9.8) | <0.001 |
| Drug | |||||
| Antibiotic usage (%) | 154 (82.8) | 92 (78.6) | 28 (100.0) | 34 (82.9) | 0.011 |
| Corticosteroids (%) | 113 (60.8) | 63 (53.8) | 24 (85.7) | 26 (63.4) | 0.006 |
| Duration of Corticosteroids | 4.00 [0.00, 9.00] | 2.00 [0.00, 7.00] | 5.00 [3.00, 7.50] | 6.00 [0.00, 11.00] | 0.018 |
| Gamma globulin | 94 (50.5) | 54 (46.2) | 15 (53.6) | 25 (61.0) | |
| Contact history | |||||
| Huanan Contact (%) | 9 (4.8) | 5 (4.3) | 1 (3.6) | 3 (7.1) | 0.703 |
| Contact to others (%) | 48 (25.7) | 31 (26.5) | 4 (14.3) | 13 (31.0) | 0.278 |
| Laboratory parameters | |||||
| PCT (mmol/L) | 0.05 [0.02, 0.13] | 0.03 [0.02, 0.07] | 0.17 [0.09, 0.50] | 0.07 [0.03, 0.13] | <0.001 |
| K+ (mmol/L) | 3.67 [3.30, 4.00] | 3.68 [3.30, 3.98] | 3.68 [3.29, 4.11] | 3.66 [3.36, 4.01] | 0.763 |
| WBC (G/L) | 5.57 [4.20, 7.96] | 5.12 [3.82, 7.07] | 6.97 [4.58, 9.93] | 6.13 [5.08, 10.83] | 0.002 |
| Neutrophil (G/L) | 4.09 [2.64, 6.57] | 3.43 [2.35, 5.30] | 6.14 [3.29, 8.36] | 5.41 [3.31, 10.05] | <0.001 |
| Lymp (G/L) | 0.88 [0.55, 1.27] | 0.93 [0.65, 1.37] | 0.56 [0.32, 0.94] | 0.86 [0.50, 1.18] | 0.001 |
| Monocyte (G/L) | 0.36 [0.26, 0.52] | 0.39 [0.29, 0.54] | 0.32 [0.21, 0.44] | 0.34 [0.23, 0.52] | 0.146 |
| Inflammatory cytokines | |||||
| SAA (mg/L) | 213.24 [62.85, 301.00] | 142.49 [30.72, 300.00] | 301.00 [264.18, 301.00] | 301.00 [166.95, 301.00] | <0.001 |
| CRP (mg/L) | 32.69 [7.83, 82.44] | 16.91 [4.04, 51.37] | 91.00 [52.56, 122.38] | 62.71 [18.24, 109.55] | <0.001 |
| IL-1β (pg/mL) | 4.90 [4.90, 5.43] | 4.90 [4.90, 5.74] | 4.90 [4.90, 9.85] | 4.90 [4.90, 4.90] | 0.002 |
| IL-10 (pg/mL) | 6.33 [4.90, 12.70] | 6.29 [4.90, 11.85] | 9.80 [7.41, 24.40] | 5.50 [4.90, 7.42] | 0.024 |
| IL-6 (pg/mL) | 14.70 [4.50, 28.60] | 14.65 [4.24, 27.00] | 29.80 [14.60, 63.89] | 11.15 [5.15, 30.38] | 0.037 |
| T lymphocyte subsets | |||||
| Lymphocyte | 628.00 [356.50, 841.00] | 691.00 [518.00, 1008.00] | 293.00 [190.00, 518.00] | 548.50 [301.75, 738.25] | <0.001 |
| CD4+/CD8+ | 1.75 [0.99, 2.46] | 1.65 [0.96, 2.48] | 1.71 [1.17, 2.20] | 1.96 [1.02, 2.51] | 0.503 |
| CD3+ T (/uL) | 357.00 [202.50, 556.50] | 445.00 [296.00, 646.00] | 154.00 [99.75, 236.25] | 271.50 [168.25, 464.00] | <0.001 |
| CD4+ T (/uL) | 222.00 [107.50, 323.50] | 256.00 [159.00, 334.00] | 84.50 [59.75, 148.00] | 175.00 [97.75, 278.50] | <0.001 |
| CD8+T (/uL) | 116.00 [75.50, 205.00] | 161.00 [95.00, 258.00] | 76.50 [31.25, 103.25] | 92.00 [50.25, 211.25] | <0.001 |
| B cell (/uL) | 89.00 [53.00, 140.50] | 99.00 [69.00, 157.00] | 54.00 [32.00, 96.50] | 84.50 [52.75, 131.00] | 0.001 |
| NK cell (/uL) | 85.00 [49.50, 140.00] | 93.00 [53.00, 153.00] | 55.00 [28.75, 89.50] | 84.50 [55.25, 138.75] | 0.008 |
| BUN (mmol/L) | 5.19 [3.82, 6.91] | 4.76 [3.59, 6.08] | 6.76 [5.72, 10.54] | 5.44 [3.77, 7.81] | <0.001 |
| Serum creatinine (umol/L) | 72.60 [59.10, 90.30] | 68.90 [57.10, 85.50] | 93.70 [71.98, 103.78] | 73.55 [60.20, 89.50] | 0.001 |
| Urea acid (umol/L) | 260.30 [211.45, 316.00] | 254.80 [200.80, 295.50] | 308.30 [237.35, 408.20] | 264.50 [210.03, 333.40] | 0.021 |
| ALT (U/L) | 22.00 [12.00, 41.00] | 21.00 [12.00, 41.00] | 25.50 [14.50, 48.50] | 19.00 [12.50, 37.50] | 0.599 |
| AST (U/L) | 27.00 [19.00, 44.00] | 24.00 [18.00, 35.00] | 38.50 [25.00, 56.50] | 29.00 [21.00, 43.75] | 0.005 |
| CK (U/L) | 89.00 [49.00, 148.50] | 83.00 [43.00, 132.50] | 137.00 [103.00, 187.00] | 80.00 [43.00, 150.00] | 0.018 |
| CK-MB (U/L) | 15.00 [10.00, 19.00] | 13.00 [10.00, 18.00] | 20.00 [14.50, 28.00] | 16.00 [11.75, 21.25] | 0.001 |
| D-Dimer (mg/L) | 0.74 [0.39, 2.01] | 0.56 [0.35, 0.96]↑ | 1.22 [0.66, 19.21] | 1.12 [0.61, 6.19] | <0.001 |
| PT (sec) | 12.90 [12.00, 13.80] | 12.60 [11.80, 13.30] | 13.70 [12.55, 14.70] | 13.40 [12.70, 14.10] | <0.001 |
| TNF (fmol/mL) | 11.10 [8.20, 15.30] | 11.30 [8.46, 16.30] | 12.20 [10.40, 14.90] | 9.62 [7.02, 13.38] | 0.113 |
| IL-1β (pg/mL) | 4.90 [4.90, 5.43] | 4.90 [4.90, 5.74] | 4.90 [4.90, 9.85] | 4.90 [4.90, 4.90] | 0.002 |
| IL-10 (pg/mL) | 6.33 [4.90, 12.70] | 6.29 [4.90, 11.85] | 9.80 [7.41, 24.40] | 5.50 [4.90, 7.42] | 0.024 |
| IL-6 (pg/mL) | 14.70 [4.50, 28.60] | 14.65 [4.24, 27.00] | 29.80 [14.60, 63.89] | 11.15 [5.15, 30.38] | 0.037 |
Note: HP: hypertension, CCD: cardio-cerebrovascular disease.
Increased inflammatory cytokines and suppressed T cell-mediated immunity in COVID-19
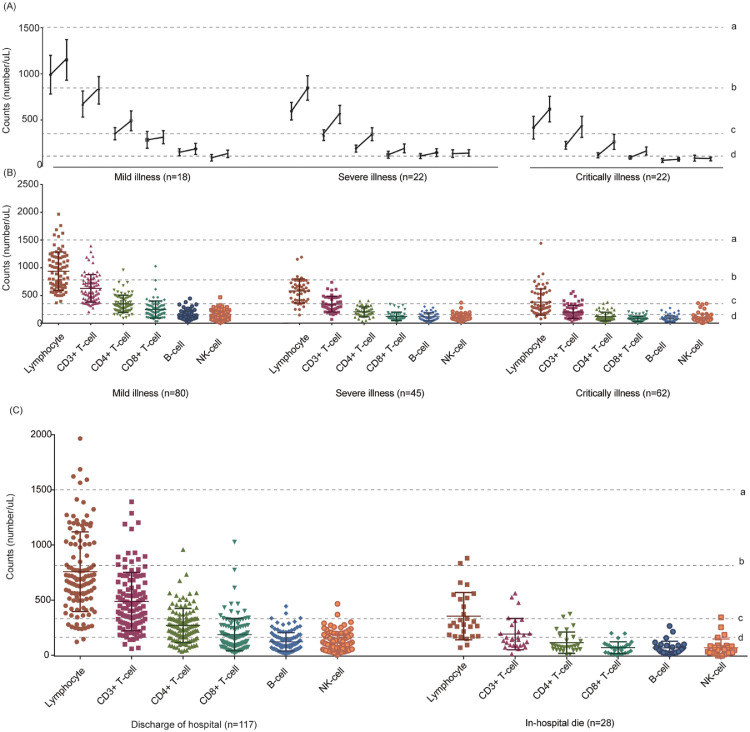
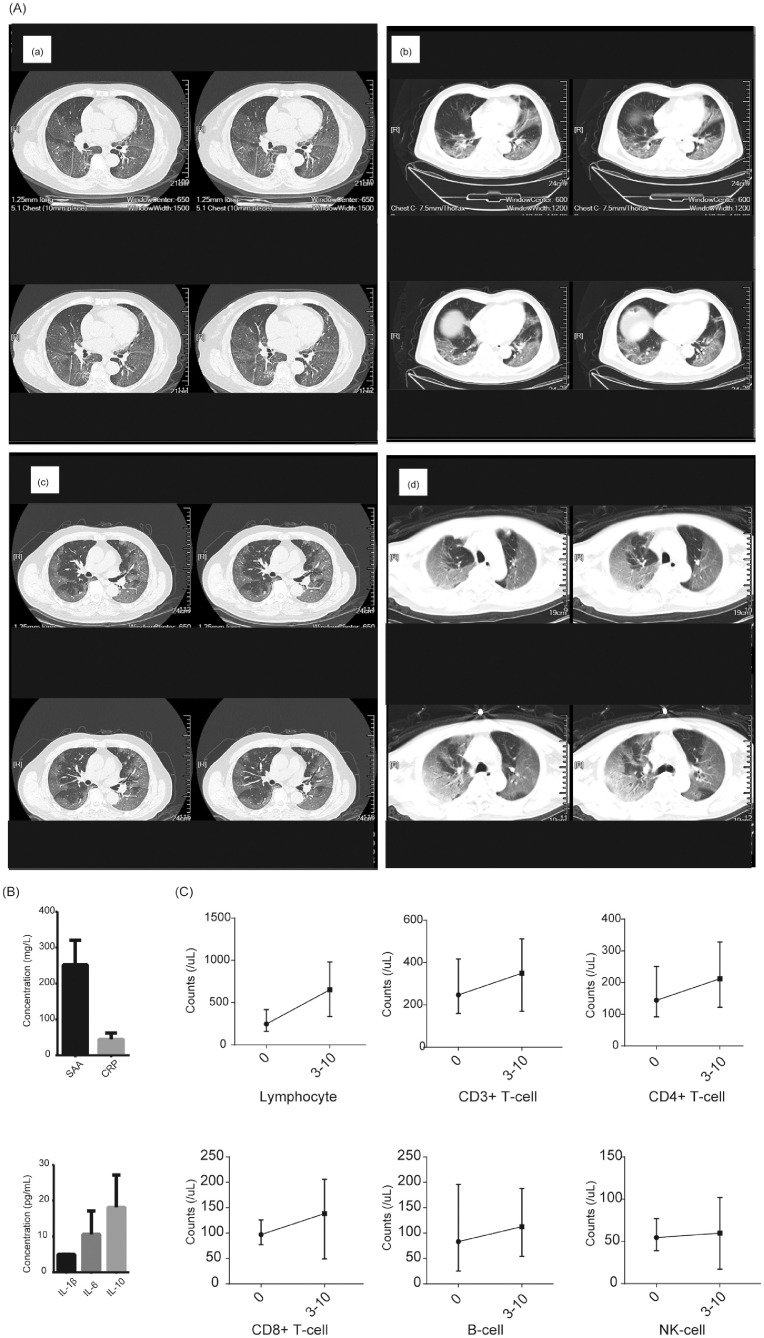
The counts of lymphocyte (median=628.00, IQR [356.50, 841.00]), T lymphocyte subset lymphocyte (median=628.00, IQR [356.50, 841.00]), CD3+ T-cell (median=357.0, IQR [202.5,556.5]), CD4+ T-cell (median=222.0, IQR [107.5,237.5]), CD8+ T-cell (median=116.0, IQR [75.5,205.0),B-cell (median=89, IQR [53,140.5]), and NK-cell (median=85, IQR [49.5,140.0]) were all decreased to half of normal values, and statistical differences found among different severity of illness and survival states (P<0.001). The lowest values of lymphocyte, CD3+ T-cell, CD4+ T-cell, CD8+ T-cell, and B-cell were 76, 9, 6, 9 and 2, respectively. Meanwhile, the levels of T lymphocyte subsets varied according to different severity of illness (Table1-2 and Fig. 2 ). 62 patients assessed T lymphocyte subsets more than once, including mild-ill (n = 18), severe-ill (n = 22) and critically-ill (n = 22). After 5–14 days in-hospital treatment, the counts of T lymphocyte subsets slightly increased than disease onset (Fig. 2A). Meanwhile, we presented CT scan images and temporal changes of T lymphocyte subsets, SAA and CRP of 4 critically-ill patients which have results more than once in Fig. 3 (Detail information in Table 2 ).
Table 1-2.
Baseline characteristics of included patients (categorized by severity of illness).
| Mild-ill (n = 80) | Severe-ill (n = 45) | Critically-ill (n = 62) | P | |
|---|---|---|---|---|
| Gender (M%) | 30 (37.5) | 31 (68.9) | 42 (67.7) | <0.001 |
| Age | 56.00 [44.00, 67.00] | 60.00 [46.00, 67.00] | 70.00 [60.25, 76.75] | <0.001 |
| Disease history | ||||
| Health (%) | 48 (60.0) | 24 (53.3) | 20 (32.3) | 0.004 |
| HP (%) | 15 (18.8) | 11 (24.4) | 24 (38.7) | 0.026 |
| CCD (%) | 3 (3.8) | 3 (6.7) | 13 (21.0) | 0.002 |
| Kidney (%) | 0 (0.0) | 0 (0.0) | 6 (9.7) | 0.002 |
| Lung (%) | 2 (2.5) | 1 (2.2) | 0 (0.0) | 0.466 |
| Metabolic (%) | 7 (8.8) | 6 (13.3) | 8 (12.9) | 0.648 |
| Outcome | <0.001 | |||
| Stay in hospital | 12 (15.0) | 8 (17.8) | 22 (35.5) | |
| Discharge | 68 (85.0) | 37 (82.2) | 12 (19.4) | |
| Death | 0 (0.0) | 0 (0.0) | 28 (45.2) | |
| Length in hospital | 16.00 [11.00, 20.00] | 18.00 [14.00, 21.00] | 18.00 [12.00, 25.00] | 0.007 |
| Clinical symptoms | ||||
| Fever (%) | 63 (79.7) | 40 (88.9) | 50 (80.6) | 0.418 |
| Cough (%) | 60 (75.9) | 37 (82.2) | 48 (77.4) | 0.766 |
| Sign (%) | 43 (54.4) | 37 (82.2) | 57 (91.9) | <0.001 |
| Chest pain (%) | 5 (6.3) | 3 (6.7) | 5 (8.1) | 0.934 |
| Muscle soreness (%) | 15 (19.0) | 10 (22.2) | 9 (14.5) | 0.562 |
| Headache (%) | 2 (2.5) | 3 (6.7) | 3 (4.8) | 0.474 |
| Dizziness (%) | 5 (6.3) | 2 (4.4) | 10 (16.4) | 0.075 |
| Sore throat (%) | 5 (6.3) | 2 (4.4) | 2 (3.2) | 0.764 |
| Diarrhea (%) | 16 (20.3) | 14 (31.1) | 13 (21.0) | 0.348 |
| Nausea (%) | 10 (12.7) | 3 (6.7) | 10 (16.1) | 0.35 |
| Poor appetite (%) | 59 (74.7) | 37 (82.2) | 55 (88.7) | 0.097 |
| Weakness (%) | 63 (79.7) | 37 (82.2) | 59 (95.2) | 0.02 |
| Complicated infection | ||||
| Bacterial infection (%) | 2 (2.5) | 4 (8.9) | 17 (27.4) | <0.001 |
| Fungi infection (%) | 0 (0.0) | 0 (0.0) | 2 (3.2) | 0.167 |
| Treatment | ||||
| Antibiotic usage (%) | 51 (64.6) | 41 (91.1) | 62 (100.0) | <0.001 |
| Corticosteroids (%) | 29 (36.7) | 29 (64.4) | 55 (88.7) | <0.001 |
| Duration of Corticosteroids | 0.00 [0.00, 5.00] | 5.00 [0.00, 7.00] | 9.00 [4.00, 12.00] | <0.001 |
| Gamma globulin | 29 (36.7) | 22 (48.9) | 43 (69.4) | |
| Contact history | ||||
| Huanan Contact (%) | 2 (2.5) | 3 (6.7) | 4 (6.5) | 0.46 |
| Contact to others (%) | 29 (36.2) | 8 (17.8) | 11 (17.7) | 0.02 |
| Laboratory parameters | ||||
| PCT (mmol/L) | 0.02 [0.02, 0.04] | 0.06 [0.03, 0.09] | 0.14 [0.07, 0.32] H | <0.001 |
| K+ (mmol/L) | 3.68 [3.30, 4.00] | 3.50 [3.19, 3.84] | 3.70 [3.40, 4.17] | 0.124 |
| WBC (G/L) | 5.00 [3.80, 6.00] | 6.02 [4.54, 9.30] | 7.00 [4.50, 11.31] H | <0.001 |
| Neutrophil (G/L) | 3.12 [2.18, 4.20] | 4.87 [3.43, 7.64] | 6.14 [3.33, 10.15] H | <0.001 |
| Lymp (G/L) | 1.25 [0.90, 1.59] | 0.82 [0.59, 1.01] | 0.52 [0.37, 0.88] | <0.001 |
| Monocyte (G/L) | 0.39 [0.31, 0.52] | 0.42 [0.29, 0.61] | 0.32 [0.18, 0.44] | 0.005 |
| Inflammatory cytokines | ||||
| CRP (mg/L) | 8.77 [2.62, 24.75]H | 51.37 [22.07, 82.41] H | 83.23 [40.12, 118.20] H | <0.001 |
| SAA (mg/L) | 83.62 [11.59, 202.50] H | 292.32 [153.03, 301.00] H | 301.00 [258.48, 301.00] H | <0.001 |
| IL-1β (pg/mL) | 4.90 [4.90, 5.21] H | 4.90 [4.90, 5.42] H | 4.90 [4.90, 5.61] H | 0.97 |
| IL-10 (pg/mL) | 5.00 [4.90, 7.16] | 8.52 [4.90, 15.80] H | 9.56 [5.14, 14.80] H | <0.001 |
| IL-6 (pg/mL) | 14.60 [4.21, 22.30] H | 11.30 [4.34, 28.43] H | 17.40 [7.18, 50.20] H | 0.072 |
| T lymphocyte subsets | ||||
| CD4+/CD8+ | 1.68 [0.96, 2.18] | 1.96 [1.02, 2.70] | 1.75 [1.06, 2.28] | 0.454 |
| CD3+ T (/uL) | 894.50 [662.75, 1192.00] | 593.00 [412.00, 725.00] | 287.50 [240.50, 528.50] | <0.001 |
| CD4+ T (/uL) | 573.50 [426.75, 771.00] | 299.00 [249.00, 460.00] L | 168.50 [125.25, 255.00] L | <0.001 |
| CD8+T (/uL) | 323.50 [232.75, 448.75] | 188.00 [134.00, 274.00] L | 92.50 [70.75, 141.50] L | <0.001 |
| B cell (/uL) | 213.50 [152.25, 314.25] | 97.00 [74.00, 162.00] L | 73.00 [36.50, 101.75] L | <0.001 |
| NK cell (/uL) | 107.50 [82.75, 149.75] | 93.00 [60.00, 161.00] L | 55.50 [32.00, 91.25] L | <0.001 |
| BUN (mmol/L) | 4.09 [3.18, 5.45] | 5.19 [3.89, 7.07] | 6.95 [5.34, 9.62] | <0.001 |
| Serum creatinine (umol/L) | 63.60 [54.55, 78.98] | 72.60 [61.00, 97.10] | 87.25 [67.22, 102.72] | <0.001 |
| Urea acid (umol/L) | 255.20 [206.28, 290.65] | 255.20 [216.70, 305.60] | 296.40 [214.40, 378.07] | 0.03 |
| ALT (U/L) | 15.00 [9.00, 31.50] | 32.00 [15.00, 61.00] | 25.50 [15.25, 44.75] | <0.001 |
| AST (U/L) | 21.00 [16.00, 28.00] | 35.00 [22.00, 54.00] | 32.00 [23.25, 54.75] | <0.001 |
| CK (U/L) | 78.00 [41.25, 119.25] | 91.00 [42.00, 135.50] | 121.00 [62.25, 182.50] H | 0.041 |
| CK-MB (U/L) | 12.00 [8.75, 15.00] | 17.00 [11.50, 21.50] | 18.00 [13.00, 23.00] | <0.001 |
| D-Dimer (mg/L) | 0.42 [0.34, 0.85] H | 0.75 [0.50, 1.71] H | 1.22 [0.55, 11.54] H | <0.001 |
| PT (sec) | 12.40 [11.70, 13.10] | 13.10 [12.00, 14.05] | 13.75 [12.70, 14.60] | <0.001 |
Fig. 2.
T lymphocyte subsets of different severity of illness and survival conditions in patients on admission with COVID-19
(A) Changes of T lymphocyte subsets from disease onset to 5–14 days later in COVID-19 patients (median with IQR).
(B) Differences of T lymphocyte subsets among mild-ill, severe-ill and critically-ill patients (mean with SD).
(C) Differences of T lymphocyte subsets between survivor and non-survivor (mean with SD).
(D) Dotted line a: lower limit of normal value of lymphocyte (1500/uL); dotted line b:lower limit of normal value of CD3+ T-cell (723/uL); dotted line c: lower limit of normal value of CD4+ T-cell (404/uL), dotted line d: lower limit of normal value of CD8+ T-cell (220/uL).
Fig. 3.
CT scan images and changes of T lymphocyte subsets of 4 critically-ill patients.
a: representative CT scan images; b: concentrations of SAA and CRP; c: temporal changes of T lymphocyte subsets between 0 and 3–10 days. (mean with range).
Table 2.
Detail information of 4 critically-ill patients.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | |
|---|---|---|---|---|
| Gender | Female | Male | Male | Male |
| Age | 73 | 69 | 57 | 75 |
| Length in hospital | 23 | 5 | 7 | 10 |
| Disease history | Hypertension | No | No | Hypertension, atrial fibrillation |
| Severity of illness | Critically | Critically | Critically | Critically |
| Survivor | No | No | No | No |
| T lymphocyte on admission | ||||
| Lymphocyte | 567 | 300 | 273 | 224 |
| CD3+T-cell | 282 | 189 | 169 | 159 |
| CD4+T-cell | 172 | 144 | 92 | 93 |
| CD8+T-cell | 79 | 44 | 77 | 83 |
| B-cell | 105 | 55 | 32 | 25 |
| NK-cell | 168 | 56 | 77 | 40 |
| Inflammatory cytokines | ||||
| SAA | 250.42 | >300.00 | 155.73 | >300 |
| CRP | 43.66 | 68.81 | 25.97 | 38.94 |
| IL1β | 4.9 | 5 | 7 | 5 |
| IL-6 | 6.58 | 7.1 | 12.4 | 18.1 |
| IL-10 | 21.6 | 24.9 | 30.2 | 7.8 |
| Treatment | Anti-virus: Ribavirin, Arbidol; Antibiotics: cephalosporin, meropenem | Anti-virus: Ribavirin, Arbidol; Antibiotics: moxifloxacin | Anti-virus: Arbidol; Antibiotics: cephalosporin | Anti-virus: Ribavirin, Arbidol; Antibiotics: moxifloxacin, meropenem |
| Respiratory support | Non-invasive mechanical ventilation | |||
| Immune globulin | No | Yes | No | Yes |
| Corticosteroids | methylprednisolone | No | methylprednisolone | No |
| T lymphocyte after 3–10 days | ||||
| Lymphocyte | 273 | 593 | 336 | 578 |
| CD3+T-cell | 165 | 368 | 169 | 392 |
| CD4+T-cell | 123 | 241 | 122 | 166 |
| CD8+T-cell | 42 | 111 | 49 | 206 |
| B-cell | 64 | 68 | 105 | 54 |
| NK-cell | 39 | 129 | 82 | 17 |
In critically-ill patients, the median counts of lymphocyte, CD3+ T-cell, CD4+ T-cell, and CD8+ T-cell were almost decreased to a third of median counts of mild-ill patients, which is equal to nearly half of median counts of severe-ill patients (P<0.001).(Table 1–2 and Fig. 2B) Among critically ill patients, 28 patients died in hospital, the median CD3+ T-cell, CD4+ T-cell, CD8+ T-cell, B-cell, and NK-cell were 154.00 (IQR, [99.75, 236.25]), 84.50 (IQR, [59.75,148.00]), 76.50 (IQR, [31.25, 103.25]), 54.00 (IQR, [32.00, 96.50]) and 55.00 (IQR,[28.75, 89.50]), respectively, which were obviously lower than other critically-ill patients (P<0.001) (Fig. 2C).
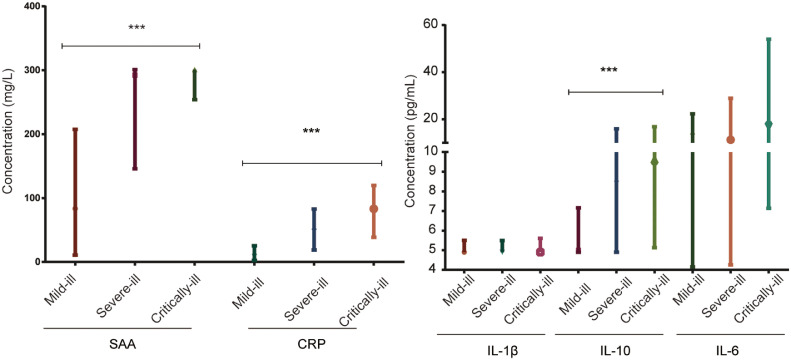
Meanwhile, we measured the concentrations of SAA, CRP and cytokines including IL-6 and IL-10 in plasma which were produced by T cells. With the severity of illness getting worse, the concentrations of SAA, CRP and IL-10 getting much higher (P<0.001) (Fig. 4 ). To be specific, the median concentration of SAA in critically-ill was nearly 4-fold to mild-ill patients. And the median concentration of CRP in critically-ill patients was nearly 10-fold to mild-ill patients. The concentrations of IL-6 and IL-10 in COVID-19 patients were higher than normal values (Fig. 4).
Fig. 4.
Concentration of inflammatory cytokines (SAA, CRP, IL-1β, IL-6, IL-10) in different severity of patients with COVID-19. (median with IQR)
(***P<0.001).
Possible associations of suppressed T lymphocyte subsets with in-hospital death
We adopted univariate logistic regression to measure the OR with 95% CI of potential association of T lymphocyte subsets with in-hospital death (Table 3 ). 145 patients in our study discharged or died in hospital (had outcomes). We found lower T lymphocyte subsets levels were associated with higher risk of in-hospital death. The risk of in-hospital death was statistically lower in those who had higher counts of lymphocyte (OR=0.99, 95% CI [0.992,0.997]), CD3+ T-cell (OR=0.99,95% CI [0.98,0.99], P<0.001), CD4+ T-cell (OR= 0.99, 95% CI [0.98,0.99], P<0.001), CD8+ T-cell (OR= 0.98, 95% CI [0.97,0.99], P<0.001), B-cell (OR=0.99, 95% CI [0.98, 0.99], P = 0.007), and NK-cell (OR= 0.99, 95% CI [0.98,0.99], P = 0.034). Furthermore, we categorized T lymphocyte subsets into different groups based on their counts. We found lower T lymphocyte subsets lymphocyte (<500/uL), CD3+T-cell (<200/uL), CD4+ T-cell (<100/uL), CD8+ T-cell (<100/uL) and B-cell (<50/uL) counts were exhibit significantly higher risk of in-hospital death (Table 3).
Table 3.
T lymphocyte subsets as risk factors of in-hospital death (n = 145).
| (/uL) | Univariable OR | 95% CI | P value | Cut-off point | AUC | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|
| Lymphocyte | 0.99 | 0.992–0.997 | 0.00 | 559 | 0.84 | 0.73 | 0.82 |
| >500 | Ref | ||||||
| <500 | 11.61 | 4.67–30.98 | 0.00 | ||||
| CD4+T/CD8+T | 1.05 | 0.745–1.42 | 0.75 | – | |||
| CD3+T | 0.99 | 0.98–0.99 | 0.00 | 235 | 0.86 | 0.86 | 0.75 |
| >500 | Ref | ||||||
| <100 | 56.00 | 9.37–526.40 | 0.00 | ||||
| 100–200 | 29.33 | 6.56–212.96 | 0.00 | ||||
| 200–300 | 5.05 | 0.91–38.69 | 0.07 | ||||
| 300–400 | 3.43 | 0.53–27.51 | 0.19 | ||||
| 400–500 | 1.41 | 0.06–15.67 | 0.78 | ||||
| CD4+T | 0.99 | 0.98–0.99 | 0.00 | 104 | 0.82 | 0.88 | 0.68 |
| >300 | Ref | ||||||
| <100 | 30.54 | 7.65–207.38 | 0.00 | ||||
| 100–200 | 3.33 | 0.61–25.22 | 0.18 | ||||
| 200–300 | 2.18 | 0.34–17.27 | 0.41 | ||||
| CD8+T | 0.98 | 0.97–0.99 | 0.00 | 85 | 0.81 | 0.82 | 0.68 |
| >200 | ref | ||||||
| <100 | 10.88 | 2.89–71.41 | 0.00 | – | |||
| 100–200 | 2.41 | 0.52–17.13 | 0.30 | – | |||
| B-cell | 0.99 | 0.98–0.99 | 0.01 | 82 | 0.70 | 0.67 | 0.71 |
| >200 | ref | ||||||
| <50 | 6.50 | 1.54–45.11 | 0.02 | ||||
| 50–100 | 2.25 | 0.52–15.68 | 0.33 | ||||
| 100–200 | 1.08 | 0.19–8.29 | 0.93 | ||||
| NK-cell | 0.99 | 0.98–0.99 | 0.03 | – | – | – | – |
| >200 | ref | ||||||
| <50 | 3.85 | 0.91–26.69 | 0.10 | ||||
| 50–100 | 2.44 | 0.57–16.99 | 0.28 | ||||
| 100–200 | 0.42 | 0.05–3.76 | 0.41 |
Note: Patients discharged from hospital or dead in hospital were included in this univariate logistic regression.
The warning values of T lymphocyte subsets to predict in-hospital death of COVID-19
We adopted ROC curve to find the warning values of T lymphocyte subsets lymphocyte, CD3+ T-cell, CD4+ T-cell, CD8+ T-cell, and B-cell which showed statistical difference between fatal and recover COVID-19. The warning values (/uL) to predict in-hospital death of lymphocyte, CD3+ T-cell, CD4+ T-cell, CD8+ T-cell, and B-cell were 559, 235, 104, 85 and 82, respectively. Lower than warning values of T lymphocyte subsets indicated a significantly higher risk of in-hospital death (Table 3).
Discussion
In this study, all patients, especially in severe-ill and critically-ill including dead patients, exhibited a significant drop of T lymphocyte subsets counts with remarkably increasing concentrations of SAA and CRP compared to normal values. In critically-ill patients, the median counts of lymphocyte, CD3+ T-cell, CD4+ T-cell, and CD8+ T-cell were almost decreased to a third of median counts of mild-ill patients, which is equal to nearly half of median counts of severe-ill patients. 28 patients died in hospital, the median lymphocyte, CD3+ T-cell, CD4+ T-cell, CD8+ T-cell, B-cell, and NK-cell were obviously lower than other critically-ill patients. Meanwhile, as the severity of illness getting worse, the concentrations of inflammatory cytokines getting higher. The median concentration of SAA in critically-ill patients was nearly 4-fold to that of mild-ill patients. The median concentration of CRP in critically-ill patients was nearly 10-fold to that of mild-ill patients. Lower counts (/uL) of T lymphocyte subsets lymphocyte (<500), CD3+T-cell (<200), CD4+ T-cell (<100), CD8+ T-cell (<100), and B-cell (<50) were associated with higher risk of in-hospital death. The warning values of lymphocyte, CD3+ T-cell, CD4+ T-cell, CD8+ T-cell, and B-cell were 559, 235, 104, 85 and 82, respectively.
T cell-mediated immunity is the central element of the adaptive immune system and includes a primary response by naïve T cells. Each T lymphocyte expresses a unique T cell receptor (TCR) on the surface.14 Mature T lymphocytes known as naïve T cells, circulate through blood and the lymphatic system and reside in secondary lymphoid organs.15 All T lymphocytes have the T-cell receptor (TCR) and pan-T-cell co-receptor CD3. CD4+ T cells recognize MHC 1 bound antigens, while CD8+ T cells recognize the MHC 1 bound antigens. Both CD4+ T cells, and CD8+ T cells have the co-receptor CD3. And the sum total counts of CD4+ T cells and CD8+ cells are almost equal to or slightly more than CD3+ cells. Damage to T lymphocytes, impressed immunity and related high levels of inflammatory cytokines might be important factor to exacerbation and mortality of patients. Previous SARS-COV infection stimulated cytokines IL-1, IL-10 etc., and T lymphocyte subsets CD4+T-cell and CD8+T-cell were decreased after onset of the disease as well.16 T-cell mediated immunity is an adaptive process of developing antigen-(Ag) specific T lymphocytes to eliminate SARS-COV-2 invasion. Since CD4+ T cells are central in the origin and regulation of autoimmunity, emphasis has been placed on the characterization of Th subsets and their possible roles in the inflammatory process. IL-1β serves as the major mediator of inflammatory diseases, including autoimmune, infectious and degenerative ones. In the central nervous system (CNS), it acts as endogenous pyrogen inducing fever, and systemically, it induces the acute phase response, either directly or indirectly via the induction of IL-6 production. IL-6 is one of the key mediators regulating inflammation. Upon activation by infectious agents like SARS-COV-2, IL-6 is produced by macrophages, endothelial cells, and T cells and activate many tissue cells, including endothelial cells and parenchymal cells to produce effector molecules of inflammation.17 IL-10 is mainly produced by monocytes and T cells and the most potent anti-inflammatory cytokine, which reduces inflammatory processes and thus limits collateral inflammation-induced tissue damage.18 , 19 On one hand, SARS-COV-2 invasion activated T cell-mediated immunity which resulted in increasing production of inflammatory cytokine. However, inhibition cytokines such as IL-10 can suppress T cell activation conversely. On the other hand, SAA and CRP are both of acute-phase proteins in response to inflammatory cytokines related by activated monocytes/macrophages after infections. High levels of SAA and CRP could reflect the severity of inflammation and result in a “cytokine storm”, during which inflammation spreads throughout the body via the circulation. Several studies revealed that a substantial decrease in total number of lymphocytes indicates that coronavirus consumes many immune cells and inhibits the body's cellular immune function.20 , 21 SARS causes similar decrease of T lymphocyte, severe lung injury and spleen shrinkage, which can progress to acute respiratory distress syndrome (ARDS).22, 23, 24 Until now, no antiviral treatment for coronavirus infection has been proven to be effective. Now, we faced on urgent need to elucidate what are the risk factors for severe illness or death. Therefore, our study tries to partly answer this question to help with the clinical practice and better know this disease.
The study has several limitations. First, we adopted this research in a retrospective method, which may limit the strength and reliability of our results. Second, all enrolled patients were from one central with inevitable selective bias. Third, not all laboratory tests were done in all patients, including IL-1β, IL-6 and IL-10. Therefore, we did not assess the association of these cytokines with in-hospital death. Fourth, considering our limited sample size, we did not include other potential risk factors such as gender and age into multivariate logistic regression to assess the risk factors of T lymphocyte subsets. This may limit the interpretations of our results. However, since we are the first hospital to admit COVID-19 patients and reported it to Chinese CDC, we believe our study population and findings are representative and reliable.
To our knowledge, this is the first clinical study to exhibit significant suppressed T cell subsets and its possible association to in-hospital death and severity of COVID-19. In addition, we assessed and analyzed inflammatory cytokines produced by T cells on admission. In summary, we find a significant decrease of T lymphocyte subset is positive correlated with in-hospital death and severity of illness. Lower counts (/uL) of T lymphocyte subsets lymphocyte (<500), CD3+T-cell (<200), CD4+ T-cell (<100), CD8+ T-cell (<100) and B-cell (<50) counts were exhibited higher risk of in-hospital death. The warning values of CD3+ T-cell, CD4+ T-cell, CD8+ T-cell, and B-cell were 559, 235, 104, 85 and 82, respectively. The decrease levels of T lymphocyte subsets reported in our study were not common among other virus infection, which may be a possible biomarker for early diagnosis. Our findings may shed light on early warning of high risk of mortality and help early intervention and treatment.
Declaration of Competing Interest
We declare no competing interests.
Acknowledgments
Funding project
National science and technology emergency project-Clinical study on prevention and treatment of Corona Virus Disease 2019 by integrated traditional Chinese and western medicine (2020YFC0841600).
Contributors
Wen-guang XIA and Ji-xian ZHANG had the idea for and designed the study and had full access to all of the data in the study. Bo XU and Cun-yu FAN take responsibility for the integrity of the data. Qing MIAO and An-lu WANG take responsibility of accuracy of the data analysis. An-lu WANG and other authors drafted the paper and did the analysis. All authors critically revised the manuscript for important intellectual content and gave final approval for the version to be published. Bo XU and Cun-yu FAN collected the data. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.04.012.
Appendix. Supplementary materials
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., et al. <em>Severe acute respiratory syndrome-related coronavirus: the species and its viruses – a statement of the coronavirus study group doi: 10.1101/2020.02.07.937862. [DOI]
- 2.Zhou P., Yang X.-L., Wang X.-G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q., Guan X., Wu P. Early transmission dynamics in wuhan, china, of novel coronavirus-infected pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001316. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan J.F.-W., Yuan S., Kok K.-H. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020 doi: 10.1016/S0140-6736(20)30154-9. S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization [Available from:https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/.
- 7.Chen H., Guo J., Wang C. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. The Lancet. 2020 doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [published Online First: 01/30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X.-W., Wu X.-X., Jiang X.-G. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W.J., Zhao M., Liu K. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-COV. Antiviral Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauls R.S., McCausland C., Taylor B.N. StatPearls Publishing; Treasure IslandFL: 2020. Histology, T-Cell Lymphocyte. StatPearls. [PubMed] [Google Scholar]
- 14.Rossjohn J., Gras S., Miles J.J. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [published Online First: 12/10] [DOI] [PubMed] [Google Scholar]
- 15.Sun H., Sun C., Xiao W. Tissue-resident lymphocytes: from adaptive to innate immunity. Cell Mol Immunol. 2019;16(3):205–215. doi: 10.1038/s41423-018-0192-y. [published Online First: 01/11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C.K.-f., Wu H., Yan H. T cell responses to whole SARS coronavirus in humans. J Immunol. 2008;181(8):5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones S.A., Jenkins B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat Rev Immunol. 2018;18(12):773–789. doi: 10.1038/s41577-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 18.Saraiva M., O'Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10(3):170–181. doi: 10.1038/nri2711. [published Online First: 02/15] [DOI] [PubMed] [Google Scholar]
- 19.Iyer S.S., Cheng G. Role of interleukin 10 transcriptional regulation in inflammation and autoimmune disease. Crit Rev Immunol. 2012;32(1):23–63. doi: 10.1615/critrevimmunol.v32.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y.-H., Dong J.-H., An W.-M. Clinical and computed tomographic imaging features of Novel Coronavirus Pneumonia caused by SARS-CoV-2. J Infect. 2020 doi: 10.1016/j.jinf.2020.02.017. S0163-4453(20)30100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J.-J., Dong X., Cao Y.-Y. Clinical characteristics of 140 patients infected by SARS-CoV-2 in Wuhan, China. Allergy. 2020 doi: 10.1111/all.14238. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 22.Zhan J., Deng R., Tang J. The spleen as a target in severe acute respiratory syndrome. FASEB J. 2006;20(13):2321–2328. doi: 10.1096/fj.06-6324com. [DOI] [PubMed] [Google Scholar]
- 23.Li T., Qiu Z., Zhang L. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. 2004;189(4):648–651. doi: 10.1086/381535. [published Online First: 02/04] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y.-D., Sin W.-Y.F., Xu G.-B. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recover from SARS. J Virol. 2004;78(11):5612–5618. doi: 10.1128/JVI.78.11.5612-5618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.