Abstract
After the construction of genomic libraries with yeast artificial chromosomes in the late 1980's for gene isolation and expression studies in cells, human artificial chromosomes were then a natural development in the 1990's, based on the same principles of formation requiring centromeric sequences for generating functional artificial chromosomes. Over the past twenty years, they became a useful research tool for understanding human chromosome structure and organization, and important vectors for expression of large genes and gene loci and the regulatory regions for full expression. Now they are being modified and developed for gene therapy both ex vivo and in vivo. The advantages of using HAC vectors are that they remain autonomous and behave as a normal chromosome. They are attractive for therapy studies without the harmful consequences of integration of exogenous DNA into host chromosomes. HAC vectors are also the only autonomous stable vectors that accommodate large sequences (>100 kb) compared to other vectors. The challenges of manipulating these vectors for efficient delivery of genes into human cells is still ongoing, but we have made advances in transfer of gene expressing HAC vectors using the helper free (HF) amplicon vector technology for generating de novo HAC in human cells. Efficient multigene delivery was successfully achieved following simultaneous infection with two HF amplicons in a single treatment and the input DNA recombined to form a de novo HAC. Potentially several amplicons containing gene expressing HAC vectors could be transduced simultaneously which would increase the gene loading capacity of the vectors for delivery and studying full expression in human cells.
Keywords: Human artificial chromosome (HAC), Herpes simplex virus-1 (HSV-1) amplicons, Hypoxanthine guanine phosphoribosyltransferase (HPRT) gene, Multigene delivery, Gene therapy
1. Introduction
Gene expression studies in mammalian and human cells have been important for advancing our knowledge in generating disease gene models in animals and ultimately for developing human gene therapy studies. Complementation of the disease gene in human cells by correcting the genetic defect following gene introduction into the relevant cell type is the aim for gene therapy of monogenic disorders [1]. Understanding the requirements for full gene expression first is key to this effect. The technology to introduce genes effectively into cells developed rapidly over the last 20 years for transient and stable gene expression. This included modifying viruses to efficiently and safely transfer genes, and more recently developing non-viral mechanisms as transfer vehicles. There is not a universal gene delivery method for all cell types, or a gene therapy for all diseases. The method of gene entry utilised in each case has depended on a number of factors including the type and size of DNA used, the host cell type for introduction and the ease of producing and/or using the method of choice for gene transfer depending on viral or non-viral methods [2]. Recent focus on targets for gene therapy include cancers and monogenic diseases including neurological, metabolic and cardiovascular disorders [3].
Viral methods as DNA carriers became popular when cDNA cloning developed in the late 1970's. Genes as cDNAs could be easily incorporated into an expression transfer vector containing the essential elements of the particular viral genome required for infecting the cell. Efficient viruses included retrovirus, adenovirus, adeno-associated virus, lentivirus, alphavirus, baculovirus, pox virus, Epstein-Barr virus and herpes virus [4]. Initial problems arose over the production of an immune response in cells and integration of DNA randomly into the host cell genome following some viral delivery. This lead to insertional mutagenesis and loss of function or inappropriate gene expression. Although adenovirus vectors formed episomal molecules in the host cells, genes were sometimes silenced from the vectors. The effects lead to oncogenesis and in some cases resulted in fatal consequences [3]. However, viral vector systems still prove an efficient method for gene targeting since more safety regulations are followed and administration of strict dosage requirements are monitored stringently in clinical trials.
Non-viral methods included introducing DNA by standard methods of transfection with calcium phosphate, cationic liposomes, peptides, polymers nanoparticles; or mechanically via electroporation, microinjection, sonoporation, magnetofection and gene gun delivery. In some cases, the addition of transposases, drugs, antioxidants or enzymes enhanced the effect to navigate the DNA intact across the cells membrane. With the advent of site specific nucleases and clustered regularly-interspaced short palindromic repeats (CRISPR) for genome editing, advances on the delivery methods of these sequences into cells are currently being developed and will improve the technologies [5].
Both types of mechanisms were useful for small gene/DNA delivery, but transfer of large genes or genomic loci into cells was problematic. With the development of yeast and bacterial and P1 artificial chromosomes (YAC, BAC, PAC) libraries [6], the genomic clones were modified as carriers of large DNA (greater than 50 kb) and whole gene loci for expression studies. However, since the larger DNA could not be accommodated in viral vector genomes for transfer to cells, research focussed on developing non-viral delivery methods for the delivery. Calcium phosphate transfection and electroporation sheared the YAC/BAC DNA on transfer across the cell membrane and required protection from degradation. Methods to coat the large YAC DNA with polyamines first and then transfer the DNA by lipofection was found to be effective and usually eliminated the degradation [7]. This ensured that the DNA remained intact in human cells, although the efficiency of transfer of large DNA was low. Although YAC and BAC were useful for gene transfer and expression studies, DNA integrated into the genome. This lead to the development of human artificial chromosomes.
1.1. Development of human artificial chromosomes
Following on from YAC and BAC vector technology, we and others developed human artificial chromosome (HAC) vectors for artificial chromosome generation in human cells [8,9]. HAC were a useful tool for understanding chromosome biology and analyzing chromosome structure and function. They were subsequently engineered as large gene transfer vectors, since they could accommodate much larger DNA and behave as a normal chromosome without DNA integrating into the host chromosomes.
The two approaches for construction included modifying existing host chromosomes into smaller mini-chromosomes or generating newly formed de novo chromosomes in human cells following transfection of the essential centromeric chromosomal sequences. Mini-chromosomes of different sizes were formed by targeted recombination of synthetic telomere-containing input DNA with complementary sequences on the host chromosome, resulting in reduction of size and replacement of a new telomeric chromosome end [10,11]. This strategy became termed as the ‘top down’ approach and the further development of mini-chromosomes has been covered elsewhere in this edition. Newly formed or ‘de novo’ chromosomes were generated in host cells by transfecting a vector assembled with the essential centromeric chromosomal sequences (isolated from genomic clones) that were required for artificial chromosome formation [12]. This was known as the ‘bottom up’ approach and the focus of our work involved developing efficient delivery methods of the HAC DNA into a range of human cells for de novo artificial chromosome generation.
De novo HAC vectors minimally need to contain sufficient alpha satellite (alphoid) sequences containing the centromere protein B binding box for centromere formation in a newly formed functional artificial chromosome within the cell nucleus [13]. They develop as circular chromosomes (not requiring telomeric sequences for function), segregate normally in the cell cycle, remain autonomous, and as vectors can be modified to accommodate larger genomic sequences. Once formed as a functional artificial chromosome then detection in cells is achieved by fluorescent in situ hybridization (FISH) with relevant DNA probes, while immunoFISH with antibodies for essential centromeric proteins identify the functional HAC. They were originally identified [12], maintained in high frequency and were mitotically stable in human fibrosarcoma cells (HT1080). The structure of the artificial chromosomes was relatively simple compared to host chromosomes.
In 2001, we successfully assembled by conventional cloning a large HAC vector (404 Kb) containing chromosome 17 alphoid DNA and the entire hypoxanthine guanine phosphoribosyltransferase (HPRT, 40 kb) gene [14]. We used polyamines and liposomes to transfect the large HAC HPRT DNA into HT1080 cells deficient for HPRT. The DNA remained intact post transfection and a functional HAC containing the HPRT gene was generated which complemented the genetic deficiency. However, the efficiency of transfer of these large molecules was consistently low (<1 clone in 106 cells), and a drawback to generating HAC routinely in cells. Since the method was challenging for large gene delivery, we developed an alternative more efficient method using Herpes Simplex Virus 1 amplicons. Amplicon delivery substantially increased the efficiency of HAC formation in cells due to the enhanced passage of intact DNA across the cell membrane and direct targeting of the DNA to the nucleus.
1.2. Herpes simplex virus amplicon technology
The strategy of HSV-1 amplicons was originally developed in the 1980's by Niza Frenkel's group [15]. They analyzed defective forms of the HSV virus that were spontaneously derived yet had a high multiplicity of infection (MOI). These viruses contained a bacterial plasmid with the HSV-1 origin of DNA replication (OriS) and the cleavage/packaging signal (Pac), for DNA replication and packaging in cells. Since the plasmids did not contain viral proteins they required the presence of helper virus to infect permissive cells to generate virions. The plasmid was termed the amplicon, as it concatemerized and amplified its sequences via rolling circle replication into a series of tandem repeats up to the size of the HSV viral genome (152 kb), before being packaged into virions. Varying forms of the amplicon were detected, some with different sized plasmids and some forms as mini circles which did not contain any bacterial sequences. The amplicons could also simultaneously carry other eucaryotic sequences which would be similarly amplified. At the time amplicons were useful for elucidating the biology and structure of the HSV virus, identifying the genes involved in its composition, replication and packaging, and shuttling eukaryotic sequences into cells.
Since the early 2000's a newer generation of amplicon vectors were engineered as gene delivery vectors, for larger genes and entire gene loci for regulated expression in the appropriate host cells, taking advantage of the capacity of the amplicon system to carry sequences up to 152 kb. Amplicon vectors for gene transfer were generated that were not dependent on helper virus for packaging virions, since it was observed that contaminating helper virus particles remaining in the amplicon stocks induced cytotoxic and inflammatory responses in host cells infected with the amplicons [16].
One strategy [17], used a co-transfection system to produce high titer amplicon stocks without helper virus contamination. The system involved co-transfecting amplicons with two other vectors which only contained the required sequences for packaging amplicon DNA but could not themselves replicate the viral genome, and were thus lost from the cells. This eliminated the helper virus function and contamination, yet the system was still efficient for delivery to permissive cells to generate virions and produce high titers of amplicons. One vector contained a 172 kb BAC with the entire HSV-1 genome but lacked the HSV-1 OriS and Pac, and was too large to be packaged itself. The other vector which also lacked the Pac, contained an essential functional ICP27 gene for transcriptional regulation of replication genes. Amplicons containing the OriS and Pac were co-transfected with the HSV-1 BAC and ICP27 plasmid into permissive cells, the DNA was packaged into virions and high titer amplicon stocks were successfully generated. Following infection into the recipient target cell there was no viral contamination, and the risk of any resulting oncogenic and inflammatory effects were eliminated. These vectors were known as helper free (HF) amplicon vectors and used to transfer BAC vectors containing the human Epstein–Barr virus (EBV) for episomal gene expression [18].
1.3. Delivery of HAC vectors by HSV-1 amplicons
Using the HF methodology, we modified the HSV-1 amplicons to contain HAC vector DNA for delivery into human cells [19]. Firstly, high titer amplicon stocks (108amplicons/ml) containing HAC DNA were generated. Briefly, amplicons containing HAC DNA, the HSV-1 BAC (fHSVΔPacΔ171+), and the ICP27 vector (pEBH ICP27, 12.2 kb) were co-transfected into permissive african green monkey (AGM) cells. The pEBH ICP27 and BAC fHSVΔPacΔ171+ vectors packaged the amplicons into virions, but not replicated or maintained in the cells. Amplicon particles containing the HAC vector DNA were then purified and used transduced into human cells. Different MOI were selected depending on the amplicon titer. Usually MOI1, 2 and 5 were used to infect cells and produce maximum positive clones. We observed that transduction of amplicons at a higher MOI into recipient cells sometimes lead to toxicity. A high transduction efficiency was usually obtained if the amplicons were concentrated by centrifugation prior to transduction.
After transduction, cells were grown under appropriate antibiotic selection to identify positive cells containing the input HAC DNA. A number of different human cells were successfully infected with the HSV-1 HAC DNA amplicons and generated functional HAC, including immortalized fibroblasts (MRC5V2), glioma cells (G16-9) and human embryonic kidney cells (293) [19]. The delivery was up to 104 times more efficient than standard delivery via non-viral methods.
A major advantage of using HSV-1 amplicons for large DNA delivery is that they can transfer up to 152 kb of DNA (the capacity of the HSV-1 genome), efficiently into the nucleus of a wide range of dividing and non-dividing human cells. Since the HAC vector requires a minimum of 40 kb centromeric alphoid DNA for artificial chromosome formation in cells, then 110 kb of the remaining amplicon can accommodate therapeutic genes and their regulatory regions for gene expression and gene therapy.
1.4. HSV-1 amplicon delivery into human embryonic stem cells
In further work, HSV-1 amplicons containing HAC vector DNA were introduced intact into a human embryonic stem cells (hESc), and genes were expressed from the HAC [20,21]. The vectors contained genes for GFP and RFP proteins as fluorescent markers to monitor delivery of HSV-1 amplicons and maintenance within the target cells as well as antibiotic resistance genes for selection of clones. Following selection for the HAC vector, positive clones were isolated and expanded into cell lines, and chromosome metaphase spreads were prepared for FISH and immunoFISH with the specific DNA probes and centromere antibodies. The efficiency of HAC formation was compared between the cultured HT1080 and hESc. In HT1080 cells, the efficiency of HAC formation was high at 80–90%, and in some cells the input HAC DNA was detected in the host genome at low frequency. In hESc the efficiency of HAC formation was up to 40% and stable in the absence of selection, but no integrated DNA was detected within the cell nucleus in all the hES HAC cell lines. This indicated that factors affecting genome stability were greater in hESc than in the HT1080 cultured cells and was an important discovery for further gene expression and therapy studies. The hES HAC cells remained pluripotent and differentiated into neuronal cells where HAC were maintained [20].
1.5. Simultaneous co-transduction of HSV-1 amplicons
The HSV-1 amplicon HAC system is ideal for large gene delivery up to approximately 100 kb in a single amplicon, since the remaining 50 kb is required for carrying alphoid DNA and selective markers. However, we investigated whether two different amplicons containing genes could be simultaneously co-transduced into host cells to increase the gene loading capacity of the system. Each amplicon would then have the capacity of accommodating over 100 kb of exogenous DNA together with the other sequence requirements. This would allow genetic disorders resulting from possible defects in larger genes, gene loci or several genes to be delivered into cells for complementation of the defect and full gene expression. Previous reports showed that human cells can be simultaneously infected with multiple viruses and sustain transient multigene expression [22].
Initially, we set up two model system studies for co-transduction. Firstly, we investigated whether two different DNA constructs in separate HSV-1 amplicons introduced simultaneously by co-transduction into cells would form a single human artificial chromosome, if the necessary sequences for artificial chromosome formation were provided in the amplicons. Two HSV-1 amplicons, one containing a 17 alphoid DNA HAC vector and RFP, and the other amplicon with a HAC vector containing 21 alphoid DNA and GFP were co-transduced into different cells including, HT1080, G16-9, hESc and iPSc. Following co-transduction of the two amplicons at an MOI 2, the efficiency was measured by RFP and GFP fluorescence of the cells 24 h post transduction in all cells. The level of both RFP and GFP was at a reasonable frequency in all cell lines tested including the hESc and IPS cells. Selected positive clones were expanded into cell lines and the metaphase chromosomes prepared for analysis by FISH and immunoFISH. Clones containing a single HAC were detected by FISH with both 17 and 21 alphoid DNA probes indicating the presence of each alphoid DNA. No HAC with either 17 or 21 alphoid DNA alone was detected in the host cells, indicating that both input alphoid DNAs had undergone recombination with each other and formed a single HAC [23]. Overall, the results showed that simultaneous co-transduction of two amplicons into cells was successful and that the cells were not affected by a secondary infection.
1.6. Multi gene delivery with HSV-1 amplicons
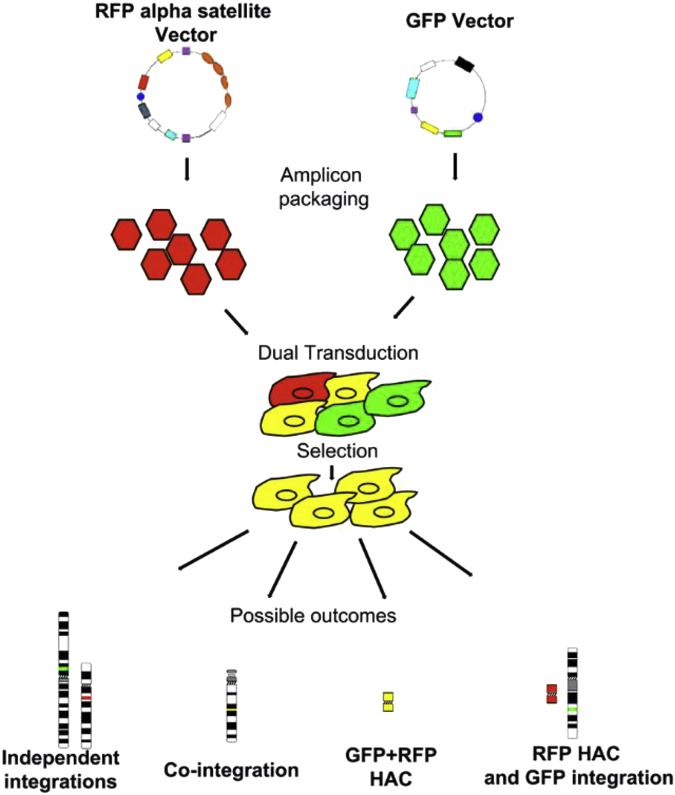
In the second study, we co-transduced two different amplicons where one contained a HAC vector with chromosome 17 alphoid DNA, RFP marker, and the blasticidin resistance gene, and the other amplicon contained a vector lacking alphoid DNA but containing the HPRT minigene, GFP marker and G418 resistance gene. Following on from previous experiments in the first study, we expected the two amplicons to recombine, to form a single HPRT carrying HAC. Alternative outcomes would have been the formation of two wholly independent HAC, or integration events (Fig. 1).
Fig. 1.
A schematic showing the possible outcomes of the delivery of two independent amplicons containing gene expressing HAC vectors after co-transduction into human cells. One amplicon vector contains 17 alphoid DNA and RFP (red), and the other amplicon vector lacking alphoid DNA and GFP (green). After selection of positive cells (yellow), a functional HAC containing both input genes (yellow) is generated (GFP + RFP HAC). The other outcomes show integrations of either input vector DNA (red or green, independent integrations) into host chromosomes, or co-localized on the same chromosome (yellow, co-integration), or a HAC containing 17 alphoid DNA only (red, RFP HAC) and GFP integration into a host chromosome (green).
The two amplicons were co-transduced into HT1080 HPRT-, G16-9, hESc and iPS cells and 24 h post transduction monitored by GFP and RFP fluorescence. The RFP and GFP fluorescence showed a high co-localization frequency in all cell lines. Positive stable clones were selected with both selection agents and in HT1080 HPRT-cells via HAT selection for the HPRT gene. Metaphase chromosome cells from positive clones were analyzed by FISH. HAC were detected in positive HT1080 HPRT-cells containing 17 alphoid DNA and the HPRT gene [23]. The HPRT HAC frequency in the cells ranged from 35 to 72%. One cell line, LWB5 was analyzed in further detail as it contained the HPRT HAC at a high frequency (70%). Western and QRT-PCR data derived from LWB5 cells showed expression of the HPRT gene in the HT1080 HPRT deficient cells compared to none detected in the parental cells. This indicated that the HPRT deficiency was complemented in LWB5, and the gene appropriately expressed from the HAC. The results showed that transduction of two different amplicons, where only one amplicon containing a vector with the chromosome 17 alphoid centromeric DNA, recombined with DNA from the second amplicon lacking alphoid DNA to form a single functional HAC with the genes from both amplicons present and expressed from the HAC in human cells [23]. This indicated that only one amplicon containing HAC centromeric DNA is required to form an artificial chromosome in a dual transduction procedure. The dual transduction system therefore increased the capacity of DNA to be transferred into cells and allows larger and more genes to be loaded into the amplicon vectors.
2. Conclusion
HAC offer long-term gene expression and the complementation of genetic deficiencies without significant risk of insertional mutagenesis and have great potential for ex vivo gene therapy. The amplicon system for gene HAC delivery is a more efficient method of gene transfer than other current methods for large DNA into different human cell types. We showed that gene expressing artificial chromosomes were generated in a human cell line following transduction by two separate HSV-1 amplicons containing different input constructs: one containing the essential alpha satellite sequences for generating a functional HAC and the other a model gene. The dual amplicon transduction was efficient, and the separate constructs recombined to form a single gene-expressing HAC. This technique eliminated the need to incorporate essential HAC forming elements and transgenes within a single amplicon HAC vector construct which can be laborious and challenging, and limits the size of the gene to be accommodated and transferred to cells for expression. It is possible that three or four different amplicon containing HACs could be introduced simultaneously in triple and quadruple transductions. This would allow the transfer of several large constructs or multiple genes in different amplicons into a single cell in one treatment. This has the potential for gene therapy of diseases that involve more than one gene (polygenic), or transfer of larger genes or the entire gene loci and offers exciting prospects for ex and in vivo gene therapy.
Declaration of competing interest
The authors declare that they have no conflicts of interest.
Acknowledgments
We thank all our colleagues for their contributions to our studies mentioned in this work. DM and ZLM were funded by the Wellcome Trust (Grant 075491/Z/04), UK and NIH (NIAMS Grant 1R21AR066938-01), USA.
References
- 1.High K.A., Roncarolo M.G. Gene Therapy Engl J Med. 2019;381:455–464. doi: 10.1056/NEJMra1706910. [DOI] [PubMed] [Google Scholar]
- 2.Nayerossadat N., Maedeh T., Abas Ali P. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012;1:27. doi: 10.4103/2277-9175.98152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufmann K.B., Galy A., Schambach A., Grez M. Gene therapy on the move. EMBO Mol. Med. 2013;5(11):1642–1661. doi: 10.1002/emmm.201202287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouard D., Alazard-Dany N., Cosset F.-L. Viral vectors: from virology to transgene expression. Br. J. Pharmacol. 2009;157:153–165. doi: 10.1038/bjp.2008.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lino C.A., Harper J.C., Carney J.C., Timlin J.A. Delivering CRISPR: a review of the challenges and approaches. 2018;25(1):1234–1257. doi: 10.1080/10717544.2018.1474964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monaco A.P., Larin Z. YACs, BACS, PACs and MACs: artificial chromosomes as research tools. Trends Biotechnol. 1994;12:280–286. doi: 10.1016/0167-7799(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 7.Marschall P, Malik N, Larin Z. Transfer of YACs up to 2.3 Mb into human cells with polyethylenimine. Gene Ther. 6:1634-1637. [DOI] [PubMed]
- 8.Grimes B.R., Monaco Z.L. Artificial and engineered chromosomes:developments and prospects for gene therapy. Chromosoma. 2005;4:230–241. doi: 10.1007/s00412-005-0017-5. [DOI] [PubMed] [Google Scholar]
- 9.Kouprina N, Earnshaw WC, Matsumoto H, and Larionov V. A new generation of human artificial chromosomes for functional genomics and gene therapy. Cell. Mol. Life Sci.. 70:1135-1148. doi: 10.1007/s00018-012-1113-3. [DOI] [PMC free article] [PubMed]
- 10.Farr C.J., Bayne R.A.L., Kipling D., Mills W., Critcher R., Cooke H.J. Generation of a human X-derived minichromosome using telomere-associated chromosome fragmentation. EMBO J. 1995;14:5444–5454. doi: 10.1002/j.1460-2075.1995.tb00228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heller R, Brown KE, Burgtorf C, and Brown WRA. Mini-chromosomes derived from the human Y chromosome by telomere directed chromosome breakage 1996 Proc. Natl. Acad. Sci. U.S.A. 93:7125-7130. [DOI] [PMC free article] [PubMed]
- 12.Harrington J.J., Van Bokkelen G., Mays R.W., Gustashaw K., Willard H.F. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 13.Ohzeki J., Nakano M., OkadaT, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mejia J.E., Willmott A., Levy E., Earnshaw W.C., Larin Z. Functional complementation of a genetic deficiency with human artificial chromosomes. Am. J. Hum. Genet. 2001;69:315–326. doi: 10.1086/321977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frenkel N. The history of the HSV amplicon: from naturally occurring defective genomes to engineered amplicon vectors. Curr. Gene Ther. Jun 2006;3:277–301. doi: 10.2174/156652306777591992. [DOI] [PubMed] [Google Scholar]
- 16.Zaupa C., Revol-Guyot V., Epstein A.L. Improved packaging system for generation of high-level noncytotoxic HSV-1 amplicon vectors using Cre-loxP site-specific recombination to delete the packaging signals of defective helper genomes. Hum. Gene Ther. 2003;14:1049–1063. doi: 10.1089/104303403322124774. [DOI] [PubMed] [Google Scholar]
- 17.Saeki Y., Breakefield X.O., Chiocca E.A. Improved HSV-1 amplicon packaging system using ICP27-deleted, oversized HSV-1 BAC DNA. Methods Mol. Med. 2003;76:51–60. doi: 10.1385/1-59259-304-6:51. [DOI] [PubMed] [Google Scholar]
- 18.Wade-Martins R., Smith E.R., Tyminski E., Chiocca E.A., Saeki Y. An infectious transfer and expression system for genomic DNA loci in human and mouse cells. Nat. Biotechnol. 2001;19:1067–1070. doi: 10.1038/nbt1101-1067. [DOI] [PubMed] [Google Scholar]
- 19.Moralli D., Simpson K.M., Wade-Martins R., Monaco Z.L. A novel human artificial chromosome gene expression system using herpes simplex virus type 1 vectors. EMBO Rep. 2006;7:911–918. doi: 10.1038/sj.embor.7400768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandegar M.A., Moralli D., Khoja S., Cowley S., Chan D.Y., Yusuf M., Mukerherjee S., Blundell M.P., Volpi E.V., Thrasher A.J., James W., Monaco Z.L. Functional artificial chromosomes are generated and stably maintained in human embryonic stem cells. Hum. Mol. Genet. 2011;20:2905–2913. doi: 10.1093/hmg/ddr144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moralli D., Monaco Z.L. Developing de novo human artificial chromosomes in embryonic stem cells using HSV-1 amplicon technology. Chromosome Res. 2015;23:105–110. doi: 10.1007/s10577-014-9456-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansouri M., Bellon-Echeverria I., Rizk A., Ehsaei Z., Cosentino C.C., Silva C.S., Xie Y., Boyce F.M., Davis M.W., Neuhauss S.C.F., Taylor V., Ballmer-Hofer K., Berger I., Berger B. Highly efficient baculovirus-mediated multigene delivery in primary cells. Nat. Commun. 2016;7:11529. doi: 10.1038/ncomms11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan D.Y.L., Moralli D., Wheatley L., Jankowska J.D., Monaco Z.L. Multigene human artificial chromosome vector delivery with herpes simplex virus 1 amplicons. Exp. Cell Res. 2020;388(2):111840. doi: 10.1016/j.yexcr.2020.111840. [DOI] [PMC free article] [PubMed] [Google Scholar]