Summary
Kinetochores mediate chromosome segregation during cell division. They assemble on centromeric nucleosomes and capture spindle microtubules. In budding yeast, a kinetochore links a single nucleosome, containing the histone variant Cse4CENP-A instead of H3, with a single microtubule. Conservation of most kinetochore components from yeast to metazoans suggests that the yeast kinetochore represents a module of the more complex metazoan arrangements. We describe here a streamlined protocol for reconstituting a yeast centromeric nucleosome and a systematic exploration of cryo-grid preparation. These developments allowed us to obtain a high-resolution cryo-EM reconstruction. As suggested by previous work, fewer base pairs are in tight association with the histone octamer than there are in canonical nucleosomes. Weak binding of the end DNA sequences may contribute to specific recognition by other inner kinetochore components. The centromeric nucleosome structure and the strategies we describe will facilitate studies of many other aspects of kinetochore assembly and chromatin biochemistry.
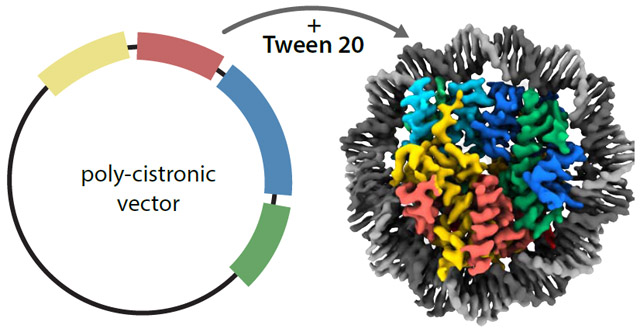
Graphical Abstract
Introduction
Centromeres are the specialized chromosomal regions that connect with mitotic spindle microtubules. They recruit specialized nucleosomes, in which a centromeric histone variant (CENP-A in metazoans; Cse4 in yeast) replaces the canonical histone H3 (Westhorpe and Straight, 2015). Human centromeres comprise long stretches of repetitive-sequence DNA, several megabases in length, with multiple CENP-A containing nucleosomes. In yeast, however, a single Cse4-containing nucleosome assembles on a sequence-specific centromere of about 125 bp (Furuyama and Biggins, 2007). Despite these differences, kinetochore components on metazoan centromeres are homologs of yeast counterparts (Hinshaw and Harrison, 2018a). Because of its simplicity, the yeast centromere is a suitable model for its more complex metazoan counterparts; reconstitution and atomic structure determination of a Cse4-containing nucleosome are thus starting points for mechanistic and structural studies of kinetochore - nucleosome complexes.
Early studies of chromatin used endogenous sources such as chicken erythrocytes or rat liver (4, 5), but more recent standard protocols use expression of individual Xenopus laevis histones as inclusion bodies, denaturing purification, and refolding of a histone octamer. These approaches have led to various high resolution x-ray crystal structures of canonical and centromeric nucleosome (e.g., (Luger, 1997; Luger et al., 1997; Tachiwana et al., 2011)) as well as to cryo-EM structures at moderate resolution of centromeric nucleosomes associated with certain kinetochore components (Chittori et al., 2018; Pentakota et al., 2018; Tian et al., 2018; Zhou et al., 2019). We have now shown that we can co-express in bacteria the four histones of a budding-yeast centromeric nucleosome and purify a soluble octamer in good yield. Several other groups have recently made related advances in co-expression of X. laevis or Homo sapiens H2A/H2B dimers, H3/H4 tetramers, or complete octamers (Anderson et al., 2010; Black et al., 2014; Fang et al., 2016; Guse et al., 2019; Klinker et al., 2014; Lee et al., 2015; Shim et al., 2012; Tian et al., 2018).
Straightforward expression of centromeric histone octamers, which readily bind a 147 bp DNA fragment with the "Widom 601" sequence (Lowary and Widom, 1998), selected for high octamer affinity, has in turn allowed us to explore systematically the conditions for obtaining homogenous fields of particles for high resolution cryo-EM structure determination. We have in particular succeeded in overcoming the strong tendency for the DNA of nucleosomes to unwrap from the histone core during cryo-EM sample preparation. We found that the extent to which DNA unwrapped or dissociated varied with ice thickness and that addition of certain non-ionic detergents both stabilized the particles and decreased the degree of preferential orientation. We have thus circumvented having to add stabilizing ligands to achieve sub-3 Å resolution, such as the single-chain variable-domain antibody fragment (scFv) used recently to stabilize a human CENP-A nucleosome wrapped with a native, α-satellite DNA (Zhou et al., 2019). While the binding of this scFv overcomes several described limitations, the epitope is a conserved acidic patch, important for recruiting many nucleosome binding proteins and thus limiting its use for larger protein complexes.
In this paper, we describe coexpression of a soluble, budding yeast centromeric histone octamer containing S. cerevisiae Cse4 and its reconstitution into a nucleosome with Widom 601 DNA. We further describe the systematic trials needed to avoid loss of DNA from the nucleosome and to determine its atomic structure at 2.7Å resolution. The results will facilitate high-resolution cryo-EM structure determination of both canonical and centromeric nucleosomes bound with protein complexes.
Results
Coexpression of yeast histone octamer containing Saccharomyces cerevisiae Cse4
Previous work has shown that recombinant expression of many kinetochore proteins from Kluyveromyces lactis gives much higher yields than similar procedures with their S. cerevisiae homologs (Cho and Harrison, 2011a; 2011b; Dimitrova et al., 2016). Likewise, while we were unable to coexpress all four components of the S. cerevisiae centromeric octamer, we could obtain more robust expression from a construct encoding the four K. lactis orthologs (Figure S1A, B and Table 1). We therefore expressed histone octamer containing K. lactis H2A, H2B, and H4 and S. cerevisiae Cse4 with a polycistronic coexpression system (ScKl 2) containing a single T7 promoter and ribosome binding site (RBS) for each histone (Figure 1A and Table 1) -- that is, we substituted S. cerevisiae Cse4 into a background of K. lactis core histones H2A, H2B and H4. We also generated two additional chimeric octamer plasmids and found that we needed to retain only the K. lactis H4 to obtain soluble, centromeric octamer (Table 1 and Figure S1C, D).
Table 1.
Histone poly-cistronic vectors
| Plasmid name |
Kl 1 | ScKl 2 | ScKl 3 | ScKl 4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Description | Kl centromeric octamer | ScKl centromeric octamer | ScKl centromeric octamer | ScKl canonical octamer | ||||||||
| Species | Tag | Name | Species | Tag | Name | Species | Tag | Name | Species | Tag | Name | |
| gene 1 | Kl | His6 | H2A | Kl | His6 | H2A | Sc | His6 | H2A | Sc | - | H3 |
| gene 2 | Kl | His6 | H2B | Kl | His6 | H2B | Sc | - | H2B | Sc | His6 | H2A |
| gene 3 | Kl | - | Cse4 | Sc | - | Cse4 | Sc | - | Cse4 | Sc | - | H2B |
| gene 4 | Kl | His6 | H4 | Kl | His6 | H4 | Kl | His6 | H4 | Kl | His6 | H4 |
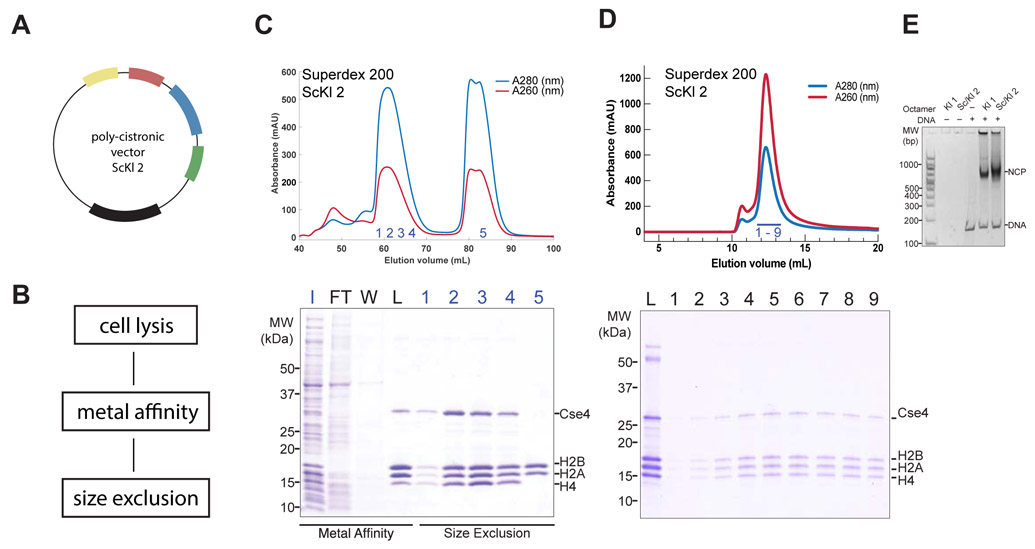
Figure 1.
Reconstitution of yeast centromeric nucleosome. A. Schematic representation of poly-cistronic expression vector ScKl 2 representing histone gene order (see details for other expression vectors in Table 1). B. Summary of histone octamer purification workflow. C. Top, size exclusion chromatogram from Superdex 200 column showing elution profile of ScKl 2. Bottom, coomassie-stained 4-12% SDS-PAGE gel with fractions corresponding to metal affinity purification (Input, Flow Through, Wash) and chromatogram on top (Load and numbered fractions). D. Top, size exclusion chromatogram from Superdex 200 column of 601 DNA wrapped centromeric nucleosome expressed from ScKl 2 vector. Bottom, coomassie-stained 4-12% SDS-PAGE gel with fractions corresponding to chromatogram on top. E. Agarose gel showing 601 DNA wrapped centromeric nucleosomes expressed from Kl 1 and ScKl 2 vectors.
We purified coexpressed octamer with a two-step purification protocol of metal-affinity and size exclusion chromatography (SEC), as outlined in detail in Methods. Purification could be accomplished in one day, a substantial improvement over the time required to produce octamer by refolding. Results for the four coexpression plasmids are in Figures 1B and S1. We assembled Nucleosome Core Particles (NCPs) from the coexpressed octamer using 147 bp 601 DNA and salt dialysis; the NCPs gave the expected EMSA band shift (Figure 1D and E).
Cryo-EM analysis of nucleosome core particles
Nucleosome DNA unwrapping and particle distribution have been described previously as a major bottleneck in obtaining high-resolution structures of the nucleosome by cryo-EM, even with the high affinity, Widom 601 DNA sequence (Cao et al., 2018; Lowary and Widom, 1998). Consistent with these findings, our centromeric nucleosome particles show DNA unwrapping and preferred orientation (top views) under standard vitrification conditions (Figure S2A).
To overcome these limitations, we subjected centromeric NCP to a matrix of anionic, nonionic and zwitterionic detergents during sample vitrification (see Methods). With concentrations close to the critical micelle concentration (CMC) of octyl glucoside (OG), n-dodecyl-β-D-maltoside (DDM), laurydimethylamine oxide (LDAO), and fluorinated Fos-choline, we observed the same degree of DNA unwrapping as with untreated samples (Figure S3), but addition of cetyl trimethyl ammonion bromide (CTAB) or to an even greater extent polysorbate 20 (Tween 20) prevented nucleosome unwrapping and improved the particle orientation distribution for centromeric NCPs (Figure S2B and S3). Similarly, we observed less DNA unwrapping with canonical nucleosomes, suggesting the addition of Tween 20 may help improve nucleosome sample preparation characteristics (Figure S2D). The sample we used to collect high resolution data of the centromeric NCP contained 0.005% Tween 20. This treatment allowed us to obtain a consistent ice thickness of ~25-35nm, measured by collecting tomograms, about 2-3 times the largest particle dimension, and essential for high resolution structure determination (Figure S2C). All data processing was carried out using the cisTEM software package (Grant et al., 2018). Multiple rounds of reference free 2D classification were used to select a total of ~500,000 particles (Figure S4). This particle stack was subclassified using 3D multi model refinement to obtain a total of 266,607 particles, which were used to determine the structure of the centromeric NCP at a resolution of 2.7Å (FSC = 0.143 criterion) (Figure 2A and S4).
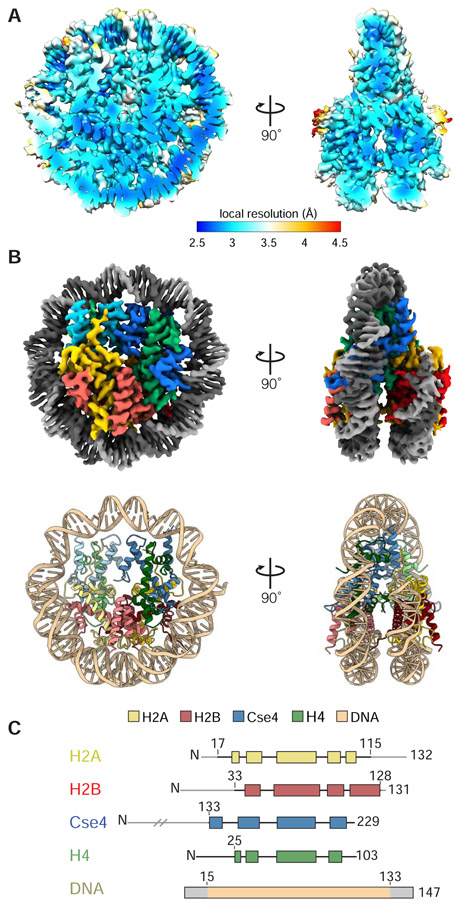
Figure 2.
Cryo-EM structure of yeast centromeric NCP. A. Local resolution map of centromeric NCP based on non-sharpened map. B. Top, cryo-EM density map of centromeric NCP at 2.7Å resolution (unsharpened). Bottom, cartoon representation of centromeric NCP model. (PDB ID: 6UPH) C. Domain representation of histones and 601 Widom DNA where light grey represents regions that have not been built in the model.
Structure of centromeric nucleosome core particles
The 3D reconstruction and the model of a yeast centromeric nucleosome built into that map were consistent with other nucleosome structures (Figure 2B) (Luger et al., 1997; Tachiwana et al., 2011; White et al., 2001a). A (Cse4:H4)2 tetramer associates with two H2A:H2B dimers to form the octameric core. The histone N-terminal tails are disordered and not resolved in the map, and the histone polypeptide chains become visible around helix 1 of each (Figure 2B and C). We built models into density for residues 17–115 of H2A, 33–128 of H2B, 133–229(end) of Cse4, 25–103(end) of H4, and 119 bps of DNA. The map showed clear features for side chains and resolved individual DNA bases. Figure 3B shows representative segments of the map for the DNA and Cse4 (Figure 3A and B).
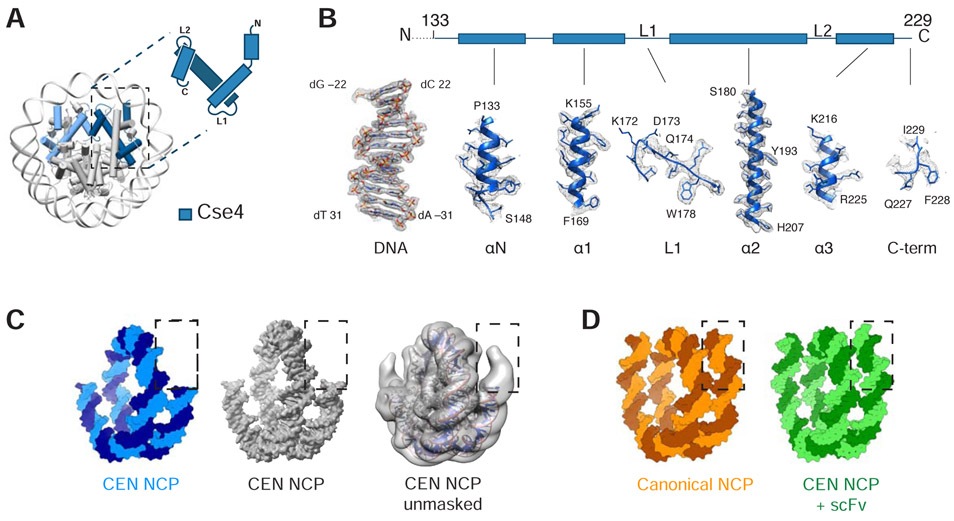
Figure 3.
Structural details of centromeric histone Cse4 and DNA ends. A. Cartoon representation of centromeric (CEN)-NCP and schematic representation of Cse4 histone. B. Density map of centromeric histone Cse4 and a representative DNA region. Dashed line indicates unmodeled N-terminus of Cse4 (residues 1-132). C. Left two, surface representation of 601 DNA from CEN-NCP structure in blue and masked density map of the CEN-NCP at 2.7Å resolution in grey. Right, unmasked density map of the CEN-NCP filtered to low resolution in grey with cartoon representation of the CEN-NCP model (structures from this paper). D. Surface representation of 601 DNA from crystal structure of canonical NCP (PDB ID: 1ID3) and of native apha-satellite DNA from cryo-EM structure of CEN-NCP+scFv (PDB ID: 6E0P) (21, 29).
K. lactis and S. cerevisiae histones H2A, H2B, and H4 differ by 3-7 residues each (H2A: 5 H2B: 7 H4: 3). Six of these substitutions are visible in the structure (H2A: 1 H2B: 2 H4: 3). The backbone at each of these positions has the same conformation as it does in the crystal structure of a canonical S. cerevisiae histone octamer. The RMSD of all atoms (backbone, side chain, DNA) between this structure and the yeast canonical nucleosome (PDB ID: 1ID3) is 1.1 Å (White et al., 2001a). We conclude that the co-expressed octamers can be used to make nucleosome core particles that are identical to ones made by refolding and that using K. lactis H2A, H2B, and H4 in place of their equivalents from S. cerevisiae does not generate any detectable perturbations.
The yeast centromeric histone octamer binds strongly to fewer base pairs of DNA than does a canonical octamer, as previously seen in an MNase protection assay using in vitro reconstituted Cse4-nucleosome (Kingston et al., 2011). Strong density was present in our map for only 119 bp of DNA, compared with 146 bp in the yeast canonical nucleosome crystal structure (Figure 3C) (White et al., 2001a). Additional DNA base pairs are seen in the low resolution unmasked map, although the weak density precludes molecular modeling (Figure 3C). A similar difference has also been reported for the crystal structure of the human centromeric nucleosome, whereas extended DNA ends have been observed in recent single particle cryo-EM structures of centromeric nucleosomes in the presence of a binding partner or a chaperone, CENP-N and scFv, respectively (Figure 3D) (Chittori et al., 2018; Pentakota et al., 2018; Tachiwana et al., 2011; Tian et al., 2018; Zhou et al., 2019).
Cse4 L1 loop and C-terminus
The histone fold domains of Cse4 and H3 (defined as residues 130-end and 40-end, respectively) are only 59% identical (83% similar) at corresponding positions in their amino-acid sequences. Although residue differences between the two are distributed throughout the histone fold domain, two areas visible in the structure are known binding sites for partners of Cse4: (i) the CATD (CENP-A Targeting Domain consisting of the L1 loop and the α2 helix) and (ii) the C-terminal segment. (A third region within the N-terminal tail interacts with COMA (30) but is not visible in our structure.) Within the Cse4 CATD (L1 loop + α2 helix), the structure shows that the α2 helix has essentially the same structure as its counterpart in H3, but that the L1 loop, with three extra residues, has a distinct, well-defined conformation (Figure 4A and B). Relevant primary structure differences between Cse4 and H3 around the L1 loop are (i) substitution of T170 in Cse4 for the corresponding residue of K79 in H3 and (ii) a three-residue insertion of KDQ (residues 172-4) in the loop itself (Figure 4B). The change K79T170 does not alter the position of the Cα. The Cα atoms of the longer L1 loop protrude 6-7Å further outward from the nucleosome surface than they do on H3. The density for the backbone is clear. The side chains of K172 and D173 are not resolved (Figure 3B), so we modeled their most likely rotamers. The solvent-accessible, L1 loop is probably an interaction site for Chl4: its counterpart in the human centromeric nucleosome contacts CENP-N, the Chl4 ortholog (Fang et al., 2015).
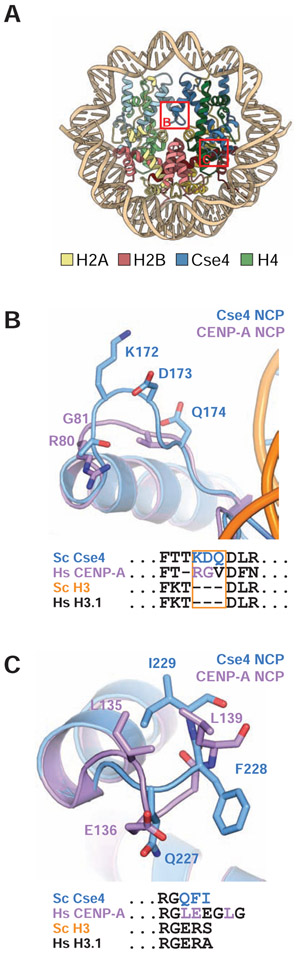
Figure 4.
Structural details of Cse4 L1 loop and C-terminus. A. Model of centromeric nucleosome indicating zoomed in regions represented in B and C. B. Overlay of L1 loop from S. cerevisiae Cse4 (blue) and human CENP-A (purple). Sequence alignment of this region is shown below. C. Overlay of S. cerevisiae Cse4 (blue) and human CENP-A (purple) C-terminus with sequence alignment of the region shown below.
The three C-terminal residues of Cse4, QFI (replacing H3 counterparts ERS), create a hydrophobic patch, important for Mif2 binding (Kato et al., 2013). Their structure is consistent with the conformation seen in a previous crystal structure of a K. lactis Cse4/H4 tetramer (Figure 4C) (Cho and Harrison, 2011b). Because of some ambiguity in density for F228 and I229, we modeled these residues as in that crystal structure, with F228 buried in the octamer core and I229 projecting outward at the nucleosome surface. Other residue changes are scattered throughout the histone fold domain of Cse4, but we could find no Cα positional shifts from positions of the corresponding atoms in H3.
Discussion
A detailed structural characterization of the nucleosome and understanding its interaction with nucleosome binding protein complexes are essential starting points for investigating the structural and functional properties of chromatin associated molecular machines. The strategy we used, in which a codon-optimized K. lactis H4 was co-expressed with the other three histones of the S. cerevisiae centromeric octamer, was critical for the high-level expression suitable for structure determination. The comparatively higher resolution of the reconstruction shows that our preparation and vitrification procedures have overcome many of the technical problems encountered in previous efforts to study nucleosomes by cryo-EM (Bilokapic et al., 2017; Chua et al., 2016). We cannot rule out the possibility that some factor beyond the K. lactis H4, such as gene order in the expression vector, optimized sequence, or mRNA structure also contributed to the expression difference.
Centromeric nucleosomes are prone to dissociation during cryo-EM grid preparation, yielding images cluttered with free DNA, possibly due to the disruption of nucleosome particles at the air water interface (Cao et al., 2018). We paid particular attention to optimizing the ice thickness to increase image contrast by varying sample volume, grid manufacture and blot time, and tested various detergents to preserve nucleosome structure integrity and for relief from preferential orientation. Both parameters influence the behavior of a nucleosome at the air-water interface. We measured ice thickness by collecting tomograms of individual grid regions and used only the areas with a thickness of approximately 25-35nm to determine the final reconstruction.
A recent study showed that it is possible to use a single-chain antibody fragment (scFv) to stabilize a human centromeric nucleosome containing CENP-A and a native α-satellite DNA and to determine its structure at 2.6 Å resolution (Zhou et al., 2019). The use of this scFv fragment led to a high resolution structure, perhaps because its binding stabilized the particle and mitigated effects of the air-water interface. A similar investigation in the absence of the scFv fragment could only deliver a structure of the nucleosome at 3.4 Å resolution. Because the scFv covers an acidic patch region of the nucleosome important to recruit the majority of nucleosome binding proteins, it cannot be used for high resolution structures of these nucleosome bound targets. The technical developments reported here have led to a comparable resolution in the absence of a stabilizing scFv fragment, and we therefore expect that they will be valuable for high resolution structural investigation of nucleosomes bound with chromatin remodeling complexes and nucleosome modifying enzymes.
Structures of the human centromeric nucleosome show that this nucleosome has fewer well-ordered DNA base pairs than do canonical nucleosomes containing histone H3 (16, 17, 34). Our structure shows that the same is also true for yeast. Two features of the centromeric histone contribute to unwrapping at the ends of the DNA: fewer interactions of DNA with the N-terminal tail and amino acid differences between H3 and Cse4CENP-A in the αN helix. Swapping the human CENP-A N-terminal tail and αN helix into H3 is sufficient to cause partial unwrapping of the DNA (35); our structure suggests that the same would be true for the yeast proteins. A recent structure of the S. cerevisiae Ctf19 complex suggests that the unwrapping of DNA ends is probably crucial for allowing other nucleosome-proximal factors of the kinetochore to bind (hinshawStephen C Harrison, 2019). Thus, the characteristics of the centromeric histone octamer that lead to release of the DNA ends contribute to specific recognition of a Cse4 nucleosome for kinetochore assembly.
The C-terminal residues of CENP-ACse4 are important for its interaction with CENP-CMif2 (Kato et al., 2013). Moreover, a segment related to the conserved "CENP-C box" binds a nucleosome containing a chimeric H3 with C-terminal residues from CENP-A through a tyrosine-tryptophan pair in hydrophobic contact with I133 and L137 of CENP-ACse4 (Kato et al., 2013). In the conformation of the C-terminal residues of Cse4 in our structure, which is the same as its conformation in a free K. lactis Cse4:H4 tetramer, F228 and I229 are poised to make hydrophobic contacts with yeast Mif2, whose CENP-C motif is conserved with its human ortholog (Figure 4C) (Cho and Harrison, 2011b; Cohen et al., 2008).
A second component of human kinetochores known to interact directly with CENP-ACse4 is CENP-NChl4. In structures of the N-terminal domain of human CENP-N with a human centromeric nucleosome, the most extensive CENP-N interactions are with DNA, but a well-defined interaction of N-terminal residues of CENP-N with the L1 loop of CENP-ACse4 appears to determine the specific point of contact (Chittori et al., 2018; Pentakota et al., 2018; Tian et al., 2018). As expected from alignment of sequences, including those of other point-centromere yeasts, the N-terminal domain of S. cerevisiae Chl4 has essentially the same structure as that of CENP-N (Hinshaw and Harrison, 2018b). We infer that all these proteins are likely to bind similarly to the centromeric nucleosome and that the L1 loop of Cse4 probably interacts with residues near the Chl4 N-terminus. The loop is three residues longer than the corresponding loop in H3, far enough from the octamer surface to facilitate interaction with Chl4 and to differentiate Cse4 from its non-centromeric counterpart, H3.
Interaction with Chl4 need not exclude other interactions, either at the same time or at different stages of kinetochore assembly. One study in yeast indicates that the L1 loop contributes to Mif2 binding affinity (Xiao et al., 2017); the published structure referenced above, which used a chimeric H3 with only the C-terminal residues of CENP-A, does not firmly rule out a contact. Our own experiments (unpublished) do not, however, indicate any cooperativity between Chl4 and Mif2. Other proteins that must recognize Cse4 or the centromeric nucleosome selectively include Psh1, which ubiquitinates Cse4 to regulate its localization (Hewawasam et al., 2010; Ranjitkar et al., 2010). Scm3, the chaperone that associates with Ndc10 and recruits Cse4-H4 heterodimers, depends on specific contacts with helix H2 of Cse4, but does not contact the L1 loop (Cho and Harrison, 2011b).
The centromeric nucleosome structure described here extends to substantially higher resolution than those from other organisms reported previously and has been achieved without the addition of chaperones or specialized binding partners. Our finding, that use of K. lactis H4 with the three other histones from S. cerevisiae gives high yield of native octamer, also greatly facilitated the structure determination. This strategy will benefit the investigation of many other aspects of yeast chromatin biochemistry, kinetochore assembly and regulation.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Inquiries for further information may be directed to and fulfilled by Lead Contact Yoana N. Dimitrova. To request reagents please submit a form to Genentech at https://www.gene.com/scientists/mta.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All proteins were expressed in Rosetta 2 E. coli cells (Novagen) in LB medium. Expression was induced at OD600 of 0.6 – 1.0 with 0.25-0.5 mM IPTG and cells grown at 18°C for 12-18 hrs.
METHOD DETAILS
Expression and purification of histones
Genes encoding S. cerevisiae and K. lactis histones were cloned into single expression vectors with a ligation-independent cloning method (LIC). For co-expression, genes were assembled into a poly-cistronic insert with a PCR-based method and cloned into a LIC vector. Plasmids were verified by sequencing.
Proteins were expressed in Rosetta 2 E. coli cells (Novagen). Culture growth was done at 37°C with shaking at 220 rpm unless otherwise stated. ~5 colonies were streaked from a transformation plate and incubated in 5ml each LB medium with appropriate antibiotics and grown for 3-4h. The starter cultures were then transferred to 150 mL LB and grown overnight. The next morning, 12L of media were seeded 1:100 and grown to an OD600 of 0.6 – 1.0. The colonies typically required 8 – 12 hrs to reach this point. They were then induced with 0.25-0.5 mM IPTG and grown overnight at 18°C. Cells were harvested by centrifugation, resuspended in high salt buffer (HSB) (2M NaCl, 50 mM HEPES pH 7.5, 10% glycerol, 1 mM TCEP) containing protease inhibitors (aprotonin, pepstatin, leupeptin, and PSMF) and frozen directly at −80°C for storage.
Cells were resuspended in 10 mL high salt buffer per L of culture (120 mL for a 12 L prep) and lysed by sonication. The lysate was clarified by centrifugation at 40,000xg in a Beckman centrifuge with a JA-20 rotor at 4°C for 1h. The supernatant was incubated with 4 mL TALON metal affinity resin (Clontech) and incubated at 4°C for 1h with agitation, then transferred to a gravity column. After discarding the flow through and washing, the protein was eluted with HSB containing 50 mM EDTA and 400 mM Imidazole. The eluate was concentrated in an Amicon centrifugal filter (10,000 MWCO) by centrifuging at 3000xg and 4°C at 15 minute intervals until the volume was less than 2mL. This typically required 2-3h total time. The concentrated eluate was run on a 120 mL Superdex 200 size exclusion column (GE). Aliquots were frozen at −80°C. SDS-PAGE was done on Bio-Rad 4-20% polyacrylamide gels, with staining by coomassie blue. The 147 bp Widom 601 DNA sequence was produced by PCR and purified on a HiTrap Q column.
Nucleosome wrapping and CryoEM sample preparation
DNA and histone octamer were mixed in a 1.1:1.0 molar ratio. Nucleosomes were reconstituted by the salt gradient dialysis method using a Rabbit peristaltic pump and a gradient over 60h at 4°C. Excess DNA was removed by further purifying the nucleosomes over a 24 mL Superdex200 or 2.4 mL Superose6 size exclusion columns. The SEC buffer was 30 mM HEPES pH 7.4, 150 mM NaCl, and 1 mM TCEP.
After SEC, nucleosome-containing fractions were brought to a concentration of 0.5-1.2 mg/ml determined by measuring the DNA concentration on a NanoDrop spectrophotometer (Fisher), based on a predicted mass of 240 kDa for the Sc/Kl centromeric nucleosome.
The purified samples were applied on 20s glow discharged (Solarus) Quantifoil R1.2/1.3 or R2/2 gold grids (200 mesh) and flash frozen in liquid ethane on an FEI Vitrobot, using a 2.5 - 5 s blot time and blotting force of 8 at 4°C and 100% humidity. Samples were vitrified under the following detergent conditions at their CMC: Octyl Glucoside (OG), n-Dodecyl-β-D-Maltoside (DDM), Laurydimethylamine oxide (LDAO), Fluorinate-Fos-Choline, Cetyl Trimethyl Ammonion Bromide (CTAB) and Polysorbate 20 (Tween 20)
Data collection
CryoEM data were collected using a Titan Krios G2 (Thermo Fisher Scientific) operated at 300keV, with a BioQuantum energy filter equipped with a K2 Summit direct detector (Gatan, Inc. Pleasanton, CA). Movies were acquired in energy filtered mode with an energy slit of 20eV, magnification of ~165kx (pixel size of 0.849 angstroms/pixel). The electron beam had a flux rate of 4.9 electrons/(angstrom)2/s, and movies were acquired at 4 frames/s for duration of 10 seconds, for a total electron interaction of 49 electrons/(angstrom)2. A 70μm condenser aperture and a 100μm objective aperture were used while imaging. Data were collected using SerialEM automated acquisition software (Mastronarde, 2005). Micrographs for the direct comparison between CEN-NCP and H3-NCP in the presence or absence of Tween 20 were collected on a Talos F200C (Thermo Fisher Scientific) operated at 200keV with a Ceta 16M camera (Thermo Fisher Scientific).
Ice thickness determination on cryo-EM girds
The tomogram was acquired on a Titan Krios at 300keV using a Bioquantum energy filter with a K2 Summit direct electron detector (Gatan), at a pixel size of 4.39Å. Data were collected using serialEM (Mastronarde, 2005).
Tilt series was collected from −45 to +45 degrees at 3 degree increments with a defocus of −5 μM. A total dose of 100 e/A2 was used for data acquisition. The tomographic reconstruction was done using IMOD (Mastronarde and Held, 2017). The reconstruction showed an ice thickness of approximately 33 nm in the areas of single particle data acquisition.
Cryo-EM data processing
Cryo-EM data (unbinned) were processed with cisTEM without applied symmetry (C1) (Grant et al., 2018). 4926 movies were corrected for frame motion using the Unblur algorithm and the contrast transfer function (CTF) was estimated with CTFfind {Mindell:2003ib, Rohou:2015wx, Grant:2015kv}. Images were filtered based on the detected fit resolution better than 3.75 Å (Figure S4A). Particles were automatically picked using the ‘ab-initio’ algorithm and a circular blob as a template with a radius of 25 Å and an exclusion radius of 55 Å (Sigworth, 2004). 862,840 particles were extracted using a box size of 192 px and subjected to two rounds of reference free 2D classification (each using a target of 500 classes) based on the maximum likelihood algorithm (Figure S4B) (Scheres et al., 2005; Sigworth, 1998). The classes obtained were visually inspected and curated to remove ice contamination, aggregation and other false positives, yielding to a final dataset comprising 491,261 particles. Seven of these particle classes, each representing a distinct view of the nucleosome, were used to calculate an ab-initio 3D reconstruction using cisTEM software package (Grigorieff, 2016). The resulting volume was used as a starting reference for a multi model refinement using the Auto Refine algorithm (cisTEM), using an initial resolution limit of 20 Å. Particles sorted into the most represented 3D class (54.3% of all particles) were extracted and further refined in iterative rounds of manual and global refinement including masking and CTF refinement until convergence of the FSC curve (Figure S4D). The resulting map was sharpened with the following parameters: flattening from a resolution of 8 Å, applying a pre-cut-off B-factor of −90 Å2 from the origin of reciprocal space, applying a post-cut-off B-factor of −10 Å2 and applying a figure-of-merit filter (Rosenthal and Henderson, 2003). To determine whether the DNA was completely or partially unwrapped, we perform manual-refine using an unmasked sum of half-maps filtered to low resolution. We low pass filtered this volume using the gaussian algorithm provided by the Chimera Software with a standard deviation of 3.0 (Pettersen et al., 2004).
Model Building
We built the nucleosome model into the map using iterative rounds of model building and local refinement in Coot (Emsley et al., 2010) and global real space refinement in Phenix (Adams et al., 2010). We took the DNA from a crystal structure of a 601 NCP (PDB 3LZ0) (Vasudevan et al., 2010) and performed a rigid body in UCSF Chimera (Pettersen et al., 2004). The resolution of the structure allowed an unambiguous fit of this asymmetric sequence. For the core histones, we used a crystal structure of the yeast canonical nucleosome (PDB 1ID3) (White et al., 2001b), altered individual residues to correspond to the sequences of K. lactis H2A, H2B, and H4 and S. cerevisiae Cse4, and performed a rigid body fit. Further rounds of manual building in Coot and real space refinement in Phenix led to the final model (Supplemental Table S1). Graphics were generated in PyMol (DeLano, 2002), Chimera, and ChimeraX (Goddard et al., 2017; Pettersen et al., 2004). Local resolution was plotted with Resmap (Kucukelbir et al., 2013).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics generated from the cryo-EM data processing, refinement and structure validation are shown in Table S1 and Figure S4.
DATA AND SOFTWARE AVAILABILITY
Atomic coordinates have been deposited in the Protein Data Bank with accession number: 6UPH and in the Electron Microscopy Data Bank with code 20839.
DATA AND SOFTWARE AVAILABILITY
Software used in this this study has been previously published as detailed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Octyl Glucoside (OG) | Anatrace | Cat#0311 |
| n-Dodecyl-β-D-Maltoside (DDM) | Anatrace | Cat#D310 |
| Laurydimethylamine oxide (LDAO) | Anatrace | Cat#D360 |
| Fluorinate-Fos-Choline | Anatrace | Cat#F300F |
| Cetyl Trimethyl Ammonion Bromide (CTAB) | Millipoer-Sigma | Cat#52365 |
| Polysorbate 20 (Tween 20) | Millipoer-Sigma | Cat#P9416 |
| Deposited Data | ||
| Atomic coordinates, CEN-NCP structure | This study | PDB: 6UPH EMD: 20839 |
| Experimental Models: Organisms/Strains | ||
| E. coli Rosetta 2 (DE3) Singles | Novagen | Cat#71400 |
| Recombinant DNA | ||
| Kl 1 | This study | N/A |
| ScKl 2 | This study | N/A |
| ScKl 3 | This study | N/A |
| ScKl 4 | This study | N/A |
| Software and Algorithms | ||
| cisTEM | Grant et al., 2018 | https://cistem.org/ |
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org |
| PyMol | DeLano, 2002 | http://www.pymol.org/ |
| UCSF Chimera | Pettersen et al., 2004 | http://www.cgl.ucsf.edu/chimera/ |
| UCSF ChimeraX | Goddard et al., 2018 | https://www.rbvi.ucsf.edu/chimerax/ |
| Coot | Emsley et al., 2010 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
Supplementary Material
Reconstitution and atomic resolution structure determination of centromeric nucleosomes are the starting point for in-depth studies of kinetochore-nucleosome complexes. Migl et al describe the generation of high resolution cryo-EM structure of yeast centromeric nucleosome enabled by optimal grid preparation that avoids DNA unwrapping. The structure shows key kinetochore assembly sites.
Highlights.
Production of monodisperse, soluble nucleosomes using poly-cistronic vectors
Optimal grid preparation for high-resolution cryo-EM structures of nucleosomes
Cryo-EM structure of yeast centromeric nucleosome shows kinetochore assembly sites
Acknowledgments
Alexis Rohou (Genentech) for cisTEM data processing advice. Plasmids #3 and 4 were a generous gift from Stephen Hinshaw (HMS). The research was supported by the Howard Hughes Medical Institute (D.M., Y.N.D., Y.K., S.C.H.) and by NIH grant GM62580 (YK). Y.N.D, C.C., M.K. and C.A. are employees of Genentech, Inc.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst (2010). D66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M, Huh JH, Ngo T, Lee A, Hernandez G, Pang J, Perkins J, and Dutnall RN (2010). Co-expression as a convenient method for the production and purification of core histones in bacteria. Protein Expression and Purification 72, 194–204. [DOI] [PubMed] [Google Scholar]
- Bilokapic S, Strauss M, and Halic M (2017). Histone octamer rearranges to adapt to DNA unwrapping. Nat Struct Mol Biol 25, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, foltz DR, Chakravarthy S, Luger K, woods VL, and Cleveland DW (2014). Structural determinants for generating centromeric chromatin. 430, 1–5. [DOI] [PubMed] [Google Scholar]
- Cao S, Zhou K, Zhang Z, Luger K, and Straight AF (2018). Constitutive centromere-associated network contacts confer differential stability on CENP-A nucleosomes in vitro and in the cell. Molecular Biology of the Cell 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittori S, Hong J, Saunders H, Feng H, Ghirlando R, Kelly AE, Bai Y, and Subramaniam S (2018). Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science 359, 339–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho U-S, and Harrison SC (2011a). Ndc10 is a platform for inner kinetochore assembly in budding yeast. Nature Publishing Group; 19, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho U-S, and Harrison SC (2011b). Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Pnas 108, 9367–9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua EYD, Vogirala VK, Inian O, Wong ASW, Nordenskiöld L, Plitzko JM, Danev R, and Sandin S (2016). 3.9 Å structure of the nucleosome core particle determined by phase-plate cryo-EM. Nucleic Acids Res 44, 8013–8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen RL, Espelin CW, De Wulf P, Sorger PK, Harrison SC, and Simons KT (2008). Structural and Functional Dissection of Mif2p, a Conserved DNA-binding Kinetochore Protein. Molecular Biology of the Cell 16, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002). The PyMOL Molecular Graphics System, Version 1.8. Schrödinger, LLC. [Google Scholar]
- Dimitrova YN, Jenni S, Valverde R, Khin Y, and Harrison SC (2016). Structure of the MIND Complex Defines a Regulatory Focus for Yeast Kinetochore Assembly. Cell 167, 1014–1021.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Liu Y, Wei Y, Deng W, Yu Z, Huang L, Teng Y, Yao T, You Q, Ruan H, et al. (2015). Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N. Genes Dev. 29, 1058–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Q, Chen P, Wang M, Fang J, Yang N, Li G, and Xu R-M (2016). Human cytomegalovirus IE1 protein alters the higher-order chromatin structure by targeting the acidic patch of the nucleosome. eLife 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S, and Biggins S (2007). Centromere identity is specified by a single centromeric nucleosome in budding yeast. PNAS 104, 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2017). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Science 27, 14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Rohou A, and Grigorieff N (2018). cisTEM, user-friendly software for single-particle image processing. eLife 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N (2016). Frealign: An exploratory tool for single particle cryo-EM. Methods Enzymology 579, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Fuller CJ, and Straight AF (2019). A cell-free system for functional centromere and kinetochore assembly. Nature Protocols 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, and Gerton JL (2010). Psh1 Is an E3 Ubiquitin Ligase that Targets the Centromeric Histone Variant Cse4. Molecular Cell 40, 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw SM, and Harrison SC (2018a). Kinetochore Function from the Bottom Up. Trends in Cell Biology 28, 22–33. [DOI] [PubMed] [Google Scholar]
- Hinshaw SM, and Harrison SC (2018b). An Iml3-Chl4 Heterodimer Links the Core Centromere to Factors Required for Accurate Chromosome Segregation. Cell Reports 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw S, Harrison Stephen C (2019). The structure of the Ctf19c/CCAN from budding yeast. 1–21. [Google Scholar]
- Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, and Bai Y (2013). A Conserved Mechanism for Centromeric Nucleosome Recognition by Centromere Protein CENP-C. Science 340, 1110–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston IJ, Yung JSY, and Singleton MR (2011). Biophysical Characterization of the Centromere-specific Nucleosome from Budding Yeast. The Journal of Biological Chemistry 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinker H, Haas C, Harrer N, Becker PB, and Mueller-Planitz F (2014). Rapid Purification of Recombinant Histones. PLoS ONE 9, e104029–e104029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, and Tagare HD (2013). Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-T, Gibbons G, Lee SY, Nikolovska-Coleska Z, and Dou Y (2015). One-pot refolding of core histones from bacterial inclusion bodies allows rapid reconstitution of histone octamer. Protein Expression and Purification 110, 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, and Widom J (1998). New DNA Sequence Rules for High Affinity Binding to Histone Octamer and Sequence-directed Nucleosome Positioning. Journal of Molecular Biology 276, 1–24. [DOI] [PubMed] [Google Scholar]
- Luger KEA (1997). Characterization of Nucleosome Core Particles Containing Histone Proteins Made in Bacteria. 1–11. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, and Richmond TJ (1997). Crystal structure of the nucleosome core particle at 2.8 Å resolution. 389, 1–10. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. Journal of Structural Biology 152, 36–51. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN, and Held SR (2017). Automated tilt series alignment and tomographic reconstruction in IMOD. Journal of Structural Biology 197, 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentakota S, Zhou K, Smith C, Maffini S, Petrovic A, Morgan GP, Weir JR, Vetter IR, Musacchio A, and Luger K (2018). Decoding the centromeric nucleosome through CENP-N. 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, and Biggins S (2010). An E3 Ubiquitin Ligase Prevents Ectopic Localization of the Centromeric Histone H3 Variant via the Centromere Targeting Domain. Molecular Cell 40, 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, and Henderson R (2003). Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-particle Electron Cryomicroscopy. Journal of Molecular Biology 333, 721–745. [DOI] [PubMed] [Google Scholar]
- Scheres SHW, Valle M, and Carazo JM (2005). Fast maximum-likelihood refinement of electron microscopy images. Bioinformatics 21, ii243–ii244. [DOI] [PubMed] [Google Scholar]
- Shim Y, Duan M-R, Chen X, Smerdon MJ, and Min J-H (2012). Polycistronic coexpression and nondenaturing purification of histone octamers. Analytical Biochemistry 427, 190–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigworth FJ (1998). A Maximum-Likelihood Approach to Single-Particle Image Refinement. 1–12. [DOI] [PubMed] [Google Scholar]
- Sigworth FJ (2004). Classical detection theory and the cryo-EM particle selection problem. Journal of Structural Biology 145, 111–122. [DOI] [PubMed] [Google Scholar]
- Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park S-Y, et al. (2011). Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476, 232–235. [DOI] [PubMed] [Google Scholar]
- Tian T, Li X, Liu Y, Wang C, Liu X, Bi G, Zhang X, Yao X, Zhou ZH, and Zang J (2018). Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Nature Publishing Group 28, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan D, Chua EYD, and Davey CA (2010). Crystal Structures of Nucleosome Core Particles Containing the “601” Strong Positioning Sequence. 1–10. [DOI] [PubMed] [Google Scholar]
- Westhorpe FG, and Straight AF (2015). The Centromere: Epigenetic Control of Chromosome Segregation during Mitosis. Cold Spring Harb Perspect Biol 7, a015818–a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Suto RK, and Luger K (2001a). Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. Embo J. 20, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Suto RK, and Luger K (2001b). Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. Embo J. 20, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Wang F, Wisniewski J, Shaytan AK, Ghirlando R, FitzGerald PC, Huang Y, Wei D, Li S, Landsman D, et al. (2017). Molecular basis of CENP-C association with the CENP-A nucleosome at yeast centromeres. Genes Dev. 31, 1958–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B-R, Yadav KNS, Borgnia M, Hong J, Cao B, Olins AL, Olins DE, Bai Y, and Zhang P (2019). Atomic resolution cryo-EM structure of a native- like CENP-A nucleosome aided by an antibody fragment. Nature Communications 10, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates have been deposited in the Protein Data Bank with accession number: 6UPH and in the Electron Microscopy Data Bank with code 20839.
Software used in this this study has been previously published as detailed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Octyl Glucoside (OG) | Anatrace | Cat#0311 |
| n-Dodecyl-β-D-Maltoside (DDM) | Anatrace | Cat#D310 |
| Laurydimethylamine oxide (LDAO) | Anatrace | Cat#D360 |
| Fluorinate-Fos-Choline | Anatrace | Cat#F300F |
| Cetyl Trimethyl Ammonion Bromide (CTAB) | Millipoer-Sigma | Cat#52365 |
| Polysorbate 20 (Tween 20) | Millipoer-Sigma | Cat#P9416 |
| Deposited Data | ||
| Atomic coordinates, CEN-NCP structure | This study | PDB: 6UPH EMD: 20839 |
| Experimental Models: Organisms/Strains | ||
| E. coli Rosetta 2 (DE3) Singles | Novagen | Cat#71400 |
| Recombinant DNA | ||
| Kl 1 | This study | N/A |
| ScKl 2 | This study | N/A |
| ScKl 3 | This study | N/A |
| ScKl 4 | This study | N/A |
| Software and Algorithms | ||
| cisTEM | Grant et al., 2018 | https://cistem.org/ |
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org |
| PyMol | DeLano, 2002 | http://www.pymol.org/ |
| UCSF Chimera | Pettersen et al., 2004 | http://www.cgl.ucsf.edu/chimera/ |
| UCSF ChimeraX | Goddard et al., 2018 | https://www.rbvi.ucsf.edu/chimerax/ |
| Coot | Emsley et al., 2010 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |