Abstract
Merkel cell carcinoma is a rare, lethal cancer histopathologically composed of cells showing similarity with mechanoreceptor Merkel cells. Merkel cell tumors manifest in two distinct forms. While a virus called Merkel cell polyomavirus is involved in the pathogenesis of one form of Merkel tumors, the other is driven by ultraviolet (UV)-linked mutations. In this study we investigated 18 cases, from the Indian population, of Merkel cell carcinoma for immunohistochemical (IHC) expression of Merkel cell polyomavirus (MCV) T antigen, including 12 cases tested by PCR, to identify viral etiopathology. We tested the tumors with two sensitive antibodies (CM2B4 and Ab3), targeting the viral large T antigen protein and with PCR primers targeting the N terminus of T antigen. Overall, we observed 38.8% (7/18) tumors displaying positive IHC expression of Merkel cell polyomavirus T antigen and 25% (3/12) tumors showing positive results, by both, immunohistochemistry and PCR. This constitutes the first report from India showing implication of MCV in Merkel cell carcinomas. Moreover, this is one of the larger series of Merkel cell carcinomas, tested for MCV, by both immunohistochemistry and PCR, in this part of the world. These results further indicate that a slightly more number of such cases in India are likely to be caused by UV-linked damage, as opposed to Merkel cell polyomavirus mediated tumorigenesis, which is definitely implicated in a subset of cases.
Keywords: Merkel cell carcinoma, Polyomavirus, T antigen, MCC, MCV
1. Introduction
Merkel cell carcinoma (MCC) is an uncommon, primary, cutaneous, neuroendocrine neoplasm, associated with a poor prognosis [1–3]. MCC disease-associated mortality is greater than 40%, significantly higher than that of melanoma and other cutaneous cancers [2,4]. Prolonged ultra violet (UV) exposure, advanced age and loss of immune competence constitute as risk factors for development of MCC [1,2]. Tumor cells of MCC resemble mechanoreceptor cells, which are located in the basal layer of the epidermis. These cells originate from the epidermal progenitors during embryonic development and play an integral role in the sensory system of the skin [3,5–7].
As a result of its rarity and its similarity with other more common neuroendocrine tumors (such as small cell lung carcinoma) measuring the incidence of MCC has been challenging. The incidence rate of MCC is different across regions of the world. In the United States of America (USA), the incidence rate reported in the 2011 SEER (Surveillance, Epidemiology, and End Results) analysis was 0.79 per 100,000 [8]. In Europe, the incidence, as reported by the RARECARE database was 0.13 per 100,000 between 1995 and 2002 [9]. Australia (Queensland) has the highest reported age-adjusted incidence of 1.6 per 100,000 from 1993 to 2010 [10]. The incidence of this neoplasm has quadrupled over the last 20 years in America and Europe, possibly due to systemic immune-suppression, cumulative UV exposure and longer life-span [8]. A recent study reported that incidence of MCC is outpacing that of other cancers with a 95% increase from 2000 to 2013 [11,12]. The authors from that study have predicted the number of newly diagnosed cases would possibly increase from 2488 in 2013–3284 in the year 2025 [11,12].
In the year 2008, Feng et al. [13] discovered integration of polyomavirus genome in the genome of MCC tumor cells. This created a major paradigm shift in the understanding of the etiology of MCC. As a result of a strong association with immunosuppression and its epidemiology, MCC was selected and subjected to a direct sequencing-based method, known as the Digital Transcriptome Subtraction (DTS) [13,14]. An in silico subtraction of MCC tumor cDNA libraries against curated human genomic sequence databases identified candidate nonhuman sequences for further examination. A transcript with homology to other polyomaviruses was identified and led the way to the discovery of a novel virus associated with MCC [13]. The virus was named after the cancer as Merkel cell polyomavirus and is the 7th known cancercausing virus in humans [15–17].
Polyomaviruses have been previously linked to human disease [18]. However, unlike other polyomaviruses, MCV is the first polyomavirus, found to be convincingly linked to a human cancer as a causative agent [19]. Compelling evidence of causation has led to its classification as a Group 2A carcinogen by World Health Organization (WHO) International Agency for Research on Cancer (IARC) [20].
Interestingly, serological and molecular studies indicate that MCV is a near-ubiquitous, asymptomatic, life-long infection of adults [21]. MCV DNA has been detected in human skin and a variety of other tissues [13,22,23]. However, MCV DNA is found in higher copy numbers and its T antigen onco-proteins (multiply-spliced) express specifically in only MCC tumor cells, and not in neighboring healthy tissue or any other cancer cells [22].
MCV T antigens (Large T and small T) contain conserved domains present across different polyomaviruses, such as DnaJ and LXCXE retinoblastoma protein binding motifs, which are important for various tumor suppressor-targeting functions of the virus [24]. MCV wild type T antigens however, are distinct from MCV tumor derived T antigens. In tumor derived MCVs, mutations truncate the T antigens, removing the C terminal helicase and origin binding domains thereby rendering the virus replication-incompetent in the tumors [22,25].
In an attempt to distinguish the two types of MCC, several studies have shown that MCPyV-negative tumors have a higher number of chromosomal aberrations and UV-specific mutation burden [26–30]. A multivariate analysis, including age, sex and immunosuppression, of 282 MCC cases showed that MCV negative MCC patients have a significantly higher risk of disease progression (hazard ratio = 1.77) and death from MCC (hazard ratio = 1.85) [31] in comparison to MCV positive MCC patients. This suggests that virus-negative tumors may be clinically more aggressive.
Isolate cases of MCC have been reported from various regions of India [32–44]. However an estimated incidence of MCC in India is not known. A recently published study by Rekhi et al.32 describes the clinico-pathologic spectrum of the first largest series of 16 cases of MCC diagnosed at a tertiary cancer referral center in India [32]. However, none of these cases were tested for MCV as the causative agent.
In this study, we tested 18 cases of MCC for MCV T antigen by immunohistochemistry, in order to identify association of MCC with MCV in Indian cases. The cases were diagnosed as MCC using a series of specific IHC markers including positive expression of cytokeratin 20 (CK20). We used immunohistochemistry against the viral Large T antigen protein using two antibodies (CM2B4 and Ab3). Moreover, we also tested 12 of these cases for MCV virus, using PCR and applied the multimodal method (both IHC and PCR) for MCV detection [31].
2. Materials and methods
2.1. Samples
After computerized search of medical records from January 1, 2000, to March 7, 2017, 24 cases were found, in which MCC was either offered as a diagnosis or constituted as one of the differential diagnoses. The diagnosis was confirmed using Immunohistochemical staining using markers described in Table 1. Nine cases included in this study were from the earlier reported case series by Rekhi et al. [32]. Finally, paraffin blocks were available for 18 cases and those were included in this study. Conventional hematoxylin and eosin-stained slides along with immunohistochemical (IHC) stained microsections were reviewed by the specialty pathologist. Institutional Ethics Committee Approval was obtained for this study (IEC-263), from both the institutions involved in the study.
Table 1. MCC Cases Tested for MCV T antigen Expression.
| Case # | Age | Sex | Site | IHC |
Treatment | Outcome | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD56 | CK20 | Chromogranin | CK7 | Synaptophysin | Others | MCVLT (CM2B4) |
MCVLT (Ab3) |
MCVLT (PCR) |
||||||
| 1 | 59 | M | Groin | + | + | + | – | – | HMWCK- | – | – | + | NK | Metastais (LN) |
| 2 | 37 | M | Thigh | + | + | + | + | c-KIT+, MIC2- | + | + | + | Rx, Rx-R0 resection | NK | |
| 3* | 37 | F | Wrist | + | + | – | MIB1-100- | – | – | + | Excision, R0 resection | FOD(21mo) | ||
| 4 | 67 | M | AxillaLN | + | + | – | + | CK+, EMA+, AE1A3+ | + | + | + | Excision, CT(Etoposide, Carboplatin) | FOD(16mo) | |
| 5 | 74 | M | Leg | + | + | CK- | – | – | + | NK | NK | |||
| 6 | 60 | F | Scalp | + | + | + | – | + | CK+,NSE + EMA+,MIB1-80- | – | – | NT | NK | NK |
| 7 | 57 | F | Nose | + | + | EMA+,CK- | + | + | NT | Excision | NK | |||
| 8 | 64 | M | Knee | + | + | EMA+, CK+,S100P- | – | – | NT | Excision | NK | |||
| 9 | 72 | M | Neck | + | + | AE1/AE3+,c-KIT+ | – | – | + | Excision | NK | |||
| 10 | 59 | M | Thumb | + | + | – | + | c-KIT+ | – | NT | NT | Amputation & CT (Paclitaxel, Carboplatin, Etoposide, Imatinib) | Metastasis (Scalp) | |
| 11 | 66 | M | Thigh | + | + | + | – | + | HMB45-,S100P-,AE1/AE3+ | – | – | + | Supportive Care | Metastasis (LN & Lung) |
| 12 | 74 | F | Knee | + | + | S100P-, GFAP-,AE1/AE3+, EMA-,BCl2+ | – | + | – | NK | NK | |||
| 13 | 62 | F | Arm | + | AE1/AE3+, MIC-2+, Desmin-,Tdt- | – | + | + | NK | NK | ||||
| 14 | 62 | F | Thigh | + | + | + | MIC2+, AE1/AE3+, Desmin-, MyoD1-, Myogenin- | – | – | – | NK | NK | ||
| 15 | 60 | F | Neck (Submadibular) | + | + | + | + | EMA+ | – | – | NT | NK | NK | |
| 16 | 50 | M | Chest wall | + | – | + | AE1/AE3+, MIC2+, Fli1+,Desmin- | – | – | – | NK | NK | ||
| 17 | 42 | M | Upper Arm | + | + | + | AE1/AE3+, MIC2+, Fli1+,Desmin-, S100P- | + | + | – | NK | NK | ||
| 18 | 49 | M | Inguinal and Gluteal | + | + | MIC2+,Desmin- | + | NT | NT | CT | Metastais (LN) | |||
Abbreviations: F-female, M-Male,+ -positive, − -negative,NT-Not tested, HMWCK-High molecular weight cytokeratin, EMA-Epithelial membrane antigen, CK- Cytokeratin, AE1/AE3- Pan- Cytokeratin, NSE- Neuron specific enolase, MIB1-Mind Bomb1, S100P–S100 calcium-binding protein P, HMB45-Human melanoma Black, GFAP-Glial fibrillary acid protein, MIC2-Cluster of differentiation 99 (CD99), Tdt-Terminal deoxynucleotidyl transferase, Fli1- Friend Leukemia Integration, * -MCC with coexisting Bowen disease., NK-not known, CT-chemotherapy, FOD-free-of-disease, LN-Lymph Node.
2.2. Immunohistochemistry
Immunohistochemical (IHC) staining was performed using the polymer technique (Dako REAL Envision detection system, Glostrup, Denmark) including peroxidase/3-3-diaminobenzidine tetrahydrochloride (DAB). The antibodies used to test for Merkel cell polyomavirus in this study were CM2B4 (Santa Cruz, sc-136172) and Ab3, both target MCV Large T antigen. All 18 tumors were tested with CM2B4 antibody. Sixteen tumors were also stained using Ab3 [45]. Ab3 was a kind gift from Prof James Decaprio, Dana Farber Cancer Institute, Boston, USA. Various other antibodies utilized have been listed in the Supplementary Table 2. Dilutions of 1:50 were used for both Ab3 and CM2B4 antibodies. Antigen retrieval was by heating – microwave for CM2B4 and pressure cooker for Ab3.
2.3. DNA extraction and PCR
For DNA isolation, 4 FFPE tissue sections of 5 μm were cut from each tumour block (microtome, Leica) and placed in sterile 1.5 mL centrifuge tubes ready for extraction. After deparaffinizing the sections with xylene, the DNA was extracted using the QiaAmp FFPE tissue kit (Cat no 56404). Briefly, tissue sample is lysed with proteinase K under denaturing conditions followed by incubation at 90 °C. The DNA is extracted through the column membrane and eluted in ATE elution buffer. These FFPE samples derived DNA were subjected to PCR using DreamTaq Master Mix (ThermoFischer Scientific, K1081). Briefly, 10 μl of 2X DreamTaq PCR Master mix, 1 μl Forward primer (RA149), 1 μl Reverse primer (RA150), 7 μl of nuclease-free-water and 1 μl (20 ng) template DNA were mixed and used for PCR. The PCR program used is as follows: 95 °C for 2 min, 95 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min (last 3 steps for 35 cycles) followed by 72 °C for 5 min. The PCR products were run on a 2% Agarose (HiMedia, MB002) gel at 120 V for 30 min. Primers: RA149 – TGGCAACATCCCTCTGATGAA and RA150-TGCTGGAATTTGCTCCAAAGG, Expected PCR product size – 120bp.
2.4. Real-time quantitative polymerase chain reaction (qPCR)
qPCR was performed with the SYBR Green method using DNA extracted from MCC FFPE samples as template, 0.25 μM gene-specific primer pairs and Maxima SYBR Green/ROX qPCR 2X Master Mix (K0221, Thermo Fisher Scientific) in a 10 μl reaction. qPCR reactions were performed in duplicate using the ViiA 7 Real-Time PCR System (Applied Biosystems) with the following thermal cycling conditions: 50 °C for 2 min, initial denaturation at 95 °C for 10 min, then 40 cycles of denaturation (95 °C, 15 s), annealing (55 °C, 45 s) and extension (72 °C, 45 s). Melt curve analysis of every qPCR was conducted after each cycle. Primers used were: For T antigen - (RA149) forward- TGGCAACATCCCTCTGATGAA and (RA150) reverse- TGCTGGAATTTGCTCCAAAGG; for GAPDH – (RA322) forward – CCCTTCATTGACCTCAACTACAT and (RA323) reverse- TGGAAGATGGTGATGGGATTT. GAPDH was amplified as internal control. Copy numbers were established from standard curves of Ct values from serial dilutions of known concentrations of MCV DNA using Plasmid pLenti.puro.MCV.ER.RAZ2. (Addgene #114382) and MKL-1 genomic DNA. Water was used as control to detect template contamination. GAPDH was used to determine cell genome copy number. Results were expressed as numbers of viral copies per cell calculated from Ct values of viral and cellular gene standards. Cellular viral DNA copy number below 1.0 × 10−3 per cell was considered negative.
3. Results
Eighteen primary MCCs tested in this study occurred in 11 men and 7 women (M/F = 1.57:1), with their age ranging from 37 to 74 years (average = 58; median = 60). Site-wise, the tumors were frequently observed in the lower extremities (9, 50%), head and neck sites (4, 22.2%), followed by the upper extremities (4, 22.2%) and chest wall (1, 5.55%) (Table 1). A single case presented with a coexisting Bowen's disease. Cases 1, 10 and 12 had high blood sugar levels and were suspected diabetic. Tumor size, known in 4 cases, varied from 2.5 cm to 16 cm. Therapeutically, surgical resection/excision was performed in 7 cases; chemotherapy was offered in 3 cases and one patient was given supportive care. Two of the patients who received excision, one along with chemotherapy (Etoposide, Carboplatin) were free of disease at 16 months and 21 months respectively. Despite treatment, 4 patients developed metastasis at lymph nodes, lung and the scalp. The Fitzpatrick scoring for skin type was done where relevant information was available. Cases 1,2, 9,10, 11 are of score 3–6. Case 3 was of score 2. Though Case 2 was clearly MCV positive and had a higher Fitzpatrick score, the other cases were 3,9,10 and 11 were negative for MCV via immunostaining and there is no clear link to Fitzpatrick skin type.
Histopathologically, almost all tumors were composed of malignant round to oval cells with scanty cytoplasm, found in diffuse, trabecular, cord-like, nesting and occasionally pseudoglandular patterns. Variable amounts of necrosis in the tumor tissue and prominent apoptotic and mitotic cells were seen.
The tumors were characterized as MCC using immunohistochemical staining for CK20, CD56, chromogranin, and synaptophysin. All 15 tumors, tested for CK20, displayed a characteristic ‘dot-like’ paranuclear-staining pattern. Remaining 3 tumors, not tested for CK20, were distinctly positive for neuroendocrine markers, namely chromogranin, as well as other epithelial markers and other pan-cytokeratin stainings (CK and AE1/AE3). Two of those tumors were also positive for another neuroendocrine marker, namely synaptophysin. Possibility of metastatic neuroendocrine carcinomas was ruled out, based on clinical and imaging results. Other immunohistochemical markers, such as MIC2, Desmin, etc. were used to differentiate these tumors from their diagnostic mimics.
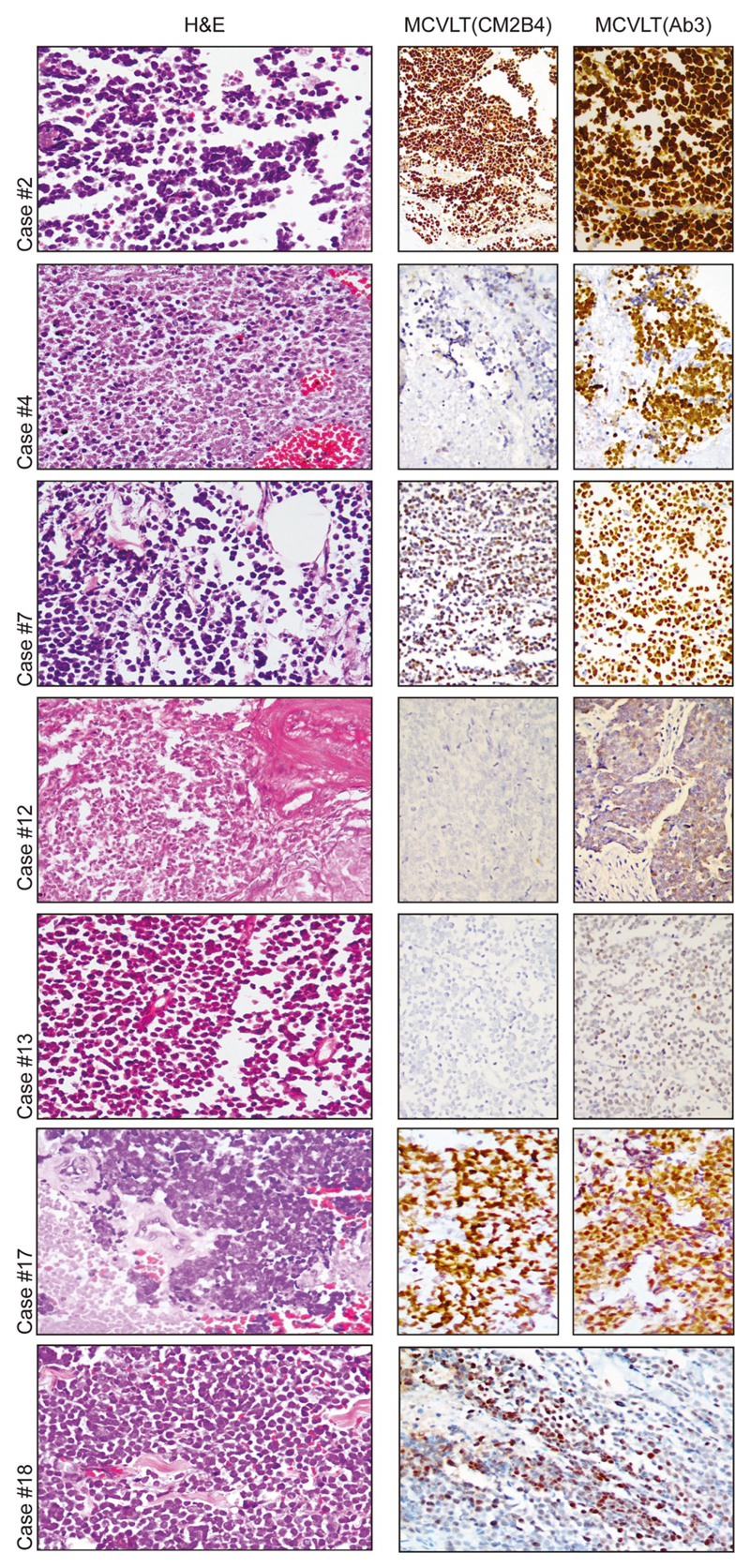
MKL-1 cells embedded in a paraffin block were used as a positive control. Sections stained without primary antibody were used as a negative control. Seven out of 18 tumors (38.8%) showed positive staining for MCV LT using either CM2B4 or Ab3 antibodies. The staining pattern ranged from diffuse intranuclear staining in cases #2, #4, #7,#17 and #18 to weak and focal staining in cases #12 and #13 (Fig. 1). Two of these cases (case#12 and #13) displayed negative immunostaining with CM2B4 antibody, but showed positive immunostaining with Ab3.
Fig. 1. Merkel cell polyomavirus positive Merkel cell carcinoma.
7 cases of Merkel cell carcinoma were found positive for Merkel cell polyomavirus. H&E staining shows epidermis with a tumor below composed of malignant round cells, medium power. Tumor cells show MCV Large T antigen positivity ranging from strong positivity (cases #2, #4, #7, #17 and #18) to weak/focal positivity (cases #12 and #13). Image for Cases #18 is of CM2B4 staining, as it was not tested with Ab3.
We also tested 14 other skin tumors by immunostaining with both CM2B4 and Ab3 antibodies. All 5 basal cell carcinomas, 5 melanomas and 3 of 4 squamous cell cutaneous carcinomas were negative for both the antibody IHC stains (Supplementary Fig. 2). A single case squamous cell carcinoma was negative with CM2B4, however showed focal weak immunostaining with Ab3. These observations further testify high specificity of both the antibodies.
To further evaluate the presence of MCV in these MCC samples, we extracted DNA from these FFPE samples and subjected them to PCR. We were able to obtain good quality DNA for only 12 MCC cases. For Case #2 we extracted DNA from 2 samples, tumor excision and post tumor excision with no residual tumor. We performed PCR using primers that target 120bp of the T antigen region, common to both small and large T. A total of 8 samples were positive (some weakly, 66.7%) (Supplementary Fig. 1). Further, qPCR was done to determine cellular viral genome copy number, using GAPDH as normalizing control. The MCC cases showed cellular viral copy numbers of the range of 2.8–9.7 (Supplementary Table 1). When analyzed together with the IHC results (Table 1 and Supplementary Table 1), for a multimodal approach, 2 cases (Case #2 and #4) was consistently positive using all assays, and 1 sample (Cases #13) was positive for at least two of the 3 assay conditions. We did see variation between the IHC positive and PCR positive cases, and five of the IHC - negative cases showed PCR positivity. Although these cases cannot be excluded as being MCV positive, this detection could be attributed to coincidental infection, low-tumor purity and infectious wild-type MCV in the adjacent nonmalignant skin. Hence, of the 12 cases tested via both IHC and PCR, 3 (25%) were determined to be MCV positive via the multimodal approach.
4. Discussion
This study is the first and largest series of 18 MCCs from our country tested for the presence of MCV as a causative agent, including 9 cases from an earlier reported series by Rekhi et al. [32]. Various reports have shown that the proportion of MCV-positive MCCs varies with the geographic location [46,47]. Although, reports from United States and Europe revealed nearly 80% viral positive cases of MCC; studies from Australia described the same in only 24% cases [46,47]. Studies from Japan, show a 76.9%–88.5% positivity for MCV in Japanese MCC patients [48]. Ours is the first such study from India, wherein we observed a relatively lesser number of MCV positive MCCs (38.8% by IHC and 25% by IHC and PCR). More cases are likely to yield a better picture of the viral positive Merkel tumor prevalence in India. Limited sensitivity of the CM2B4 antibody, used in this study, can be another reason for this difference.
We noted 7 out of 18 (38.8%) cases of MCC, positive for MCV large T antigen protein. The antibodies used to detect MCV presence in this study was the CM2B4 antibody [22], with a specificity of 0.943 and sensitivity of 0.882 [31], and Ab3 [45], with a specificity of 0.453 and a sensitivity of 0.983 [31]. We observed a range of very strong positive staining to weak/focal positive staining that is reflective of the level of T antigen present in the different MCC patients. Such a range depends on the specific truncation of the T antigen in the MCC tumor and thereby its protein levels, as has been seen with MCC cell lines [49,50]. We identified a single case that showed negative immunostaining with CM2B4 antibody, but showed positive immunostaining with Ab3, though weak and focal. This could be explained by the higher sensitivity of this antibody.
PCR based detection for MCV, although a widely used easy-to-use diagnostic method is not as sensitive or specific as IHC [31,51,52]. Moshiri et al. recently reported a qPCR that showed 82.5% sensitivity and 81.1% specificity for identifying intratumoral MCV [33]. In our study a total of 8 samples were positive for PCR of the 12 tested. However, only 3 cases were positive for both IHC and PCR. Besides low sensitivity, PCR contamination and false-positivity are major concerns of using PCR diagnostically. In the case of MCV detection, background, wild-type MCV infection can also complicate interpretation. Low purity tumor samples might be confounded by background signals from skin and/or other non-MCC tumors and hence coincidental MCV is often detected in these cases [51].
Hence, a multimodal approach, to convincingly determine MCV positivity, using both IHC and PCR testing assays, has recently been suggested [31]. In our study, using this multimodal method, we observed that only 3 of the 12 cases (25%) tested by both IHC and PCR were positive for MCV by both tests, compared to when using only IHC to determine the same (7/18, 38.8%).
Viral positive MCCs did not show any specific clinical profile in comparison to the viral negative cases. Both forms of MCC, as described, occurred in sun-exposed, as well as un-exposed regions of the body and stained similarly with IHC markers. Hence, unless tested for MCV oncoprotein expression, to a pathologist both types of MCC are indistinguishable. However, our study results concurred with that of Pasternak et al.’s recent immunochemical profile of MCC subsets. Few of our MCV positive MCC cases fit the “classical” CK20+, Chromogranin+, CK7- and TTF1-profiles, (where all 4 markers were tested) as they described, however we did not test for Neurofilament [53,54].
Although information related to patient outcomes was not available for all the cases, of the 4 cases that showed metastatic growth,3 were negative for MCV immunostaining. Also, of the 2 cases reported to be free of disease, one was strongly viral positive via all tests. These observations support Moshiri et al. data [31] relating to better prognosis for virus positive patients.
Lately, studies based on Next-generation sequencing (NGS) of whole genomes and transcriptomes have shown distinction between the MCV positive and MCV negative MCC tumors [28–30]. MCV is clearly the primary etiological agent of virus-associated tumors and these have minimally altered genomes. On the other hand, non-virus associated tumors or virus negative tumor genomes are dominated by UV-mediated mutations. These mutations have been mostly observed in p53 signaling, cell-cycle and DNA-damage response pathways. Collectively, all these studies indicate that MCV has a dominant control and drives tumorigenesis to the same severity as all the somatic genome alterations seen in MCCs2, 28.
Most importantly, these distinct mutagenic processes, operative in viral and non-viral MCCs affect disease outcomes. Furthermore, Moshiri et al.31 observed that MCV negative MCCs showed a clinically more aggressive phenotype and also a lower progression free survival in comparison to polyomavirus positive MCC [31]. Hence, MCV T antigen expression in MCC is an important determinant not just of causation of the disease but also has implications for the effective treatment of these tumors and determining subgroup-related prognosis. Our work has direct impact on both diagnostic practice and prognostic prediction and we hope this kind of testing can be incorporated into clinical use.
5. Conclusions
The present study constitutes the first study on analysis of MCCs from the Indian subcontinent, showing implication Merkel cell polyomavirus (MCV) as the etiologic agent in a significant number of Merkel tumors.
Supplementary Material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micpath.2019.103778.
Acknowledgments
This study was performed via collaboration between Dr. Reety Arora, NCBS, Bangalore and Dr. Bharat Rekhi at Tata Memorial Hospital, Mumbai, India. We are grateful for the Immunohistochemistry support from Dr. Bharat Rekhi's laboratory at Tata Memorial Hospital, Mumbai. We would like to extend our thanks to Ms Anjali Vijaykumar and Ms Komal Gupta for their assistance. We would also like to thank and acknowledge both NCBS and TMH administration teams for their assistance with MOU preparation and processing. RA would like to thank Zaina, Ziyah and Karan for their love, strength and support.
Funding
This work was supported by the Wellcome Trust/DBT India Alliance (Early Career Award IA/E/14/1/501773 to RA).
Footnotes
Ethical approval
Institutional Ethics Committees of TMH, Mumbai and NCBS, Bangalore approved this study (IEC-263).
Declaration of competing interest
The authors declare no conflict of competing interests related to this work.
References
- [1].Hodgson NC. Merkel cell carcinoma: changing incidence trends. J Surg Oncol. 2005;89:1–4. doi: 10.1002/jso.20167. [DOI] [PubMed] [Google Scholar]
- [2].Becker JC, Stang A, DeCaprio JA, Cerroni L, Lebbe C, Veness M, et al. Merkel cell carcinoma. Nat Rev Dis Prim. 2017;3:17077. doi: 10.1038/nrdp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Schadendorf D, Lebbe C, Zur Hausen A, Avril MF, Hariharan S, Bharmal M, et al. Merkel cell carcinoma: epidemiology, prognosis, therapy and unmet medical needs. Eur J Cancer. 2017;71:53–69. doi: 10.1016/j.ejca.2016.10.022. [DOI] [PubMed] [Google Scholar]
- [4].Lemos B, Nghiem P. Merkel cell carcinoma: more deaths but still no pathway to blame. J Investig Dermatol. 2007;127:2100–2103. doi: 10.1038/sj.jid.5700925. [DOI] [PubMed] [Google Scholar]
- [5].Haeberle H, Fujiwara M, Chuang J, Medina MM, Panditrao MV, Bechstedt S, et al. Molecular profiling reveals synaptic release machinery in Merkel cells. Proc Natl Acad Sci U S A. 2004;101:14503–14508. doi: 10.1073/pnas.0406308101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lucarz A, Brand G. Current considerations about Merkel cells. Eur J Cell Biol. 2007;86:243–251. doi: 10.1016/j.ejcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- [7].Van Keymeulen A, Mascre G, Youseff KK, Harel I, Michaux C, De Geest N, et al. Epidermal progenitors give rise to Merkel cells during embryonic development and adult homeostasis. J Cell Biol. 2009;187:91–100. doi: 10.1083/jcb.200907080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fitzgerald TL, Dennis S, Kachare SD, Vohra NA, Wong JH, Zervos EE. Dramatic increase in the incidence and mortality from Merkel cell carcinoma in the United States. Am Surg. 2015;81:802–806. doi: 10.1177/000313481508100819. [DOI] [PubMed] [Google Scholar]
- [9].van der Zwan JM, Trama A, Otter R, Larranaga N, Tavilla A, Marcos-Gragera R, et al. Rare neuroendocrine tumours: results of the surveillance of rare cancers in Europe project. Eur J Cancer. 2013;49:2565–2578. doi: 10.1016/j.ejca.2013.02.029. [DOI] [PubMed] [Google Scholar]
- [10].Youlden DR, Soyer HP, Youl PH, Fritschi L, Baade PD. Incidence and survival for Merkel cell carcinoma in Queensland, Australia, 1993-2010. JAMA Dermatol. 2014;150:864–872. doi: 10.1001/jamadermatol.2014.124. [DOI] [PubMed] [Google Scholar]
- [11].Paulson KG, Park SY, Vandeven NA, Lachance K, Thomas H, Chapuis AG, et al. Merkel cell carcinoma: current US incidence and projected increases based on changing demographics. J Am Acad Dermatol. 2018;78:457–463 e2. doi: 10.1016/j.jaad.2017.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Voelker R. Why Merkel cell cancer is garnering more attention. J Am Med Assoc. 2018 doi: 10.1001/jama.2018.7042. [DOI] [PubMed] [Google Scholar]
- [13].Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Feng H, Taylor JL, Benos PV, Newton R, Waddell K, Lucas SB, et al. Human transcriptome subtraction by using short sequence tags to search for tumor viruses in conjunctival carcinoma. J Virol. 2007;81:11332–11340. doi: 10.1128/JVI.00875-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chang Y, Moore PS, Weiss RA. Human oncogenic viruses: nature and discovery. Philos Trans R Soc Lond B Biol Sci. 2017;372 doi: 10.1098/rstb.2016.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Moore PS, Chang Y. Common commensal cancer viruses. PLoS Pathog. 2017;13:e1006078. doi: 10.1371/journal.ppat.1006078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat Rev Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gjoerup O, Chang Y. Update on human polyomaviruses and cancer. Adv Cancer Res. 2010;106:1–51. doi: 10.1016/S0065-230X(10)06001-X. [DOI] [PubMed] [Google Scholar]
- [19].DeCaprio JA. Merkel cell polyomavirus and Merkel cell carcinoma. Philos Trans R Soc Lond B Biol Sci. 2017;372 doi: 10.1098/rstb.2016.0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bouvard V, Baan RA, Grosse Y, Lauby-Secretan B, El Ghissassi F, Benbrahim-Tallaa L, et al. Carcinogenicity of malaria and of some polyomaviruses. Lancet Oncol. 2012;13:339–340. doi: 10.1016/s1470-2045(12)70125-0. [DOI] [PubMed] [Google Scholar]
- [21].Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernandez-Figueras MT, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int J Cancer. 2009;125:1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, et al. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int J Cancer J Int du Cancer. 2009;125:1250–1256. doi: 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pipas JM. Common and unique features of T antigens encoded by the polyomavirus group. J Virol. 1992;66:3979–3985. doi: 10.1128/jvi.66.7.3979-3985.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Arora R, Chang Y, Moore PS. MCV and Merkel cell carcinoma: a molecular success story. Curr Opin Virol. 2012;2:489–498. doi: 10.1016/j.coviro.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Goh G, Walradt T, Markarov V, Blom A, Riaz N, Doumani R, et al. Mutational landscape of MCPyV-positive and MCPyV-negative Merkel cell carcinomas with implications for immunotherapy. Oncotarget. 2016;7:3403–3415. doi: 10.18632/oncotarget.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Paulson KG, Lemos BD, Feng B, Jaimes N, Penas PF, Bi X, et al. Array-CGH reveals recurrent genomic changes in Merkel cell carcinoma including amplification of L-Myc. J Investig Dermatol. 2009;129:1547–1555. doi: 10.1038/jid.2008.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Starrett GJ, Marcelus C, Cantalupo PG, Katz JP, Cheng J, Akagi K, et al. Merkel cell polyomavirus exhibits dominant control of the tumor genome and transcriptome in virus-associated Merkel cell carcinoma. mBio. 2017;8 doi: 10.1128/mBio.02079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Harms PW, Collie AM, Hovelson DH, Cani AK, Verhaegen ME, Patel RM, et al. Next generation sequencing of Cytokeratin 20-negative Merkel cell carcinoma reveals ultraviolet-signature mutations and recurrent TP53 and RB1 inactivation. Mod Pathol. 2016;29:240–248. doi: 10.1038/modpathol.2015.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Harms PW, Vats P, Verhaegen ME, Robinson DR, Wu YM, Dhanasekaran SM, et al. The distinctive mutational spectra of polyomavirus-negative Merkel cell carcinoma. Cancer Res. 2015;75:3720–3727. doi: 10.1158/0008-5472.CAN-15-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Moshiri AS, Doumani R, Yelistratova L, Blom A, Lachance K, Shinohara MM, et al. Polyomavirus-negative Merkel cell carcinoma: a more aggressive subtype based on analysis of 282 cases using multimodal tumor virus detection. J Investig Dermatol. 2017;137:819–827. doi: 10.1016/j.jid.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rekhi B, Kane SV, Jambhekar NA. Clinicopathological spectrum of a series of Merkel cell carcinomas diagnosed at a tertiary cancer referral center in India, with current concepts. Ann Diagn Pathol. 2015;19:341–346. doi: 10.1016/j.anndiagpath.2015.07.006. [DOI] [PubMed] [Google Scholar]
- [33].Anand MS, Krishnamurthy S, Ravindranath S, Ranganathan J. Merkel cell carcinoma with seborrheic keratosis: a unique association. Indian J Pathol Microbiol. 2018;61:101–102. doi: 10.4103/IJPM.IJPM_659_16. [DOI] [PubMed] [Google Scholar]
- [34].Kini JR, Tapadia R, Shenoy S. Merkel cell carcinoma metastasis to stomach: an infrequent culmination of a rare neoplasm. J Gastrointest Cancer. 2018;49:78–80. doi: 10.1007/s12029-016-9852-2. [DOI] [PubMed] [Google Scholar]
- [35].Basu S, Ranade R. Favorable response of metastatic Merkel cell carcinoma to targeted 177Lu-DOTATATE therapy: Will PRRT evolve to become an important approach in receptor-positive cases? J Nucl Med Technol. 2016;44:85–87. doi: 10.2967/jnmt.115.163527. [DOI] [PubMed] [Google Scholar]
- [36].Gaopande VL, Joshi AR, Khandeparkar SG, Deshmukh SD. Merkel cell carcinoma of the abdominal wall. Indian Dermatol Online J. 2015;6:269–273. doi: 10.4103/2229-5178.160262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Ganjre AP, Bagul N, Sarode G. Merkel cell carcinoma - beast with two Backs. Oral Oncol. 2015;51:880–881. doi: 10.1016/j.oraloncology.2015.05.011. [DOI] [PubMed] [Google Scholar]
- [38].Roy S, Das I, Nandi A, Roy R. Primary Merkel cell carcinoma of the oral mucosa in a young adult male: report of a rare case. Indian J Pathol Microbiol. 2015;58:214–216. doi: 10.4103/0377-4929.155318. [DOI] [PubMed] [Google Scholar]
- [39].Khairwa A, Dey P, Bal A. Fine-needle aspiration cytology of recurrent Merkel cell carcinoma of eye-brow. J Cytol. 2014;31:179–180. doi: 10.4103/0970-9371.145666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gupta N, Samra SS, Nimbran V, Gupta RK, Kallianpur AA, Khurana U. Merkel cell carcinoma-A rare primary neuroendocrine skin tumor: case report and discussion. J Cancer Res Ther. 2014;10:437–439. doi: 10.4103/0973-1482.136683. [DOI] [PubMed] [Google Scholar]
- [41].Bhattacharjee PK, Halder SK, Ray RP. Merkel cell carcinoma: a rare presentation. J Cancer Res Ther. 2014;10:399–400. doi: 10.4103/0973-1482.136670. [DOI] [PubMed] [Google Scholar]
- [42].Medhi S, Purandare NC, Dua SG, Gujral S. Bilateral renal metastases in a case of Merkel cell carcinoma. J Cancer Res Ther. 2010;6:353–355. doi: 10.4103/0973-1482.73365. [DOI] [PubMed] [Google Scholar]
- [43].Prabhu S, Smitha RS, Punnya VA. Merkel cell carcinoma of the alveolar mucosa in a young adult: a rare case report. Br J Oral Maxillofac Surg. 2010;48:48–50. doi: 10.1016/j.bjoms.2008.12.005. [DOI] [PubMed] [Google Scholar]
- [44].Biswanath Paul BM, Pal Mallika, Saha Tarak Nath, Maiti Ashok. Merkel cell carcinoma : an Indian experience. Glob J Med Res: C Microbiol Pathol. 2014;14 [Google Scholar]
- [45].Cheng J, Rozenblatt-Rosen O, Paulson KG, Nghiem P, DeCaprio JA. Merkel cell polyomavirus large T antigen has growth-promoting and inhibitory activities. J Virol. 2013;87:6118–6126. doi: 10.1128/JVI.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Colunga A, Pulliam T, Nghiem P. Merkel cell carcinoma in the age of immunotherapy: facts and hopes. Clin Cancer Res. 2018;24:2035–2043. doi: 10.1158/1078-0432.CCR-17-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Garneski KM, Warcola AH, Feng Q, Kiviat NB, Leonard JH, Nghiem P. Merkel cell polyomavirus is more frequently present in North American than Australian Merkel cell carcinoma tumors. J Investig Dermatol. 2009;129:246–248. doi: 10.1038/jid.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hattori T, Takeuchi Y, Takenouchi T, Hirofuji A, Tsuchida T, Kabumoto T, et al. The prevalence of Merkel cell polyomavirus in Japanese patients with Merkel cell carcinoma. J Dermatol Sci. 2013;70:99–107. doi: 10.1016/j.jdermsci.2013.02.010. [DOI] [PubMed] [Google Scholar]
- [49].Velasquez C, Amako Y, Harold A, Toptan T, Chang Y, Shuda M. Characterization of a Merkel cell polyomavirus-positive Merkel cell carcinoma cell line CVG-1. Front Microbiol. 2018;9:713. doi: 10.3389/fmicb.2018.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, et al. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Harms PW, Harms KL, Moore PS, DeCaprio JA, Nghiem P, Wong MKK, et al. The biology and treatment of Merkel cell carcinoma: current understanding and research priorities. Nat Rev Clin Oncol. 2018;15:763–776. doi: 10.1038/s41571-018-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Miner AG, Patel RM, Wilson DA, Procop GW, Minca EC, Fullen DR, et al. Cytokeratin 20-negative Merkel cell carcinoma is infrequently associated with the Merkel cell polyomavirus. Mod Pathol. 2015;28:498–504. doi: 10.1038/modpathol.2014.148. [DOI] [PubMed] [Google Scholar]
- [53].Pasternak S, Carter MD, Ly TY, Doucette S, Walsh NM. Immunohistochemical profiles of different subsets of Merkel cell carcinoma. Hum Pathol. 2018 doi: 10.1016/j.humpath.2018.07.022. [DOI] [PubMed] [Google Scholar]
- [54].Kervarrec T, Tallet A, Miquelestorena-Standley E, Houben R, Schrama D, Gambichler T, et al. Diagnostic accuracy of a panel of immunohistochemical and molecular markers to distinguish Merkel cell carcinoma from other neuroendocrine carcinomas. Mod Pathol. 2018 doi: 10.1038/s41379-018-0155-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.