Abstract
Background
Understanding how growth state influences Mycobacterium tuberculosis responses to antibiotic exposure provides a window into drug action during patient chemotherapy. In this article, we describe the transcriptional programs mediated by isoniazid (INH) during the transition from log-phase to nonreplicating bacilli, from INH-sensitive to INH-tolerant bacilli respectively, using the Wayne model.
Results
INH treatment did not elicit a transcriptional response from nonreplicating bacteria under microarophilic conditions (NRP2), unlike the induction of a robust and well-characterized INH signature in log-phase bacilli.
Conclusion
The differential regulation (between drug-free NRP2 and log-phase bacilli) of genes directly implicated in INH resistance could not account for the abrogation of INH killing in nongrowing bacilli. Thus, factors affecting the requirement for mycolic acids and the redox status of bacilli are likely responsible for the reduction in INH efficacy. We speculate on additional mechanisms revealed by transcriptome analysis that might account for INH tolerance.
While effective at resolving active Mycobacterium tuberculosis (TB) disease, current tuberculosis chemotherapy requires multiple drugs to be taken over 6 months to clear infection of genotypically drug-sensitive bacilli. This long treatment period exerts huge demands on healthcare infrastructure and finances, and risks the emergence of drug resistance through poor patient compliance. The search for a more effective shortened drug regimen is complicated by the presence of persistent organisms, likely phenotypically drug-tolerant bacilli that require prolonged drug treatment to clear [1,2].
Phenotypic drug tolerance, unlike genetically defined resistance, is a transient bacterial state in which many antibiotics are ineffective. Tolerance is hypothesized to be dependent upon pre-existing subpopulations of genetically susceptible bacteria in a slow/nonmultiplying state that are refractory to drug action [3]. This small fraction of phenotypically distinct bacteria enable bacterial populations to survive dramatic changes in the environment [4]. In addition to stochastic mechanisms that create subpopulations of nongrowing cells in a phenotypically heterogeneous bacterial community, recent evidence suggests that tolerant bacterial subpopulations are generated after antibiotic stress [5]. In TB clinical settings, phenotypically tolerant bacilli may be responsible for the persistence of M. tuberculosis bacilli through the drug-treatment period [6]. We speculate that phenotypically tolerant bacilli in patients may exist as a result of:
-
□
Stochastic processes;
-
□
Exposure of bacilli to multiple microenvironments imposing individual constraints on growth that may be temporally and spatially separated;
-
□
Prolonged antibiotic stress.
There is sparse information defining subpopulations of M. tuberculosis, but both clinical data and the demonstration of subpopulations from in vitro culture and animal models [7,8] indicate that subpopulations exist. Further in vivo evidence is provided by quantitative variations in lipid inclusion body staining of human sputum-derived bacilli [9] and the observation that bacilli in different locations of human lung tissues display unique gene-expression patterns [10]. Furthermore, murine models of chronic infection show increased tolerance to isoniazid (INH), suggesting slow growth rates are associated with reduced drug efficacy [11].
Isoniazid is a front-line drug in TB treatment and prophylaxis. Bactericidal activity during the first 2 days of TB treatment is almost entirely due to INH eliminating approximately 90% of bacilli [12]. However, minimal killing is observed after exposure of nonreplicating M. tuberculosis bacilli to INH in vitro [13], so it is unlikely that INH contributes to the clearance of persistent bacilli in patients during the extended treatment phase. INH is a prodrug that enters the cell by passive diffusion where it is converted by a katG-encoding catalase–peroxidase to an active INH–NAD adduct (reviewed elsewhere [14]). The primary target of INH–NAD is InhA, an enoyl-ACP reductase, part of the FASII cycle, which is required for mycolic acid synthesis and maintenance of the mycobacterial cell wall structure [15,16]. The INH–NAD(P) adduct(s) are likely to inhibit multiple secondary targets, such as the NADPH-dependent dihydrofolate reductase, which may also contribute to INH-mediated killing [17]. Resistance to INH is most frequently determined by mutations in katG, inactivating the catalase–peroxidase required for INH prodrug conversion. Mutations in the promoter of inhA, resulting in the overexpression of the target (InhA) or affecting the InhA active site also confer resistance [18]. Mutations in a number of other genes have been identified in INH-resistant M. tuberculosis strains [19]. However, approximately 22–29% of INH-resistant strains do not possess mutations in genes known to affect INH-resistance [20,21], indicating that there may be unexplored mechanisms that influence INH efficacy in M. tuberculosis.
Transcriptional profiling by microarray analysis has been used extensively to define the M. tuberculosis genome-wide responses to antibiotic exposure (classifying drug action and identifying possible resistance mechanisms [22–24]) and to fluctuations in environmental factors such as limited oxygen [25–27] or nutrient starvation [28,29] are reviewed in [30]. In one such model of mycobacterial persistence, developed by Wayne and colleagues, oxygen levels are gradually reduced by stirring liquid cultures in sealed tubes [31]. Under these conditions, bacilli enter a physiological state known as nonreplicating persistence (NRP), which is characterized by two distinct stages. The first, NRP1, occurs in microaerophilic conditions when the concentration of oxygen in the medium descends to 1%. During this phase, bacilli shut down DNA synthesis and cell division, restrict biosynthetic activity and use alternative energy sources to stabilize and protect essential cell components. The second NRP2 stage, where general metabolism is reduced further and replication ceases, occurs in anaerobic conditions when the dissolved oxygen concentration drops below 0.06% [13]. Nonreplicating bacteria may become tolerant to antibiotic exposure [32,33]; bacilli in NRP2 exhibit some definite but reduced killing with rifampicin and are highly tolerant to INH and ciprofloxacin [31]. Furthermore, the absence of an INH-inducible transcriptional signature has been used to characterize in vitro and in vivo M. tuberculosis dormant states [34].
In an attempt to understand why nonreplicating bacilli are refractory to killing by INH and, thus, to identify putative INH-tolerance mechanisms mediated by changing transcript abundance, we defined the transcriptional responses to INH exposure in log-phase and Wayne model NRP2 bacilli. Having contrasted the changing pattern of gene expression through the transition from log-phase to NRP2 with the INH-induced transcriptome signature, we also discuss INH tolerance in nongrowing populations and consider mechanisms that may influence drug tolerance.
Experimental
■. Aerobic log-phase & Wayne model cultures
Seed cultures of M. tuberculosis H37Rv were incubated in 10 ml Dubos liquid medium (BD, Franklin Lakes, USA) supplemented with Dubos medium albumin, with shaking at 37°C for 6 days to an optical density 580 nm of 0.4. Both aerobic and NRP cultures were set up in glass tubes (125 × 20 mm) with screw-top lids containing 1.5 mm magnetic stir bars and stirred at 120 rpm as characterized by Wayne [35]. Log-phase bacilli were incubated for 3 days in 10 ml Dubos medium in tubes with loosened caps. The Wayne model cultures were incubated for 6 days (NRP1) or 21 days (NRP2) in 17 ml Dubos medium in tubes with sealed lids (with a resulting head-space ratio of 0.5). Growth was monitored by measuring optical density every 24 h. RNA was isolated from aerobic log-phase and Wayne model cultures on two independent occasions.
■. INH treatment
Cultures were treated with 0.2 μg/ml INH final concentration (from 100 μg/ml stock solution, Sigma, Saint Louis, USA) at days 6 and 21 in the Wayne model (for NRP1 and NRP2, respectively), and at day 3 in log-phase aerobic cultures (Supplementary Figure 1 [available at www.future-science.com/toc/fmc/2/8]). An equivalent volume of sterile water was added to drug-free carrier control samples. Mycobacterial RNA was extracted after exposure to INH for 4 h. Prior to addition, syringes containing INH (or sterile water) were pre-incubated in an anaerobic cabinet overnight, to eliminate oxygen from the solutions and prevent the reintroduction of dissolved oxygen to the NRP cultures.
■. Mycobacterial RNA isolation
RNA was extracted using the GTC/TRIzol method [36,37]; at each timepoint the culture volume (eight tubes per condition) was added to four volumes of 5 M guanidine thiocyanate solution and the bacteria harvested by centrifugation. The bacilli were lyzed in TRIzol using a reciprocal shaker and the nucleic acid extracted with chloroform, before isopropanol precipitation. The RNA samples were purified and DNase I treated on RNeasy columns (Qiagen, Hilden, Germany). Total RNA concentration was determined by spectrophotometry (NanoDrop ND-1000 3.1, Thermo Fisher Scientific, Waltham, MA, USA) and size distribution assessed using an Agilent 2100 Bioanalyser (Agilent Technologies, Santa Clara, USA).
■. Microarray hybridization
Whole-genome microarrays generated by the Bacterial Microarray Group at St George’s (ArrayExpress accession number A-BUGS-23 [101]) were hybridized as described previously [37, 38]. Briefly, 5 μg of cDNA derived from each RNA sample and 2 μg of M. tuberculosis H37Rv genomic DNA were labeled with Cy5-dCTP and Cy3-dCTP, respectively. RNA extracted from two biological replicates were hybridized in triplicate, resulting in six hybridizations per condition. Microarrays were scanned at 532 and 635 nm corresponding to Cy3 and Cy5 excitation maxima, respectively, using the Affymetrix 428™ Array Scanner (MWG Biotech, Ebersberg, Germany). Comparative spot intensities from the images were calculated using Imagene 5.5 (BioDiscovery, El Segundo, USA) and the data imported into GeneSpring GX 7.3.1 (Agilent Technologies, Santa Clara, USA) for analysis. The data were normalized to the 50th percentile of all genes detected to be present and filtered to include genes flagged to be present in 80% of the arrays.
■. Transcriptional profiling analysis
Genes that were significantly differentially expressed between growth states or after INH treatment were identified using a t-test with Benjamini and Hochberg multiple testing correction (p < 0.05) and a minimum fold-change of 1.5. Genes were hierarchically clustered using Cluster and the results displayed using Treeview [39]. The hypergeometric distribution was used to determine if previously identified gene-expression signatures or genomic functional categories were significantly enriched in each comparison [40]. Significantly differentially expressed genes are detailed further in Supplementary Tables 1 & 2 (Available at www.future-science.com/toc/fmc/2/8). Fully annotated microarray data from this study have been deposited in BμG@Sbase (accession number: E-BUGS-104 [102]) and ArrayExpress (accession number: E-BUGS-104).
■. Quantitative RT-PCR
Primers and probes for cydA, efpA, icl, inhA, iniA, kasA, katG, narX, ndh and tgs1 were designed using the Primer Express 1.0 software (Life Technologies, Carlsbad, USA) and are detailed further in Supplementary Table 3 (Available at www.future-science.com/toc/fmc/2/8). All probes were labeled with 5-carboxyfluoroscein (FAM) at the 5´, and N,N,M’,N´-tetramethyl-6-carboxyrhodamine (TAMRA) at the 3´. An internal control, sigA (labeled with VIC), was used to normalize mRNA levels. Mycobacterial RNA (5 μg) was reverse transcribed in a total volume of 25 μl using random primers (Life Technologies, Carlsbad, USA) according to the manufacturer’s instructions. A total of 1 μl of template cDNA was used for qRT-PCR with AmpliTaq Gold polymerase, alongside no template and no RT negative controls. qRT-PCR was performed using an ABI PRISM 7700 Sequence detector system (Life Technologies, Carlsbad, USA), in amplification conditions of 95°C for 10 min followed by 50 cycles at 95°C for 15 s and at 60°C for 60 s. Each reaction was performed in triplicate. The ΔΔCt method [41] was used to determine changes in relative gene expression.
Results & discussion
■. Gene-expression programs illustrate the transition from log-phase to nonreplicating persistence
To examine the relationship between growth rate and phenotypic drug tolerance, we used genome-wide microarray RNA profiling and qRT-PCR to map the transcriptional status of M. tuberculosis with and without exposure to INH in log-phase and in the Wayne model of NRP. We hypothesize that transcriptome patterns may reveal mechanisms that explain INH tolerance, such as changes in expression levels for genes involved in drug efflux, alteration in target levels (inhA) or reduced katG expression (activation of drug). We first compared drug-free NRP bacilli to log-phase bacilli (drug-free NRP2 vs drug-free log-phase) to describe the changes in gene expression that occur as oxygen and nutrients become limiting, as defined by Wayne and Hayes [31].
As mycobacterial growth rate slows and then plateaus, 944 genes were significantly differentially expressed in NRP1 (323 induced and 621 repressed) and 1451 genes differentially regulated in NRP2 (649 induced and 802 repressed) compared with aerobic log-phase growth (Supplementary Table 1). As expected, there were highly significant overlaps in geneexpression patterns with previous studies characterizing the transcriptional response of bacilli to stationary phase growth [26], oxygen limitation [25], nutrient starvation [28] and the Wayne model itself [42], with hypergeometric p-values for enrichment of NRP2-induced genes of 7.17 × 10-38, 3.44 × 10-18, 4.23 × 10-16 and 9.36 × 10-31, respectively. Genes with functional classifications such as aerobic respiration, ribosomal protein synthesis and ATP-proton motive force were repressed in NRP2 compared with aerobic axenic growth (hypergeometric p-values: 5.14 × 10-8, 8.52 × 10-6 and 4.34 × 10-5, respectively) [43]. Conversely, the dosR (hypoxia and NO-responsive dormancy regulon) [44], mprA (two-component response regulator) [45] and kstR (cholesterol metabolism) [46] regulons were significantly induced in NRP2 compared with log-phase growth (hypergeometric p-values: 5.85 × 10-28, 6.10 × 10-5 and 1.20 × 10-4, respectively); as were the alternative sigma factors sigB, sigE, sigF and sigG, and the regulator of the stringent response, relA. The transition from rapidly multiplying to nonreplicating bacilli in this Wayne model of dormancy mirrors previously identified transcriptional changes, reflecting a change in respiratory state from aerobic to anaerobic as oxygen becomes limited, a switch to lipid-based carbon sources as nutrients become scarce and a modification of cell-wall composition and a slowing of metabolism as growth terminates [27,47].
■. Nonreplicating bacilli do not mount a transcriptional response to INH
We then asked whether the transcriptional responses to INH would differ between these two physiologically distinct populations of bacilli. The hypothesis being that drug-induced responses reveal genes involved in potential resistance mechanisms or compensatory responses that reduce the effect of the drug and, hence, may underpin tolerance. We defined genes differentially expressed after 4 h exposure to 0.2 μg/ml INH in log-phase and NRP2 bacilli (in INH-treated log-phase vs drug-free log-phase, and INH-treated NRP2 vs drug-free NRP2 comparisons).
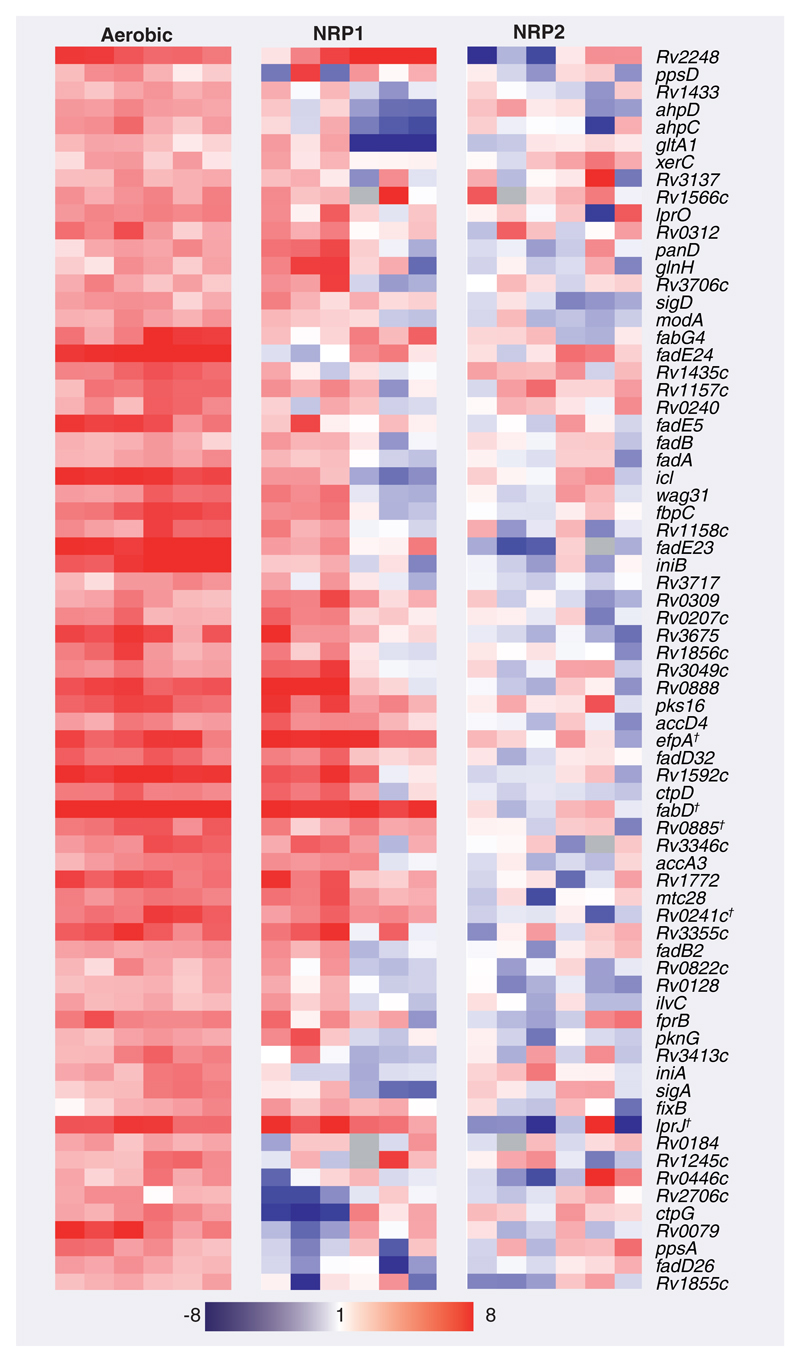
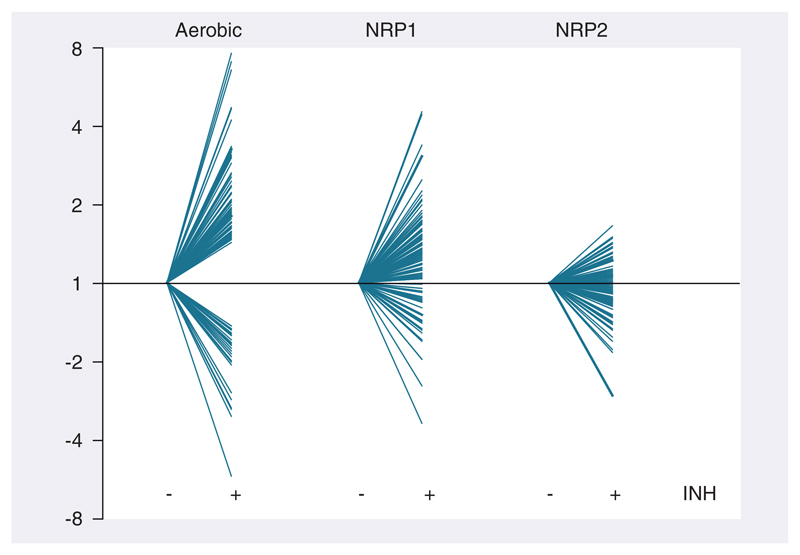
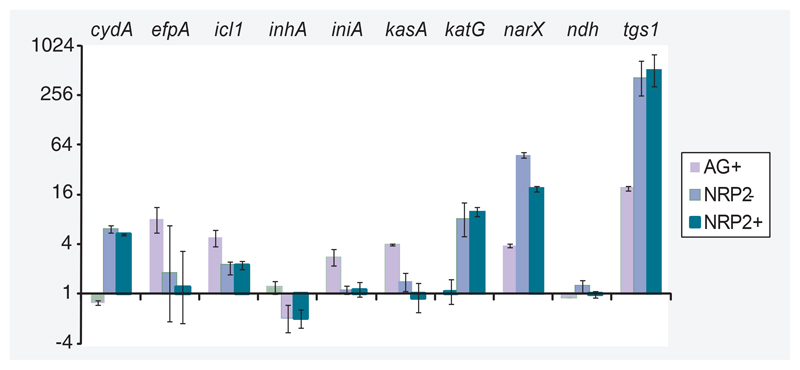
In rapidly multiplying log-phase bacilli, 100 genes were significantly differentially regulated after 4 h INH treatment; with 71 genes induced and 29 genes repressed relative to drug-free log-phase bacilli (Supplementary Table 2). There was a high degree of correlation between INH-responsive genes identified in this investigation and previous studies examining INH-mediated transcriptional programs; hypergeometric p-values: 2.15 × 10-24 [48], 5.79 × 10-16 [22], 3.54 × 10-12 [23] and 5.20 × 10-10 [49]. Genes indicative of cell wall inhibitors such as accA3, accD4, ahpC, ahpD, efpA, fadD32, fbpC, htdX, iniA, iniB, lprJ, pks16 and Rv3717 were significantly induced [23]. By comparison, no genes were significantly differentially expressed after INH treatment of NRP2 bacilli relative to drug-free NRP2 bacilli. To ensure that the absence of differentially regulated genes after INH treatment in NRP2 bacilli was not an artifact of significance testing, we plotted the expression pattern of log-phase INH-responsive genes in both log-phase and NRP conditions (Figures 1 & 2). Figure 1 shows a heat map of six replicates of all genes significantly upregulated by INH in log-phase bacilli and the gradual diminution of transcriptional response as transition proceeds through NRP1 towards NRP2. This confirmed that genes comprising the INH transcriptional signature of log-phase bacilli were not differentially regulated after INH treatment in NRP2 bacilli. These observations were verified by quantitative RT-PCR using a panel of genes selected to be indicative of mycobacterial growth state and implicated in INH resistance (Figure 3). Thus, increased tolerance of NRP2 bacilli to INH seems not to be dependent on the initiation of a transcriptional program after exposure to INH that renders the drug ineffective.
Figure 1. Transcriptional programs induced by isoniazid exposure in log-phase, NRP1 and NRP2 growth states.
The expression pattern of 71 genes induced by isoniazid (INH) treatment in aerobic conditions are clustered alongside the transcriptional activity of these genes after INH exposure in NRP1 and NRP2. The genes are displayed as rows, growth conditions as columns. No genes were significantly differentially expressed in NRP2 after INH treatment. Red coloring signifies induction; blue denotes repression relative to the respective drug-free growth state controls.
†Genes that were also INH-responsive in NRP1. NRP: Nonreplicating persistence.
Figure 2. Diminished transcriptional response to isoniazid exposure in NRP2.
Gene expression profiles of 100 genes (71 induced, 29 repressed) significantly differentially expressed after INH treatment in aerobic conditions are plotted (±INH) alongside the expression pattern of these genes in NRP1 and NRP2. The expression ratios are presented as fold-change relative to drug-free growth state controls.
INH: Isoniazid; NRP: Nonreplicating persistence.
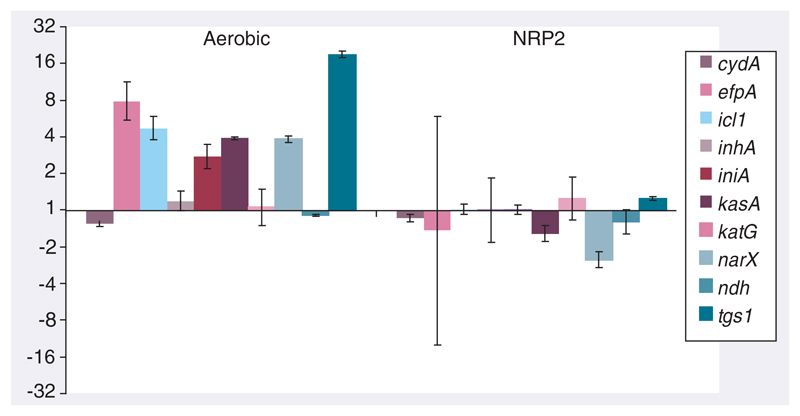
Figure 3. The absence of a transcriptional response to isoniazid in NRP2 bacilli, verified by quantitative RT-PCR with a panel of genes chosen to exemplify the isoniazid transcriptional signature and the physiological status of bacilli.
Expression ratios, marked in fold-change, were calculated relative to sigA and drug-free growth controls using the ΔΔCt method. Standard deviations from triplicate samples are marked. Red–pink coloring highlights genes selected to assay INH mechanism of action; blue coloring indicates genes chosen to illustrate the metabolic and respiratory state of bacilli.
INH: Isoniazid; NRP: Nonreplicating persistence.
The absence of a transcriptional response to INH indicates no activity of the drug in NRP2 bacilli. This could be explained in several ways:
-
□
Isoniazid has not entered the cell due to alterations in cell wall structure, or has been exported rapidly;
-
□
INH is not converted to the toxic product by KatG, or there is reduced availability of NAD to form the INH–NAD adduct;
-
□
The enzyme target(s) of INH–NAD outnumber the toxic adducts generated, or the inhibition of pathways affected by INH are no longer functionally significant for bacilli in NRP2.
We have utilized the gene-expression signatures of NRP2 and INH as a framework to explore these hypotheses further.
■. Differential regulation of genes identified to harbor mutations in INH-resistant M. tuberculosis strains do not explain INH tolerance in NRP2
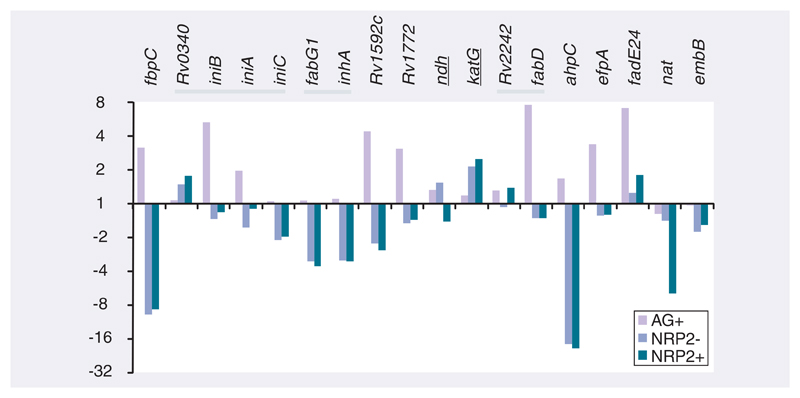
Following the hypothesis that differences in transcript abundance between log-phase and NRP2 bacilli might influence INH tolerance, we asked whether the differential regulation of genes associated with INH resistance might explain INH tolerance in NRP2 bacilli. We used the assumption that differential expression of these genes (drug-free NRP2 vs drug-free log-phase) may relate to changes in protein (or drug) activity resulting in tolerance, even though these genes may not be directly involved in the mechanism of drug action. Therefore, we compared the expression profiles of 18 genes with mutations in INH drug-resistant strains, collated on the TB drug-resistance mutation database [19] and present in the filtered microarray dataset used in this study (Figure 4). Mutations that result in the overexpression of inhA (increased target) lead to INH resistance by overwhelming the active INH–NAD adduct present in exposed bacilli; however, both inhA and the upstream gene fabG1 were repressed in NRP2 relative to log-phase bacilli [14]. An alternative mechanism of drug resistance might be caused by the increased activity of arylamine N-acetyltransferase, which inactivates INH by acetylation leading to resistance. However, the induction of nat was not observed in NRP2 compared with log-phase bacilli. The catalase–peroxidase activity encoded by katG is required for the formation of the INH–NAD adduct; mutations in this gene result in INH resistance. Therefore, a downregulation of katG could be hypothesized to increase INH tolerance. However, katG expression was induced in NRP2 relative to log-phase bacilli, which might be predicted to increase rather than decrease the susceptibility of NRP2 bacilli to killing by INH. Conversely, the enzyme activity of this catalase–peroxidase is reduced at low oxygen levels, which might result in the formation of less INH–NAD adduct and, thus, INH tolerance [50]. Notably, this reduction of KatG activity in hypoxic conditions might also be predicted to result, through compensatory feedback mechanisms, in the induction of katG gene expression in NRP2 compared with log-phase bacilli, as was observed. The repression of ndh, leading to a reduction in NDH-attributed NADH dehydrogenase activity and an accumulation of NADH that acts as a competitive inhibitor with INH–NAD for INHA binding [51], thereby increasing drug tolerance, was not evident.
Figure 4. The changes in RNA abundance of genes associated with INH resistance in log-phase and NRP2 bacilli ± INH relative to drug-free log-phase bacilli.
Expression ratios, derived from the microarray dataset, are plotted in fold change for log-phase aerobic (AG) and NRP2 ± INH treatment. Genes are ordered by chromosome position, with the dark bars highlighting adjacent genes. Underlined gene annotations indicate that the repression of these gene products in NRP2 may be hypothesized to increase INH tolerance. Conversely, the over-expression of the remaining genes might be expected to increase INH tolerance in NRP2.
AG: Aerobic; INH: Isoniazid; NRP: Nonreplicating persistence.
Genes identified as potentially influencing INH activity because they are highly upregulated after INH treatment (such as fbpC, iniBAC, Rv1592c, Rv1772 and efpA), were not induced in NRP2 compared with log-phase growth as might be the case if these gene products contributed to INH tolerance in NRP2 (Figure 4) [22,52]. Interestingly, fadE24, a probable acyl-CoA dehydrogenase likely involved in the β-oxidation of fatty acids that is induced after INH exposure, was marginally induced in NRP2 relative to log-phase bacilli. It has been previously proposed that the induction of genes involved in lipid degradation may be responsible for recycling fatty acids that accumulate after exposure to INH and the inhibition of the FASII cycle [22]. The differential regulation of genes encoding efflux pumps may modify INH entry and exit kinetics affecting killing efficacy: efpA, mmr, Rv1819c, Rv2459 and Rv3728 have been demonstrated to be induced by INH in multidrug-resistant strains [53]. However, none of these genes were significantly induced in NRP2 relative to log-phase growth. Of the genes assigned by Cole et al. to the functional category III.A.6 efflux genes [43], three were induced in NRP2: Rv1250 (a probable drug-transport integral membrane protein, a member of the major facilitator superfamily), Rv1634 (a possible drug-efflux protein, also a member of the MFS family) and Rv2209 (a conserved integral membrane protein); four genes were repressed relative to log-phase bacilli (drrB, drrC, emrB and Rv2459). This may be a possible explanation for increased efflux-based tolerance, but little is known about the specificity of each transporter. The expression profiles of a subset of these genes were verified by qRT-PCR (Figure 5); this demonstrated excellent correlation between ratios generated from microarray and qRT-PCR techniques.
Figure 5. Quantitative RT-PCR confirming the differential regulation of isoniazid responsive genes and genes indicative of bacterial metabolic state in log-phase and NRP2 relative to drug-free log-phase bacilli.
Expression ratios ± isoniazid (INH), measured in fold-change, were calculated relative to sigA and untreated log-phase control using the ΔΔCt method. Standard deviations from triplicate samples are marked.
AG: Aerobic; NRP: Nonreplicating persistence.
■. Complex transcriptional responses reflecting the changing physiological state of bacilli in NRP2 likely affect INH efficacy
Unlike mutations to single genes that affect the primary target of INH or its modification into an active molecule that results in INH resistance, adaptations involving the differential expression of many genes involved in respiratory and metabolic pathways may influence INH tolerance in NRP2. For example, the maintenance of redox potential in NRP2 bacilli is very different to log-phase aerobically respiring bacilli. This may prohibit the conversion of INH to INH–NAD adduct (akin to the inactivation of KatG). Cytochrome P450 genes, and their putative redox partners [54], were differentially regulated in NRP2 relative to log-phase; with cyp51, cyp124, cyp125, cyp130, cyp132, fdxA, fdxC, fdxD, fprB and fprD induced and cyp121, cyp135A1, cyp136, cyp140, cyp141 and cyp143 repressed. In addition, the requirement for cytochrome F420 may differ [47], with the NADPH-dependent glucose-6-phosphate dehydrogenases (zwf1 and zwf2) downregulated, and F420-requiring glucose-6-phosphate dehydrogenases (encoded by fgd1 and fgd2) induced in NRP2. Mutations conferring INH resistance in genes coding for mycothiol biosynthesis – mshA, mshC, mshD in Mycobacterium smegmatis [55] and aphC [56] – have also been suggested to reduce the capacity for prodrug conversion, resulting in increased tolerance to INH. However, these genes were not significantly differentially expressed in NRP2 relative to log-phase bacilli. Most interestingly, recent evidence suggests that M. tuberculosis is able to maintain redox potential using the anabolism of propionate-derived lipids as a reductant sink [57]. A range of genes involved in fatty acid metabolism was differentially regulated in NRP2 relative to log-phase (Supplementary Table 1). Furthermore, modulation of lipid metabolism in NRP2 may transform the architecture of the mycobacterial cell, which has been observed to thicken in stationary-phase bacilli [58]. Changes to the cell wall structure affecting cell permeability (or the suppression of porin expression) may impact on a range of physiological processes [59]; not least in this scenario, limiting drug uptake that may result in increased tolerance to INH in nonreplicating bacilli.
Both fabG1 (mabA) and inhA were repressed in NRP2 compared with log-phase growth; inhA was downregulated two- to three-fold as determined by microarray and qRT-PCR. This reduction in INH target (inhA) is perhaps consequent on a redundant metabolic role for FASII in NRP2 due to the absence of cell wall synthesis in the nonreplicating state. As such, INH does not induce a compensatory response in NRP2 as cell wall biosynthesis is already homeostatically downregulated. In addition, genes involved in mycolic acid modification cmaA2, cmrA, mmaA2, mmaA3, mmaA4 and umaA1 were also repressed in NRP2 relative to log-phase bacilli. DNA replication encoding genes dnaA, dnaB, dnaE1, dnaE2, dnaN, gyrA and ssb were downregulated as expected in NRP2 relative to log-phase growth, as were recA, recF and recR [60]. The dihydrofolate reductase (product of dfrA), an essential precursor to DNA replication and postulated to be a target for INH, was also repressed in NRP2 [17]. It is therefore likely that it is the reduced requirement for mycolic acids and the FASII cycle, necessary for de novo cell-wall biosynthesis, in NRP2 as replication ceases that confers phenotypic tolerance to INH.
■. Use of transcriptional profiling to screen for NRP and INH-responsive genes that may influence M. tuberculosis drug tolerance
Delineating the transcriptional response of M. tuberculosis to antimicrobial agents has led to the characterization of mechanisms affecting drug tolerance. For example, the iniB/A/C genes, encoding an MDR-like pump, were identified to be induced after INH treatment in M. tuberculosis [61]. While deletion mutants were more sensitive to killing by INH; overexpression conferred resistance to both INH and ethambutol [62]. This transmembrane complex likely acts as an efflux pump maintaining cellular functions after the perturbation of cell-wall biosynthesis. We reasoned that distinguishing genes that were differentially expressed in NRP2 that were also INH-responsive might reveal novel pathways that affect INH tolerance. We used the transcriptional program initiated by exposure to INH in log-phase bacilli (in an INH-treated log-phase drug-free vs log-phase comparison) to screen the 100s of genes differentially regulated in NRP2 relative to log-phase bacilli (drug-free NRP2 vs drug-free log-phase) (Figure 6).
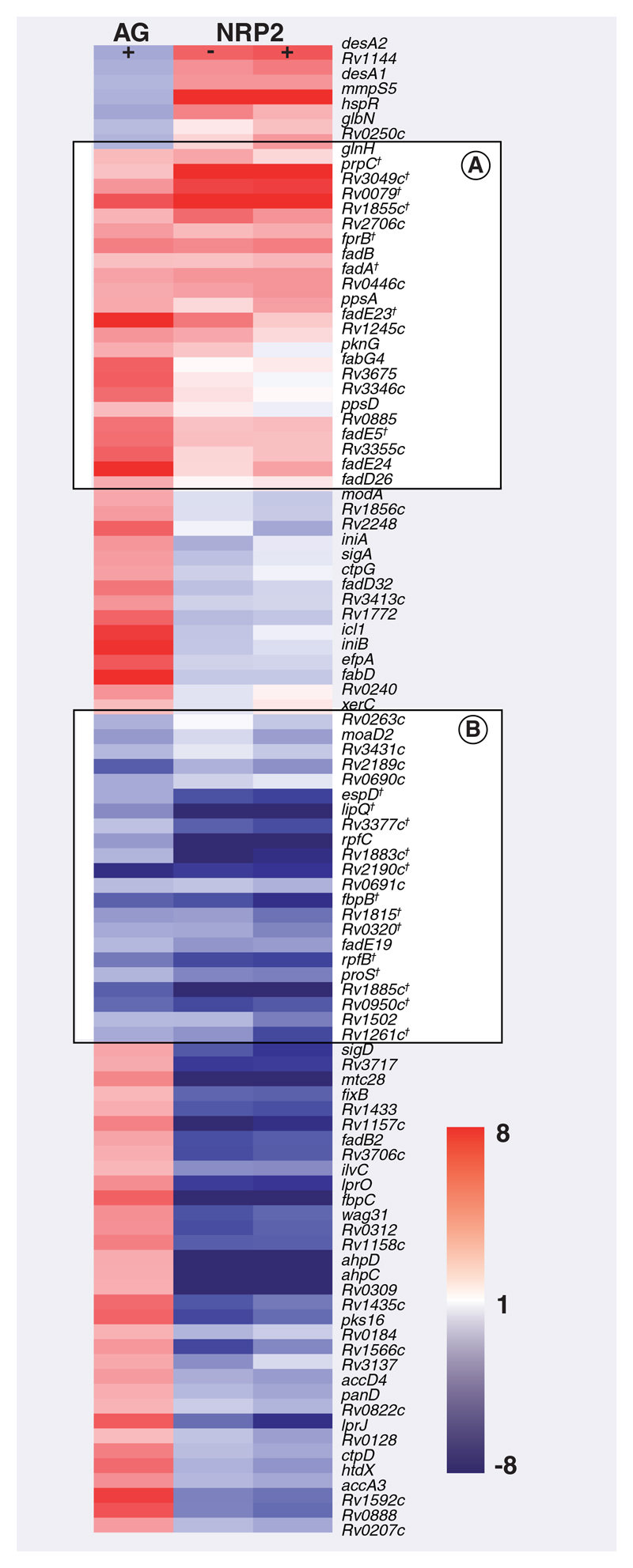
Figure 6. Expression pattern of isoniazid-responsive genes in log-phase and NRP2 bacilli ± isoniazid relative to drug-free log-phase bacilli.
A total of 100 genes, identified as significantly differentially regulated after isoniazid (INH) exposure in AG conditions, were clustered alongside the NRP2 transcriptional profiles. (A & B) show genes induced or repressed after INH exposure and transition to NRP2 compared with untreated log-phase bacilli. Genes are displayed as rows and growth conditions ± INH as columns. Red: induction; blue: repression relative to log-phase drug-free control.
†Genes significantly differentially expressed in both drug-free NRP2 and INH-treated AG bacilli compared with untreated log-phase bacilli.
AG: Aerobic; NRP: Nonreplicating persistence.
We isolated 23 genes (Figure 6A) that were significantly induced by INH exposure and that were also upregulated in drug-free NRP2 relative to untreated logphase bacilli. A total of 22 genes (Figure 6B) repressed by INH were correspondingly repressed in NRP2 compared with log-phase bacilli (Supplementary Table 4 [available at www.future-science.com/toc/fmc/2/8]). Of the genes involved in fatty acid metabolism, seven were induced in both NRP2 and INH-treated log-phase bacilli (fabG4, fadE5, fadA, fadB, prpC, fadE24 and fadE23), perhaps reflecting the redirection of lipid precursors, or metabolic shift to lipid degradation after environmental challenge. In addition, fadD26, ppsA and ppsD, part of the gene cluster responsible for the biosynthesis of phthiocerol dimycocerosate, were induced in both scenarios. Overexpression of this propionate-derived complex lipid may modify the redox status of bacilli and might also alter cell-wall permeability, thereby influencing INH tolerance. Furthermore, three genes that may also contribute to redox potential, fprB, an NADPH:aderenodoxin oxidoreductase (and the adjacent conserved hypothetical Rv0885), together with possible oxidoreducatase (Rv1855c) and putative monooxygenase (Rv3049c) were also induced. Other gene products that might influence drug kinetics, such as the predicted membrane proteins of unknown function, Rv0446c, Rv3346c, Rv3355c and Rv3675, were upregulated in NRP2 and after INH exposure. Interestingly, pknG, encoding a protein kinase associated with the regulation of M. tuberculosis metabolism [63] and the ability to survive intracellularly [64], was induced by both INH and NRP2. The loss of pknG function has been previously implicated in the increased sensitivity of M. smegmatis to multiple antibiotics [65], perhaps due to a decrease in cell-wall hydrophobicity. In addition, quantitative RT-PCR (Figure 5) revealed that icl1 (encoding isocitrate lyase, part of the glyoxylate bypass), narX (involved in nitrate reduction) and tgs1 (a triacylglycerol synthase) were all NRP2 and INH-responsive. These three genes have all been associated with the ability of M. tuberculosis to persist in vivo [42, 66–68].
Conclusion
Understanding why drugs do and do not work during TB chemotherapy should advance novel drug-design strategies, and reveal a little about the in vivo physiological states of M. tuberculosis bacilli that clearly influence the efficacy of antibiotic regimens. Transcriptional profiling is a powerful tool in this respect, mapping the expression pattern of all annotated genes in a whole-genome approach to defining the adaptations necessary for M. tuberculosis survival. This strategy does not, however, take into account processes affecting protein activity that also influence drug action. In this article, we contrasted the M. tuberculosis transcriptional response to INH in log-phase and NRP2 bacilli as a model for INH tolerance. The Wayne model was selected to represent non-growing bacilli as this model is strictly defined, well characterized and likely results in a largely homogenous M. tuberculosis population as bacilli display features of synchronous growth on exit from NRP2 [31]. Thus, a comparison between log-phase, with the predominant gene-expression signature derived from rapidly multiplying bacilli, and NRP2 bacilli should model the differences between INH-sensitive and -tolerant bacterial populations. Exposure of NRP2 bacilli to INH did not elicit a transcriptional response, and the RNA abundance of genes inducible by INH in log-phase bacilli did not change after exposure of NRP2 bacilli to INH. This correlates with findings by Karakousis and colleagues, who found the INH-mediated transcriptional program greatly diminished in oxygen- and nutrient-depleted in vitro models and murine lung tissue after 6 weeks of aerosol infection [34].
Tolerance to INH in NRP2 bacilli cannot be correlated to the changing transcriptional profiles (comparing drug-free NRP2 to log-phase bacilli) of genes directly implicated in INH resistance through resistance-conferring mutations. However, it should be noted that factors affecting protein activity, such as the reduction of KatG catalase–peroxidase activity in hypoxic conditions, have not been characterized in this study [50]. Phenotypic tolerance in NRP2 is likely the result of complex multigene adaptations to limited oxygen and diminishing carbon resources as bacterial multiplication slows and stops. The reduced requirement for mycolic acids and the FASII cycle in nonreplicating bacilli renders bacilli tolerant to the effects of INH. Other factors such as redox state and cell wall permeability may also contribute to the abrogation of killing by INH in nongrowing M. tuberculosis bacilli.
We speculate that the induction of a subset of genes involved in lipid metabolism and capable of performing oxidative functions in both NRP2 and after exposure to INH suggest that specific pathways remodelling the metabolic and respiratory state of M. tuberculosis may also influence the efficacy of INH killing. Furthermore, many of these genes have been identified to be upegulated after macrophage infection [69,70]. Could the induction of these pathways in replicating intracellular bacilli account for the significant decrease in INH sterilizing ability between axenic culture and after macrophage infection [71] or in vivo [72]? Finally, the upregulation of tgs1, involved in triacylglycerol lipid body formation, after INH exposure and in NRP2 bacilli suggests that the metabolic consequences of specific growth constraints that induce a drug-tolerant fat and lazy mycobacterial phenotype are also encountered in the human lung [42].
Future perspective
There are several technological advances that impact on our understanding of drug resistance mechanisms, which together may resolve the important phenomenon of tolerance in M. tuberculosis.
The application of high-throughput wholegenome DNA sequencing will transform M. tuberculosis resistance testing by mapping mutations to drug mode of action. Currently, in the absence of a comprehensive, genome-wide correlation of genotypic changes with phenotypic drug resistance, it is difficult to predict resistance simply by sequence analysis for single nucleotide polymorphisms (SNPs). Implementation of the soon-to-be-available, rapid and cost-effective, third-generation, highly parallel sequencing platforms will permit every M. tuberculosis isolate to be sequenced. The correlation of genomic sequence to phenotypic resistance profile of many thousands of isolates will result in complex predictive modeling of resistance-modifying SNPs. However, it would be unwise to abandon sensitivity testing completely. This SNP mapping together with consequent gene-specific studies will expose multiple primary and secondary drug targets, further elucidating mechanisms of antibiotic killing. It may also reveal new classes of mutation conferring low-level resistance, affecting features such as cell permeability or growth rate. Furthermore, such genome-wide analyses will begin to uncover linkage between resistance mutations, as well as a large number of mutations that have no effect on antibiotic resistance at all. The resulting payoff will provide clinicians with a resistance profile for problem isolates, enabling chemotherapy to be tailored effectively. Although tolerance is an adaptive process, genotypic changes revealed by sequencing will define a panel of genes and highlight complex pathways that when modified at the mRNA or protein level, may also influence antibiotic tolerance.
Predictive modeling of drug tolerance using transcriptional profiling and systems approaches to test M. tuberculosis drug-tolerant phenotypes will be instrumental in defining in vitro conditions that reflect the complex in vivo status. This will enable tolerance mechanisms to be investigated in models that accurately represent the antibiotic sensitivity profile of bacilli during natural infection and allow drug-screening programs to target persistent-like bacilli. Modeling the changing physiological state of bacilli exposed to different in vivo or intracellular environments, diverse carbon sources and varied oxidative scenarios (defining mRNA, protein or metabolite levels) will help to delineate the unique transcriptional profiles derived from human sputa or lung tissue sections.
Single cell technologies and microfluidics have enabled bacterial populations to be visualized and differentiated; further advances in these fields will allow responses of individual bacteria to the changing microenvironment to be assayed. The identification and characterization of subpopulations of phenotypically diverse bacteria and the factors that influence their creation promises to reveal much about persisters, and their potential roles during infection. This, together with transcriptionally defined drug-screening targeted to specific bacterial metabolic and respiratory states, should pave the way for discovery of novel compounds that kill nonreplicating persistent bacteria. The continued support and patience of funding agencies will be required to smelt these silver bullets with the hope of eliminating the hidden monster of persistent infection.
Supplementary Material
Supplementary information accompanies this paper at www.future-science.com/toc/fmc/2/8.
Executive summary.
-
□
Effective tuberculosis chemotherapy is hampered by prolonged treatment, which is necessary to remove persistent bacilli, most likely because most front-line antibiotics primarily kill replicating bacilli.
-
□
An understanding of how and why Mycobacterium tuberculosis bacilli become phenotypically tolerant to some antibiotics would illuminate current drug-design programs, which are aimed at reducing duration of treatment from months to weeks.
-
□
Transcriptional profiling may be used to reveal important metabolic and physiological adaptations associated with M. tuberculosis infection in vivo and underpinning phenotypic tolerance to antibiotics. This genome-wide approach is also valuable in highlighting genes of interest that have no predicted function and that may not have been identified by gene-specific assays. Novel insights into underlying mechanisms may thus be discovered.
-
□
M.tuberculosis bacilli in nonreplicating persistence state 2 do not respond transcriptionally to isoniazid (INH) exposure and are tolerant to INH. Tolerance to INH in nongrowing bacilli is therefore not a result of INH-inducible adaptive responses, but rather a reflection of the underlying metabolic state in which INH has minimal effect.
-
□
Altered transcript abundance of key genes implicated in INH resistance (involved in drug activation or target manipulation) does not explain why nonreplicating persistence state 2 bacilli become tolerant to INH.
-
□
The reduced requirements for mycolic acids and disparate redox state of nongrowing bacilli probably result in tolerance to the effects of INH exposure.
-
□
Genes involved in lipid metabolism and alternative redox pathways may play functionally significant roles in INH-tolerant M.tuberculosis bacilli that persist through chemotherapy.
Financial disclosure
This work was supported by a postdoctoral fellowship to Griselda Tudó from the Ministerio de Educación y Ciencia (MEC), Spain (grant number 2005–0565) and the Spanish Network for Research in Infectious Diseases (REIPI, RD06/0008). Philip Butcher would like to thank The Wellcome Trust for funding the Bacterial Microarray Group at St. George’s (grant no. 062511). Simon Waddell is supported by the European Commission ‘New Medicines for TB – NM4TB’ program (LHSP-CT-2005–018923). Mycobacterium tuberculosis H37Rv reference DNA was kindly provided by Colorado State University (Contract No. HHSN266200400091C; NIH, NIAID N01-AI-40091). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Key Terms
- Phenotypic drug tolerance
Mechanisms by which genetically susceptible bacilli are refractory to killing with antimicrobial agents. Tolerance may be dependent on pre-existing subpopulations of physiologically distinct bacilli, or may be initiated by changes in the microenvironment or drug exposure.
- Persistence
Bacilli capable of causing active disease that survive prolonged chemotherapy, resulting in lengthy and expensive treatment for tuberculosis.
- Isoniazid
Front-line anti-tuberculosis drug targeting fatty acid synthesis and affecting the mycobacterial cell wall; responsible for the majority of mycobacterial killing within the first 2 days of treatment with standard regimen.
- Transcriptional profiling by microarray
Exploring the global responses of bacilli to the surrounding microenvironment by simultaneously quantifying mRNA abundance for all defined Mycobacterium tuberculosis genes.
- Wayne model
In vitro model where primarily oxygen is limited to mimic Mycobacterium tuberculosis nonreplicating persistence.
Footnotes
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Mitchison DA. The action of anti-tuberculosis drugs in short-course chemotherapy. Tubercle. 1985;66(3):219–225. doi: 10.1016/0041-3879(85)90040-6. [DOI] [PubMed] [Google Scholar]
- 2.Warner DF, Mizrahi V. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin Microbiol Rev. 2006;19(3):558–570. doi: 10.1128/CMR.00060-05. [▪▪ Review of the problems facing tuberculosis (TB) chemotherapy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin BR, Rozen DE. Review of the problems facing TB chemotherapy.Non-inherited antibiotic resistance. Nat Rev Microbiol. 2006;4(7):556–562. doi: 10.1038/nrmicro1445. [▪▪ Review of phenotypic tolerance in bacteria.] [DOI] [PubMed] [Google Scholar]
- 4.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [▪▪ Study into population heterogeneity identifying pre-existing persistent bacteria.] [DOI] [PubMed] [Google Scholar]
- 5.Dorr T, Lewis K, Vulic M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet. 2009;5(12):e1000760. doi: 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchison DA. The search for new sterilizing anti-tuberculosis drugs. Front Biosci. 2004;9:1059–1072. doi: 10.2741/1293. [DOI] [PubMed] [Google Scholar]
- 7.Hu Y, Coates AR, Mitchison DA. Sterilizing activities of fluoroquinolones against rifampin-tolerant populations of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003;47(2):653–657. doi: 10.1128/AAC.47.2.653-657.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhillon J, Lowrie DB, Mitchison DA. Mycobacterium tuberculosis from chronic murine infections that grows in liquid but not on solid medium. BMC Infect Dis. 2004;4:51. doi: 10.1186/1471-2334-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garton NJ, Christensen H, Minnikin DE, Adegbola RA, Barer MR. Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology. 2002;148(Part 10):2951–2958. doi: 10.1099/00221287-148-10-2951. [DOI] [PubMed] [Google Scholar]
- 10.Rachman H, Strong M, Ulrichs T, et al. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immun. 2006;74(2):1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhillon J, Fielding R, dler-Moore J, Goodall RL, Mitchison D. The activity of low-clearance liposomal amikacin in experimental murine tuberculosis. J Antimicrob Chemother. 2001;48(6):869–876. doi: 10.1093/jac/48.6.869. [DOI] [PubMed] [Google Scholar]
- 12.Jindani A, Dore CJ, Mitchison DA. Bactericidal and sterilizing activities of anti-tuberculosis drugs during the first 14 days. Am J Respir Crit Care Med. 2003;167(10):1348–1354. doi: 10.1164/rccm.200210-1125OC. [DOI] [PubMed] [Google Scholar]
- 13.Wayne LG, Sohaskey CD. Nonreplicating persistence of Mycobacterium tuberculosis. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 14.Vilcheze C, Jacobs WR., Jr The mechanism of isoniazid killing: clarity through the scope of genetics. Annu Rev Microbiol. 2007;61:35–50. doi: 10.1146/annurev.micro.61.111606.122346. [▪ Review of isoniazid mechanism of action and resistance-conferring mutations.] [DOI] [PubMed] [Google Scholar]
- 15.Banerjee A, Dubnau E, Quemard A, et al. InhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263(5144):227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 16.Rozwarski DA, Grant GA, Barton DH, Jacobs WR, Jr, Sacchettini JC. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science. 1998;279(5347):98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 17.Argyrou A, Vetting MW, Aladegbami B, Blanchard JS. Mycobacterium tuberculosis dihydrofolate reductase is a target for isoniazid. Nat Struct Mol Biol. 2006;13(5):408–413. doi: 10.1038/nsmb1089. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Yew WW. Mechanisms of drug resistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2009;13(11):1320–1330. [PubMed] [Google Scholar]
- 19.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. Tuberculosis drug resistance mutation database. PLoS Med. 2009;6(2):e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazbon MH, Brimacombe M, Bobadilla DV, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006;50(8):2640–2649. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho YM, Sun YJ, Wong SY, Lee AS. Contribution of dfrA and inhA mutations to the detection of isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2009;53(9):4010–4012. doi: 10.1128/AAC.00433-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson M, DeRisi J, Kristensen HH, et al. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA. 1999;96(22):12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE., 3rd The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem. 2004;279(38):40174–40184. doi: 10.1074/jbc.M406796200. [▪ Comprehensive description and discussion of TB transcriptional responses to numerous antibiotics.] [DOI] [PubMed] [Google Scholar]
- 24.Makarov V, Manina G, Mikusova K, et al. Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science. 2009;324(5928):801–804. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bacon J, James BW, Wernisch L, et al. The influence of reduced oxygen availability on pathogenicity and gene expression in Mycobacterium tuberculosis. Tuberculosis (Edinb) 2004;84(3–4):205–217. doi: 10.1016/j.tube.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis(Edinb) 2004;84(3–4):218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Muttucumaru DG, Roberts G, Hinds J, Stabler RA, Parish T. Gene expression profile of Mycobacterium tuberculosis in a nonreplicating state. Tuberculosis (Edinb) 2004;84(3–4):239–246. doi: 10.1016/j.tube.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43(3):717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 29.Hampshire T, Soneji S, Bacon J, et al. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis (Edinb) 2004;84(3–4):228–238. doi: 10.1016/j.tube.2003.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butcher PD. Microarrays for Mycobacterium tuberculosis. Tuberculosis (Edinb) 2004;84(3–4):131–137. doi: 10.1016/j.tube.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64(6):2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wayne LG, Sramek HA. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38(9):2054–2058. doi: 10.1128/aac.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paramasivan CN, Sulochana S, Kubendiran G, Venkatesan P, Mitchison DA. Bactericidal action of gatifloxacin, rifampin and isoniazid on logarithmic- and stationary-phase cultures of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2005;49(2):627–631. doi: 10.1128/AAC.49.2.627-631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karakousis PC, Williams EP, Bishai WR. Altered expression of isoniazid-regulated genes in drug-treated dormant Mycobacterium tuberculosis. J Antimicrob Chemother. 2008;61(2):323–331. doi: 10.1093/jac/dkm485. [▪ Study using the absence of an isoniazid-induced transcriptional signature to characterize Mycobacterium tuberculosis dormant states.] [DOI] [PubMed] [Google Scholar]
- 35.Wayne LG. In vitro model of hypoxically induced nonreplicating persistence of Mycobacterium tuberculosis. In: Parish T, Stoker NG, editors. Mycobacterium tuberculosis protocols. Humana Press Inc; NJ, USA: 2001. pp. 247–269. [DOI] [PubMed] [Google Scholar]
- 36.Mangan JA, Sole KM, Mitchison DA, Butcher PD. An effective method of RNA extraction from bacteria refractory to disruption, including mycobacteria. Nucleic Acids Res. 1997;25(3):675–676. doi: 10.1093/nar/25.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waddell SJ, Butcher PD. Use of DNA arrays to study transcriptional responses to antimycobacterial compounds. In: Gillespie SH, McHugh TD, editors. Antibiotic Resistance Protocols. Vol. 642. Springer; USA: 2010. pp. 75–91. [DOI] [PubMed] [Google Scholar]
- 38.Stewart GR, Wernisch L, Stabler R, et al. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology. 2002;148(Pt 10):3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 39.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waddell SJ, Stabler RA, Laing K, Kremer L, Reynolds RC, Besra GS. The use of microarray analysis to determine the gene expression profiles of Mycobacterium tuberculosis in response to anti-bacterial compounds. Tuberculosis(Edinb) 2004;84(3–4):263–274. doi: 10.1016/j.tube.2003.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-δ-δ- C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Garton NJ, Waddell SJ, Sherratt AL, et al. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 2008;5(4):e75. doi: 10.1371/journal.pmed.0050075. [▪▪ Transcriptional profile of TB bacilli from sputum, demonstrating a unique signature that correlates with phenotypic markers of slow growth, and hence antibiotic tolerance.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 44.Park HD, Guinn KM, Harrell MI, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48(3):833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balazsi G, Heath AP, Shi L, Gennaro ML. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol Syst Biol. 2008;4:225. doi: 10.1038/msb.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kendall SL, Withers M, Soffair CN, et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol Microbiol. 2007;65(3):684–699. doi: 10.1111/j.1365-2958.2007.05827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boshoff HI, Barry CE., 3rd Tuberculosis – metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3(1):70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 48.Fu LM. Exploring drug action on Mycobacterium tuberculosis using affymetrix oligonucleotide genechips. Tuberculosis (Edinb) 2006;86(2):134–143. doi: 10.1016/j.tube.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Betts JC, McLaren A, Lennon MG, et al. Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003;47(9):2903–2913. doi: 10.1128/AAC.47.9.2903-2913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bardou F, Raynaud C, Ramos C, Laneelle MA, Laneelle G. Mechanism of isoniazid uptake in Mycobacterium tuberculosis. Microbiology. 1998;144(Part 9):2539–2544. doi: 10.1099/00221287-144-9-2539. [DOI] [PubMed] [Google Scholar]
- 51.Vilcheze C, Weisbrod TR, Chen B, et al. Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob Agents Chemother. 2005;49(2):708–720. doi: 10.1128/AAC.49.2.708-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramaswamy SV, Reich R, Dou SJ, et al. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003;47(4):1241–1250. doi: 10.1128/AAC.47.4.1241-1250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta AK, Katoch VM, Chauhan DS, et al. Microarray analysis of efflux pump genes in multidrug-resistant Mycobacterium tuberculosis during stress induced by common antituberculous drugs. Microb Drug Resist. 2019;16(1):21–28. doi: 10.1089/mdr.2009.0054. [DOI] [PubMed] [Google Scholar]
- 54.McLean KJ, Munro AW. Structural biology and biochemistry of cytochrome P450 systems in Mycobacterium tuberculosis. Drug Metab Rev. 2008;40(3):427–446. doi: 10.1080/03602530802186389. [DOI] [PubMed] [Google Scholar]
- 55.Vilcheze C, Av-Gay Y, Attarian R, et al. Mycothiol biosynthesis is essential for ethionamide susceptibility in Mycobacterium tuberculosis. Mol Microbiol. 2008;69(5):1316–1329. doi: 10.1111/j.1365-2958.2008.06365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miesel L, Weisbrod TR, Marcinkeviciene JA, Bittman R, Jacobs WR., Jr NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J Bacteriol. 1998;180(9):2459–2467. doi: 10.1128/jb.180.9.2459-2467.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh A, Crossman DK, Mai D, et al. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5(8):e1000545. doi: 10.1371/journal.ppat.1000545. [▪ Article linking the maintenance of redox potential with the regulation of complex lipid metabolism in TB.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cunningham AF, Spreadbury CL. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J Bacteriol. 1998;180(4):801–808. doi: 10.1128/jb.180.4.801-808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barry CE., 3rd Interpreting cell wall ‘virulence factors’ of Mycobacterium tuberculosis. Trends Microbiol. 2001;9(5):237–241. doi: 10.1016/s0966-842x(01)02018-2. [DOI] [PubMed] [Google Scholar]
- 60.Mizrahi V, Dawes S, Rubin EJ. DNA Replication. In: Hatfull GF, Jacobs WR Jr, editors. Molecular Genetics of Mycobacteria. ASM Press; USA: 2000. pp. 159–172. [Google Scholar]
- 61.Alland D, Kramnik I, Weisbrod TR, et al. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1998;95(22):13227–13232. doi: 10.1073/pnas.95.22.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Colangeli R, Helb D, Sridharan S, et al. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol Microbiol. 2005;55(6):1829–1840. doi: 10.1111/j.1365-2958.2005.04510.x. [DOI] [PubMed] [Google Scholar]
- 63.O’Hare HM, Duran R, Cervenansky C, et al. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol Microbiol. 2008;70(6):1408–1423. doi: 10.1111/j.1365-2958.2008.06489.x. [DOI] [PubMed] [Google Scholar]
- 64.Walburger A, Koul A, Ferrari G, et al. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. 2004;304(5678):1800–1804. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- 65.Wolff KA, Nguyen HT, Cartabuke RH, Singh A, Ogwang S, Nguyen L. Protein kinase G is required for intrinsic antibiotic resistance in mycobacteria. Antimicrob Agents Chemother. 2009;53(8):3515–3519. doi: 10.1128/AAC.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406(6797):735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 67.Honaker RW, Stewart A, Schittone S, Izzo A, Klein MR, Voskuil MI. Mycobacterium bovis BCG vaccine strains lack narK2 and narX induction and exhibit altered phenotypes during dormancy. Infect Immun. 2008;76(6):2587–2593. doi: 10.1128/IAI.01235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Low KL, Rao PS, Shui G, et al. Triacylglycerol utilization is required for regrowth of in vitro hypoxic nonreplicating Mycobacterium bovis bacillus Calmette–Guerin. J Bacteriol. 2009;191(16):5037–5043. doi: 10.1128/JB.00530-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schnappinger D, Ehrt S, Voskuil MI, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198(5):693–704. doi: 10.1084/jem.20030846. [▪ First report of the TB intracellular transcriptome, identifying temporal changes as the intracellular microenvironment shifts.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tailleux L, Waddell SJ, Pelizzola M, et al. Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages. PLoS ONE. 2008;3(1):e1403. doi: 10.1371/journal.pone.0001403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jayaram R, Shandil RK, Gaonkar S, et al. Isoniazid pharmacokinetics-pharmaco-dynamics in an aerosol infection model of tuberculosis. Antimicrob Agents Chemother. 2004;48(8):2951–2957. doi: 10.1128/AAC.48.8.2951-2957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gomez JE, McKinney JD. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis(Edinb) 2004;84(1–2):29–44. doi: 10.1016/j.tube.2003.08.003. [DOI] [PubMed] [Google Scholar]
■ Websites
- 101.BμG@Sbase. Mycobacterium tuberculosis array design. http://bugs.sgul.ac.uk/A-BUGS-23.
- 102.BμG@Sbase. microarray dataset. http://bugs.sgul.ac.uk/E-BUGS-104.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.