Abstract
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved cellular process capable of degrading various biological molecules (e.g., protein, glycogen, lipids, DNA, and RNA) and organelles (e.g., mitochondria, endoplasmic reticulum, ribosomes, lysosomes, and micronuclei) via the lysosomal pathway. Ferroptosis is a type of oxidative stress-dependent regulated cell death associated with iron accumulation and lipid peroxidation. The recently discovered role of autophagy, especially selective types of autophagy (e.g., ferritinophagy, lipophagy, clockophagy, and chaperone-mediated autophagy), in driving cells towards ferroptotic death motivated us to explore the functional interactions between metabolism, immunity, and cell death. Here, we describe types of selective autophagy and discuss the regulatory mechanisms and signaling pathways of autophagy-dependent ferroptosis. We also summarize chemical modulators that are currently available for triggering or blocking autophagy-dependent ferroptosis and that may be developed for therapeutic interventions in human diseases.
Keywords: autophagy, ferroptosis, network, regulated cell death, selective autophagy
Graphical Abstract
eTOC Blurb
Liu et al describes types of selective autophagy and discuss the regulatory mechanisms and signaling pathways of autophagy-dependent ferroptosis. A deeper understanding of the process and function of autophagy-dependent cell death is critical for creating innovative therapeutic strategies for oxidative stress-related diseases.
Introduction
Cell homeostasis is controlled by an integrated system that maintains the balance between biosynthesis (anabolism) and decomposition (catabolism). Macroautophagy (hereafter referred to as autophagy), which was first described in the 1950s, is a phylogenetically conserved catabolic process that plays a fundamental role in removing or clearing the accumulation of intracellular waste (Klionsky and Emr, 2000; Levine and Kroemer, 2019; Yang and Klionsky, 2010). After its breakdown by autophagy, cellular garbage, such as aggregated protein and damaged organelles, can be recycled for macromolecular synthesis and energy production. In addition, elements of the autophagic machinery can be used to eliminate invading pathogens or engulfed apoptotic cells via lysosomes. Thus, autophagy is generally beneficial for cellular survival in response to environmental stresses ranging from nutrient starvation to growth factor depletion, hypoxia, temperature change, and infectious pathogens (Kroemer et al., 2010; Murrow and Debnath, 2013). Increased autophagy appears to be widely observed during cell death, and the term “autophagic cell death” has been used to describe type II cell death according to morphological alterations (Kroemer and Levine, 2008). Excessive or dysfunctional autophagy can result in cellular killing. In particular, the term “autophagy-dependent cell death” was recommended by the Nomenclature Committee on Cell Death to describe a form of regulated cell death (RCD) that mechanistically depends on the autophagic machinery or components thereof (Galluzzi et al., 2018). Autophagy-dependent cell death has been implicated in several diseases such as cancer, neurodegeneration, and ischemia-reperfusion injury (Bialik et al., 2018; Denton and Kumar, 2019). Mechanistically, it involves the progressive cellular consumption or degradation of cytoprotective proteins, although the specific signaling cascade remains to be elucidated (Bialik et al., 2018; Denton and Kumar, 2019).
Cell death has been divided into two categories: accidental cell death and RCD (Galluzzi et al., 2018). Unlike accidental cell death that lacks clear molecular signals and regulators, RCD is an active process involving tightly controlled mechanisms that can be targeted by genetic manipulations or drugs. The interplay between autophagy and cell death decides cell fate by activating integrated signaling pathways and influencing gene expression programs. Autophagy can coincide with apoptosis, the most common form of the RCD, as well as with non-apoptotic forms of RCD including ferroptosis (Zhou et al., 2019). Ferroptosis was initially described as a unique type of RCD observed in oncogenetic RAS-mutated cancer cells that is distinct from apoptosis, necrosis, and autophagy at the morphological, biochemical, and genetic levels (Dixon et al., 2012). However, increasing evidence challenges these early observations and suggests that the autophagic machinery, at least under certain conditions, contributes to ferroptotic cell death (Kang and Tang, 2017; Zhou et al., 2019). Autophagy-dependent ferroptosis may contribute to anticancer therapies, inflammation responses, and tissue fibrosis (Kang and Tang, 2017; Zhou et al., 2019). In this review, we introduce the basic process of autophagy and ferroptosis, describe types of selective autophagy, and discuss recent advances in understanding the molecular mechnisms of autophagy-dependent ferroptosis and its regulation, placing special emphasis on the biomedical implications of these findings.
The molecular machinery of autophagy
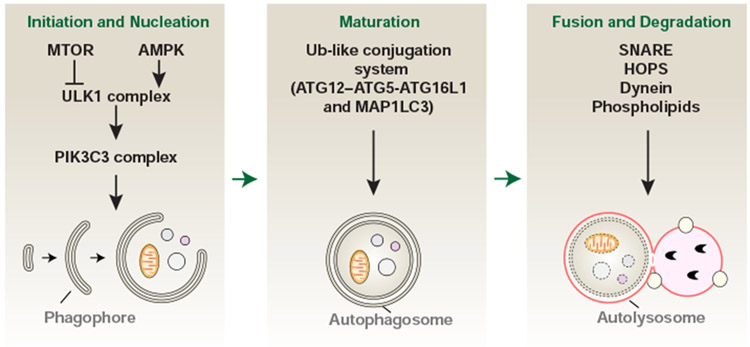
Autophagy is a dynamic process that relies on the formation and maturation of specific membrane structures such as phagophores, autophagosomes, and autolysosomes (Dikic and Elazar, 2018) (Fig. 1). The cup-shaped phagophores (also known as the isolation membrane) can be generated from multiple sources (e.g., the endoplasmic reticulum [ER], mitochondria-ER associated membrane, Golgi complex, plasma membrane, and recycling endosomes) to engulf cytoplasm (Tooze and Yoshimori, 2010). The subsequent expansion and closure of the phagophores lead to the formation of typical double-membrane structures, which are termed autophagosomes. In a subsequent step, autophagosomes fuse with lysosomes to generate autolysosomes, where the degradation of the cargo proceeds by lysosomal hydrolytic enzymes acting at low pH. Mechanistically, autophagy-related (ATG) proteins are core components of the machinery that drives the initiation, progression, and maintenance of autophagy. Genetic screens in yeast have led to the identification of over 40 ATG genes, many of which have mammalian homologs. These ATG proteins can interact with other factors governing phagophore and autophagosome formation, which may rely on multiple posttranslational modifications (Dikic and Elazar, 2018; Xie et al., 2015). Although the molecular details are still not completely understood, members of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) family (e.g., syntaxin 17 [STX17], synaptosome-associated protein 29 [SNAP29], and vesicleassociated membrane protein 8 [VAMP8]), the homotypic fusion and vacuole protein sorting (HOPS) complex, certain cytoskeleton motor proteins (e.g., dynein), and regulatory lipids (e.g., phospholipids) are involved in the formation of autolysosomes (Nakamura and Yoshimori, 2017).
Figure 1. The process of autophagy.
Autophagy is an intracellular degradation process that occurs with the formation and maturation of specific membrane structures, including phagophores, autophagosomes, and autolysosomes, which is controlled by ATG and other proteins. See also Table S1.
The ULK1 complex
The unc-51 like autophagy activating kinase (ULK) family consists of five serine/threonine-specific protein kinases: ULK1, ULK2, ULK3, ULK4, and serine/threonine kinase 36 (STK36). ULK1 and ULK2, the mammalian orthologs of yeast Atg1, drive the formation of the phagophore through binding to ATG13, RB1-inducible coiled-coil 1 (RB1CC1/FIP200, ortholog of yeast Atg17), and ATG101. Binding to ATG13 or RB1CC1 increases the stability and kinase activity of ULK, which is partly regulated by ATG101-dependent ATG13 phosphorylation (Mercer et al., 2009). Moreover, ULK1 can phosphorylate itself as well as ATG13 and RB1CC1 during autophagy induction. ULK1 and ULK2 may have some functional redundancy in autophagy induction, although they share only 52% amino acid identity (Lee and Tournier, 2011). The formation of the ULK1-ATG13-ATG101-RB1CC1 complex is determined by the status of intracellular energy and nutrients. This complex can be activated under starvation via the inhibition of the mechanistic target of rapamycin kinase complex 1 (MTORC1, containing MTOR; regulatory associated protein of MTOR complex 1 [RPTOR]; MTOR associated protein, LST8 homolog [MLST8]; DEP domain containing MTOR interacting protein [DEPTOR]; and AKT1 substrate 1 [AKT1S1/PRAS40]) or via the activation of the energy sensor 5’-AMP-activated protein kinase (AMPK) (Kim et al., 2011). Under starvation conditions, MTORC1 and AMPK directly regulate ULK1 (de)phosphorylation at multiple sites. In addition to phosphorylation, ULK1 undergoes acetylation and ubiquitination (Ub) as an alternative regulatory mechanism (Lin et al., 2012; Nazio et al., 2013). Many phosphorylation substrates of ULK1, such as ATG9A (a lipidembedded protein for phagophore membrane cycling) (Zhou et al., 2017), Beclin 1 (BECN1; a mammalian homolog of yeast Vps30/Atg6) (Park et al., 2018; Russell et al., 2013), and autophagy and beclin 1 regulator 1 (AMBRA1) (Russell et al., 2013), have been identified to regulate the formation of phagophores or autophagosomes. The recruitment of ULK1 to the phagophore assembly site may involve the Golgi-localized WW domain containing adaptor with coiled-coil (WAC), a positive regulator of autophagy (Joachim et al., 2015). Thus, the ULK1 complex is an important signal node, translating multiple protein phosphorylation signals into the initiation of autophagy.
The PIK3C3 complex
The PIK3C3 complex contains the catalytic subunit phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3, an ortholog of yeast Vps34) that converts phosphatidylinositol (PtdIns) into PtdIns-3-phosphate, BECN1, and phosphoinositide-3-kinase regulatory subunit 4 (PIK3R4, a mammalian homolog of yeast Vps15), which can be activated by the ULK1 complex. This core PIK3C3 complex can further form several subcomplexes that involve distinct co-factors, such as the PI3KC3-C1 complex (containing ATG14) (Fan et al., 2011), the PI3KC3-C2 complex (containing UV radiation resistance associated [UVRAG]) (Liang et al., 2006), and the rubicon autophagy regulator (RUBCN)-containing complex (Tabata et al., 2010). Functionally, the PI3KC3-C1 complex is responsible for the formation of the phagophore assembly site, membrane nucleation, elongation, and autophagosome formation. The PI3KC3-C2 complex enhances, whereas the RUBCN complex suppresses, autophagosome maturation. Accordingly, there are multiple positive and negative feedback loops involved in the regulation of PIK3C3 complex activity through different BECN1-binding proteins, such as ULK1 (Park et al., 2018), BCL2 apoptosis regulator (BCL2) (Pattingre et al., 2005), BCL2 like 1 (BCL2L1/BCLXL) (Oberstein et al., 2007), high mobility group box 1 (HMGB1) (Tang et al., 2010b), vacuole membrane protein 1 (VMP1) (Molejon et al., 2013), inositol 1,4,5-trisphosphate receptor type 3 (ITPR3/IP3R) (Vicencio et al., 2009), PTEN induced kinase 1 (PINK1) (Michiorri et al., 2010), baculoviral IAP repeat containing 5 (BIRC5/SURVIVIN) (Niu et al., 2010), and epidermal growth factor receptor (EGFR) (Wei et al., 2013). These findings suggest that diversity in the assembly of the PIK3C3 complex could be associated with different roles of the autophagic machinery, connecting it to other signaling networks. For example, BECN1 and its partners are implicated in the control of apoptosis and other types of RCD in both an autophagy-dependent and -independent manner, indicating the existence of a BECN1-centric network that orchestrates cell death and stress responses (Kang et al., 2011).
The ubiquitin-like conjugation systems
Two ubiquitin-like conjugation systems play key roles in the process of phagophore elongation and autophagosome formation. The first system involves the ATG12-ATG5-ATG16L1 pathway (Hanada et al., 2007). ATG12 was the first identified ubiquitin-like protein (Ubl) in autophagy and is covalently conjugated to ATG5 by the E1 enzyme ATG7 and then the E2 enzyme ATG10. The ATG12-ATG5 conjugate can further interact with ATG16L1, a homolog to yeast Atg16, to form a multimeric complex at a phagophore assembly site. Structurally, the homo-oligomers of ATG16L1 are required for the production of the ATG12-ATG5-ATG16L1 complex. The second system involves microtubule associated protein 1 light chain 3 (MAP1LC3, including MAP1LC3A, MAP1LC3B, and MAP1LC3C), the first identified protein to associate with autophagosomal structures (Kabeya et al., 2000). The turnover of MAP1LC3 between MAP1LC3-I (the cytosolic form) and MAP1LC3-II (the lipidated and membrane-associated form) is responsible for the formation of the autophagosome and the recruitment of cargo proteins destined for lysosomal degradation. This process is initiated by ATG4, the sole cysteine protease that cleaves MAP1LC3 at the C terminus to expose a glycine residue (Scherz-Shouval et al., 2007). MAP1LC3-I is further processed by the E1 enzyme ATG7 and then the E2 enzyme ATG3, finally resulting in the formation of phosphatidylethanolamine-conjugated MAP1LC3-II through the ATG12-ATG5-ATG16L1 system. As a recycling mechanism, the membrane-associated MAP1LC3-II can be cleaved by ATG4 to release free MAP1LC3-I for reuse in autophagosome formation (Nakatogawa et al., 2012). Although MAP1LC3-II is the most widely used autophagosome marker, other orthologs of yeast Atg8, such as GABA type A receptor-associated protein (GABARAP) and GABA type A receptor-associated protein like 2 (GABARAPL2/GATE-16) may play similar roles in the formation of the autophagosome in mammalian cells (Schaaf et al., 2016); the LC3 and GABARAP proteins are collectively referred to as Atg8-family proteins. Protein kinase A-mediated MAP1LC3 phosphorylation limits its autophagic activity (Cherra et al., 2010). In addition to its role in the bulk degradation process, MAP1LC3-II also interacts with cargo receptors (e.g., sequestosome 1 [SQSTM1/p62], calcium-binding and coiled-coil domain 2 [CALCOCO2/NDP52], and optineurin [OPTN]) to regulate the process of selective forms of autophagy. At the same time, MAP1LC3-II can be degraded in a lysosome-dependent manner (Mizushima and Yoshimori, 2007; Pankiv et al., 2007). Thus, autophagic flux capacity refers to the balance between the formation and degradation of autophagosomes or MAP1LC3-II during a certain time period (Yoshii and Mizushima, 2017). The measurement of autophagic flux is facilitated by the use of various inhibitors targeting the early (e.g., wortmannin, 3-methyladenine, LY294002, and spautin-1) or late (e.g., bafilomycin A1 and chloroquine) stage of autophagy (Table S1), as denoted in specific guidelines (Klionsky et al., 2016).
The types of selective autophagy
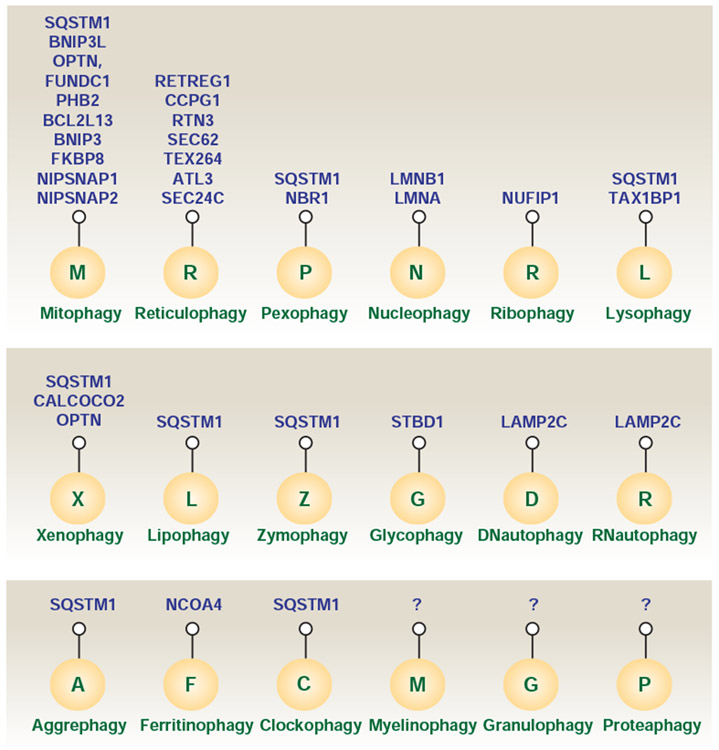
Selective autophagy serves as an intracellular quality control mechanism to mediate the degradation of specific targets, such as invading pathogens, aggregated proteins, and damaged organelles. Mechanistically, specific autophagy receptors are used for cargo recognition and degradation, and thus better fulfill the catabolic necessities of the cell in response to various types of stress, including cell death stimuli (Gatica et al., 2018). The autophagy receptors function by interacting simultaneously with cargoes and Atg8-family proteins. In general, the autophagy receptors interact with lipidated LC3 or GABARAP subfamily proteins by LC3-interacting region (LIR) motifs that bind to the LIR docking site (LDS) of Atg8 family proteins in a cargo-dependent manner (Johansen and Lamark, 2020). The LIR-LDS interaction is also modulated by the upstream regulators of autophagy, such as ULK1 and PIK3C3 (Turco et al., 2020). Here, we introduce different types of selective autophagy for the degradation of organelles in mammalian cells (Fig. 2) and emphasize the mechanisms of action of selective autophagy receptors in providing cargo specificity.
Figure 2. Selective autophagy receptors in mammalian cells.
In addition to bulk degradation in a non-elective manner, selective autophagy serves as an intracellular quality control mechanism to mediate the degradation of specific targets, such as invading pathogens, aggregated proteins, and damaged organelles through specific receptors. Different forms of selective autophagy are usually named by combining a prefix derived from the cargo and the suffix “phagy.” Note the large number of receptors found in the most-studied forms of selective autophagy (mitophagy and reticulophagy). It is unclear whether this correlates with their relative importance as homeostatic mechanisms or whether this reflects more extensive and long-lasting research of these phenomena.
Mitophagy
Mitochondria are intracellular powerhouses that convert oxygen and nutrients into adenosine triphosphate (ATP), the chemical energy "currency" of the cell. These branch-like organelles, made up of an outer membrane, an intermembrane space, an inner membrane, and a matrix, are regulated by mitochondrial dynamics (including fusion and fission), biogenesis, and degradation, as well as communication with other organelles (e.g., the ER and nucleus). Mitophagy is a well-studied type of selective autophagy that segregates and degrades mitochondria through different mechanisms, depending on the context. Mutations in PINK1 (a serine-threonine protein kinase) and parkin RBR E3 ubiquitin protein ligase (PRKN/PARK2) are implicated in the development of Parkinson’s disease. The PINK1-PRKN pathway plays a major role in the regulation of mitophagy through a Ub-dependent process. Oxidative injury and depolarization of mitochondria can increase PINK1 accumulation on the outer membrane, and subsequent PRKN recruitment from the cytosol to mitochondria. This dynamic change of PINK1 and PRKN leads to the assembly of phosphorylated Ub chains on proteins of the outer mitochondrial membrane, which in turn recruit multiple cargo receptors (e.g., SQSTM1 (Geisler et al., 2010), OPTN (Wong and Holzbaur, 2014), NBR1 (NBR1 autophagy cargo receptor) (Gao et al., 2015), CALCOCO2 (Lazarou et al., 2015), and Tax1 binding protein 1 (TAX1BP1) (Lazarou et al., 2015) for the degradation of mitochondria in a context-dependent manner. This process is significantly enhanced by the activation of TANK-binding kinase 1 (TBK1) because TBK1 can directly phosphorylate cargo receptors (e.g., SQSTM1, OPTN, CALCOCO2, and TAX1BP1), enhancing their binding to cargos and MAP1LC3 (Heo et al., 2015; Heo et al., 2018; Moore and Holzbaur, 2016). In contrast, de-ubiquitinating enzymes, such as ubiquitin specific peptidase 8 (USP8), USP15, USP30, and USP35, can limit PRKN-mediated mitophagy (Wang et al., 2015). In addition to PRKN, other E3 ubiquitin ligases (e.g., mitochondrial E3 ubiquitin protein ligase 1 [MUL1] (Yun et al., 2014), siah E3 ubiquitin protein ligase 1 [SIAH1] (Szargel et al., 2016), SMAD specific E3 ubiquitin protein ligase 1 [SMURF1] (Orvedahl et al., 2011), and autocrine motility factor receptor [AMFR/GP78]) may elicit mitophagy through poorly characterized mechanism (Fu et al., 2013). In addition to Ub-dependent receptors, Ub-independent receptors (e.g., BCL2 interacting protein 3 like [BNIP3L/NIX] (Sandoval et al., 2008), FUN14 domain containing 1 [FUNDC1] (Liu et al., 2012), BCL2 interacting protein 3 [BNIP3] (O'Sullivan et al., 2015), nipsnap homolog 1 [NIPSNAP1] (Princely Abudu et al., 2019), nipsnap homolog 2 [NIPSNAP2] (Princely Abudu et al., 2019), prohibitin 2 [PHB2] (Wei et al., 2017), two putative yeast Atg32 orthologs, BCL2 like 13 [BCL2L13] (Murakawa et al., 2015) and FKBP prolyl isomerase 8 [FKBP8] (Bhujabal et al., 2017)) and mitochondrial membrane phospholipids lipid (e.g., cardiolipin and ceramide) (Chu et al., 2013) contribute to mitophagy under different mitochondrial stresses (e.g., hypoxia or intrinsic stress signals). In addition, the inhibition of mitochondrial fission by the depletion of dynamin 1 like (DNM1L/DRP1) or fission, mitochondrial 1 (FIS1) reduces the levels of mitophagy, whereas mitophagy deficiency increases mitochondrial fragmentation (Dagda et al., 2009; Kageyama et al., 2014). Collectively, the molecular machinery involved in mitophagy is complex, and multiple cargo receptors and signals are required for the recognition of damaged mitochondria. Impaired mitophagy enhances cell death, tissue injury and inflammation in mouse models, suggesting a major homeostatic role for this type of selective autophagy (Kang et al., 2016; Li et al., 2018; Sliter et al., 2018).
Reticulophagy
The ER is a network of tubules and flattened sacs that connects to the nucleus. This organelle includes rough and smooth ER, which play a key role in the regulation of protein synthesis, folding and secretion, calcium homeostasis, and lipid biosynthesis. The ER also undergoes dynamic changes in its morphology and functional properties. ER stress results from the accumulation of misfolded proteins and causes the activation of a signaling network called the unfolded protein response (UPR). ER stress not only promotes cell survival, but also causes cell death when the UPR is overwhelmed. Autophagic structures are associated with the ER membrane. The removal of unwanted portions of the ER through autophagy, a process called reticulophagy/ER-phagy, is critical for the maintenance of ER homeostasis. The level of reticulophagy is remarkably upregulated by ER stress, starvation, MTOR inhibition, misfolded protein aggregates, or microbial infection. Up to now, seven autophagic cargo receptors, including cell cycle progression 1 (CCPG1) (Smith et al., 2018), reticulophagy regulator 1 (RETREG1/FAM134B) (Khaminets et al., 2015), reticulon 3 (RTN3) (Grumati et al., 2017), SEC62 homolog, preprotein translocation factor (SEC62) (Fumagalli et al., 2016), testis expressed 264, ER-phagy receptor (TEX264) (Chino et al., 2019), atlastin GTPase 3 (ATL3) (Chen et al., 2019), and SEC24 homolog C, COPII coat complex component (SEC24C) (Cui et al., 2019) specifically bind to the ER and autophagosomal membranes, thus contributing to reticulophagy. RETREG1 in sheets and RTN3 in tubules are essential reticulophagy receptors that promote the remodeling and scission of ER sheets through the reticulon domains (Grumati et al., 2017; Khaminets et al., 2015). SEC62 (an ER transmembrane protein) is required for the reticulophagy-mediated degradation of ER chaperones (e.g., calnexin [CANX], calreticulin [CALR], and heat shock protein family A [Hsp70] member 5 [HSPA5/BiP]), and folding enzymes (protein disulfide isomerase family A member 4 [PDIA4/ERp72] and protein disulfide isomerase family A member 3 [PDIA3/ERp57]) during cellular recovery from the UPR (Fumagalli et al., 2016). CCPG1 (an ER transmembrane protein) can deliver portions of ER carrying insoluble proteins to phagophores in pancreatic cells (Smith et al., 2018). TEX264 is an ER-resident protein responsible for nutrient starvation-induced reticulophagy and has a particular high affinity for Atg8-family proteins (Chino et al., 2019). ATL3 is a GABARAP-binding receptor (but not a MAP1LC3 receptor) and plays a major role in starvation-induced tubular ER turnover during reticulophagy (Chen et al., 2019). In contrast, SEC24C is required for the degradation of RETREG1 and RTN3 during reticulophagy (Cui et al., 2019). In-depth characterization of these reticulophagy pathways, which presumably regulate separate stress-response pathways, will be important for the development of new approaches for the treatment of ER-related disorders, such as diabetes, nonalcoholic fatty-liver disease, and Alzheimer’s disease (Smith and Wilkinson, 2017).
Pexophagy
Peroxisomes are membrane-bound organelles containing a wide variety of metabolic enzymes that regulate the redox status of specific biomolecules. Selective autophagy of peroxisomes, pexophagy, is initiated by the ubiquitination of peroxisomal biogenesis factor 3 (PEX3, a regulator of the assembly and maintenance of the peroxisomal membrane) (Yamashita et al., 2014) or PEX5 (a peroxisomal matrix protein carrying peroxisomal targeting signals) (Wang and Subramani, 2017). Unlike PEX5, which is required for peroxisome protein import, the functional contribution of PEX3 to peroxisomes remains controversial. ATM serine/threonine kinase (ATM)-mediated phosphorylation of PEX5 is required for monoubiquitination of PEX5, which in turn promotes the binding between PEX5 and SQSTM1 (Zhang et al., 2015). Either SQSTM1 (Zhang et al., 2015) or NBR1 (Deosaran et al., 2013) act as autophagic receptors in pexophagy. Other PEX members, such as PEX14 (Zutphen et al., 2008), PEX13 (Lee et al., 2017), or PEX2 (Sargent et al., 2016), may participate in pexophagy through PEX5-dependent or -independent circuitries. In particular, PEX14 can directly bind MAP1LC3 or ATG9A to trigger pexophagy (Li et al., 2017). It will be important to examine the consequences of dysfunctional pexophagy on oxidative stress-mediated human diseases.
Nucleophagy
The nucleus is the largest cellular organelle that stores genomic materials to control gene expression. The selective removal of the nucleus or nuclear components by autophagy, namely nucleophagy, was first observed in yeast under nutrient-rich conditions as well as during nitrogen deprivation. Nucleophagy also occurs in mammalian cells and can cause the degradation of nuclear lamina components upon DNA damage. Nucleophagy-associated lamin B1 (LMNB1) degradation is associated with oncogenic RAS activation, but not starvation (Dou et al., 2015). The SUMOylation of lamin A/C (LMNA) promotes molecular interactions between the lamina and MAP1LC3 during nucleophagy (Li et al., 2019). Micronuclei are nuclear bodies containing damaged chromosome fragments that appear after genomic stress or mitotic catastrophe and that are usually removed by autophagy (Rello-Varona et al., 2012). Autophagy deficiency increases the number of micronuclei, which may play a pathological role in aging, cancer, and neurodegenerative diseases (Dou et al., 2015; Papandreou and Tavernarakis, 2019). The impact of DNA sensors, such as cyclic GMP-AMP synthase (CGAS) and HMGB1, on the regulation of nucleophagy remains to be characterized.
Ribophagy
Ribophagy refers to the selective degradation of ribosomes via autophagy. MTOR plays a key role in protein synthesis and ribosome biogenesis. The inhibition of MTOR by starvation or pharmacological agents such as rapamycin or torin1 can trigger ribophagy in mammalian cells. Using an MTOR inhibition model, nuclear FMR1-interacting protein 1 (NUFIP1) was identified as a cargo receptor for ribophagy (Wyant et al., 2018). NUFIP1 and its binding partner zinc finger HIT-type containing 3 (ZNHIT3) can bind ribosomes and promote their targeting to phagophores by interacting with MAP1LC3B, but not with other MAP1LC3 isoforms or GABARAP family members (Wyant et al., 2018). Regarding the wider role of MTOR inhibition in nonselective autophagy, it is important to identify the specific downstream signals responsible for NUFIP1 translocation from the nucleus to the cytoplasm during selective autophagy. In contrast, a recent study reported that agents that induce defects in the ribosomal quality control fails to induce robust ribophagy in HEK293 cells using a Ribo-Keima reporter system (An and Harper, 2018), indicating that there may be no specific autophagy receptor for the clearance of damaged ribosomes.
Lysophagy
Lysosomes are sac-like structures delimited by a single-layer membrane. This organelle contains a variety of hydrolases for the degradation of various kinds of macromolecules. The leakage of lysosomal constituents into the cytosol can trigger “lysosomal cell death”. A membrane-damaged lysosome itself can be a target of autophagy, and this type of selective autophagy is termed lysophagy. For example, agents causing lysosomal rupture (e.g., silica, monosodium urate, and LLOMe) can induce lysophagy in an ATG5-dependent manner. Moreover, the ubiquitination of the damaged lysosomes is a biomarker and contributor to lysophagy. The cargo receptors SQSTM1 (Hung et al., 2013) and TAX1BP1 (Koerver et al., 2019) play a context-dependent role in the recognition of damaged lysosomes during lysophagy. This process can be further regulated by galectin 8 (LGALS8) (Aits et al., 2015), galectin 3 (LGALS3) (Maejima et al., 2013), ubiquitin conjugating enzyme E2 Q family like 1 (UBE2QL1) (Koerver et al., 2019; Mizushima, 2019), F-box protein 27 (FBXO27) (Yoshida et al., 2017), and valosin containing protein (VCP/P97) (Papadopoulos and Meyer, 2017) through either ubiquitination-dependent or -independent mechanisms. Further research is needed to clarify the interplay between lysophagy and lysosomal cell death in the control of lysosome homeostasis.
Other selective autophagy pathways
In addition to degrading cellular organelles as discussed above, selective autophagy also contributes to intracellular homeostasis through eliminating invading pathogens (xenophagy for bacteria or viruses) (Sharma et al., 2018), lipid droplets (lipophagy) (Singh et al., 2009), zymogen granules (zymophagy) (Grasso et al., 2011), glycogen (glycophagy) (Reichelt et al., 2013), DNA (DNautophagy) (Fujiwara et al., 2015), RNA (RNautophagy) (Fujiwara et al., 2015), myelin (myelinophagy) (Gomez-Sanchez et al., 2015), stress granules (granulophagy) (Buchan et al., 2013), proteasomes (proteaphagy) (Marshall et al., 2015), protein aggregates (aggrephagy) (Overbye et al., 2007), and specific proteins, e.g., ferritin (ferritinophagy) (Mancias et al., 2014) and aryl hydrocarbon receptor nuclear translocator like (ARNTL/BMAL1 for clockophagy) (Yang et al., 2019a). These types of selective autophagy also require specific, and in some cases unknown, receptor-cargo interactions (Fig. 2), although the precise structural basis of this interaction and the ubiquitin substrates required for autophagic targeting remain to be further explored.
The molecular machinery of ferroptosis
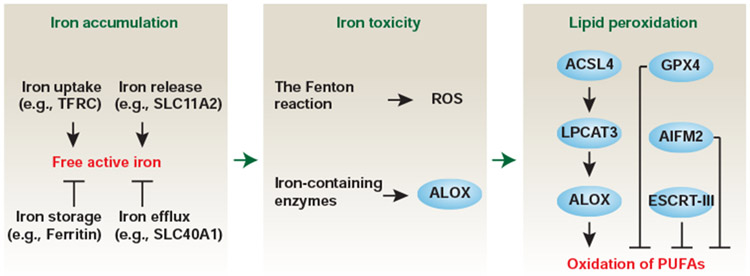
Reactive oxygen species (ROS) are a key determinant of RCD when the integrated antioxidant systems are impaired. In general terms, ferroptosis is a form of ROS-dependent regulated necrosis that occurs through multiple signals and pathways. Among them, iron accumulation and lipid peroxidation seem to play a major role in the initiation of ferroptosis, although the specific downstream effector proteins (e.g., pore-forming proteins) and the precise mechanisms remain unidentified (Stockwell et al., 2017; Xie et al., 2016) (Fig. 3). Here, we summarize the key connections between ferroptosis and autophagy, including small molecules or drugs that can be used to trigger or inhibit ferroptosis (Table S1).
Figure 3. The process of ferroptosis.
The ferroptotic response is characterized by iron accumulation and lipid peroxidation, resulting in the formation of lipid peroxides. Increased iron not only triggers the Fenton reaction to produce ROS, but also increases the activity of iron-containing enzymes (e.g., ALOX), which contributes to lipid peroxidation. The activation of the GPX4, AIFM2, or ESCRT-III pathways can limit or repair the oxidation of PUFAs that result from the activation of the ACSL4-LPCAT3-ALOX pathway. See also Table S1.
Iron overload and toxicity
Iron is an essential trace metal required by all living organisms. The basic process of iron metabolism in animals and humans includes iron absorption, in vivo distribution, the formation of various pools of protein-bound iron, iron storage, and iron excretion (Fig. 4). Many diseases are linked to abnormal iron metabolism, and altered iron metabolism itself can trigger a variety of diseases. The majority of iron come from nutritional or endogenous sources, the latter of which is mostly from hemoglobin in aging red blood cells. Most of the exogenous iron is directly absorbed into the bloodstream through the duodenal mucosal cells, and part of the iron is stored in ferritin in mucosal cells. Intestinal iron intake and endogenous iron re-use is balanced by iron excretion. When iron is deficient, the iron absorption rate and endogenous iron utilization rate increase, and iron storage decreases. When the iron level in food is high, iron absorption decreases while iron storage rises, thus allowing a homeostatic maintenance of physiological iron levels.
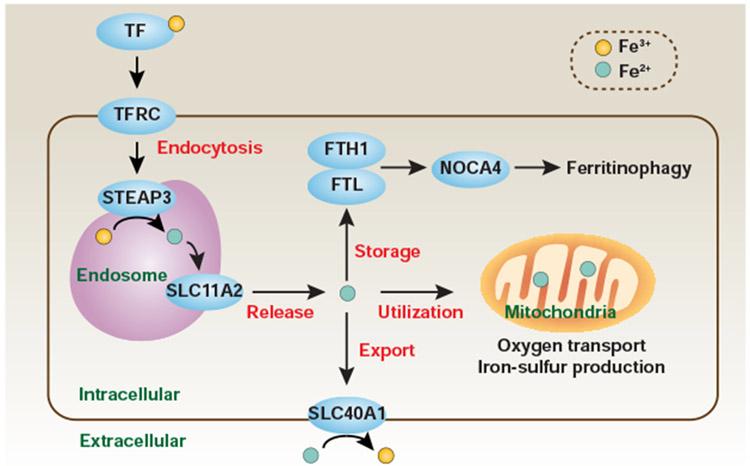
Figure 4. Role of cellular iron metabolism in ferroptosis.
Most cells acquire plasma Fe3+ via TFRC-mediated endocytosis of TF-bound iron. In the endosome, Fe3+ is reduced to Fe2+ by STEAP3 and then released from the endosome to the cytoplasm by SLC11A2. The labile Fe2+ can be stored by ferritin, including FTH1 and FTL. Fe2+ plays an important function in regulating multiple processes, such as oxygen transport and iron-sulfur production in mitochondria. The export of intracellular iron across the cell membrane requires the iron efflux pump SLC40A1, leading to the production of Fe3+ from Fe2+. In general, increased iron uptake (e.g., due to TFRC overexpression), reduced iron storage (e.g., due to the knockdown of ferritin or induction of ferritinophagy), and impaired cellular iron export (e.g., resulting from the knockdown of SLC40A1) enhance the sensitivity to ferroptosis.
The control of cellular iron levels is a dynamic process. Iron exists in two oxidation states: ferrous (Fe2+, Fe(II)) or ferric (Fe3+, Fe(III)). Transferrin (TF)-bound ferric iron from serum is recognized by transferrin receptor (TFRC/TFR1), a carrier protein for TF in cell membranes. After uptake by TFRC, ferric iron is reduced to ferrous iron by STEAP3 metalloreductase (STEAP3) in the endosome and then released from the endosome to the cytoplasm by solute carrier family 11 member 2 (SLC11A2/DMT1). Ferrous iron serves numerous important functions in regulating multiple metabolic and biochemical processes, such as oxygen transport, DNA synthesis, and iron-sulfur production. Ferritin is the primary form of iron storage protein, which includes two subunits: ferritin light chain (FTL) and ferritin heavy chain 1 (FTH1). Although the sequences of these subunits are highly homologous, FTL and FTH1 exhibit different functions in the regulation of iron storage and utilization, respectively. Ferritin degradation can occur by ferritinophagy, a type of selective autophagy (discussed below). Finally, the export of intracellular iron across the cell membrane requires the iron efflux pump SLC40A1/ferroportin-1 (solute carrier family 40 member 1), leading to the production of ferric iron from ferrous iron. Thus, iron transport across the plasma membrane determines the redox status.
Iron overload, resulting from excessive extracellular or intracellular iron, can cause cell death and tissue injury, which is involved in a number of diseases, such as neurodegeneration and cancer. In particular, intracellular iron overload can trigger ferroptotic cell death through at least two mechanisms (Fig. 3). On the one hand, iron can generate excessive ROS via the Fenton reaction, which then increases oxidative DNA and lipid damage. On the other hand, iron can increase the activity of nonheme iron-containing enzymes, such as lipoxygenases responsible for lipid peroxidation. Indeed, increased iron uptake (e.g., due to TFRC overexpression), reduced iron storage (e.g., due to the knockdown of ferritin [FTL and FTH1] or induction of ferritinophagy), and impaired cellular iron export (e.g., resulting from the knockdown of SLC40A1/ferroportin) enhance the sensitivity to ferroptosis (Bogdan et al., 2016; Du et al., 2018; Gao et al., 2016; Geng et al., 2018; Hou et al., 2016; Yang and Stockwell, 2008). In contrast, intracellular iron chelators such as desferrioxamine block ferroptosis. The transport of iron between cytosol, mitochondria, and lysosome, as well as the activation of iron responsive element binding protein 2 (IREB2, a major regulator of iron homeostasis) also affect the onset of ferroptosis (Dixon et al., 2012). Collectively, iron is an intracellular second messenger promoting ferroptosis. It remains unknown whether, under certain conditions, iron-mediated ferroptosis may amplify or diminish the susceptibility of cells to other types of RCD such as apoptosis (Yang et al., 2017), necrosis (Yang et al., 2017), and pyroptosis (Zhou et al., 2018). Moreover, the relationship between systemic iron homeostasis (e.g., dietary iron absorption and the concentration of circulating iron) and ferroptosis is largely uncharacterized, although it is known that excessive heme can induce ferroptosis (NaveenKumar et al., 2018). Iron is essential for both the host and its pathogens, and the potential impact of gut microbiota on ferroptotic cell death in host cells remains to be clarified.
Lipid peroxidation and toxicity
Lipid peroxidation plays a major role in ferroptosis. Lipids are not only the building block of cellular membranes, but also the key players in energy storage, heat insulation, cell communication, and signal transduction. Cell membrane rupture that is associated with the breakdown of membrane lipids (phospholipids, glycolipids, and cholesterol) is the hallmark of regulated necrosis, including necroptosis, pyroptosis, alkaliptosis, and ferroptosis (Tang et al., 2019). Lipid peroxidation of polyunsaturated fatty-acid-containing phospholipids (PUFA-PLs) plays a major role in promoting ferroptotic cell death (Yang et al., 2016). This process is driven by the activation of specific enzymes, including acyl-CoA synthetase long chain family member 4 (ACSL4) (Doll et al., 2017; Kagan et al., 2017; Yuan et al., 2016b) and lipoxygenase (arachidonate lipoxygenase [ALOX]) enzymes (Yang et al., 2016). ACSL4 is required for the production of PUFA-PLs, whereas lipoxygenases oxidize PUFA-PLs to the corresponding hydroperoxy derivatives, namely lipid peroxides or lipid oxidation products. The mammalian ALOX family, consisting of six members (ALOXE3, ALOX5, ALOX12, ALOX12B, ALOX15, and ALOX15B), plays a tissue- or cell-dependent role in the induction of ferroptosis (Hinman et al., 2018; Liu et al., 2015; Shintoku et al., 2017; Wenzel et al., 2017). ACSL4 is not only a contributor to ferroptosis, but also may be a biomarker for it (Yuan et al., 2016b). Increased ACSL4 expression, but not that of other members of the ACSL family, has been observed in ferroptotic cancer cell death (Yuan et al., 2016b). In addition, the expression of lysophosphatidylcholine acyltransferase 3 (LPCAT3), a downstream effector of ACSL4, is upregulated during ferroptosis (Dixon et al., 2015); however, there is no direct evidence showing that the inhibition of LPCAT3 blocks ferroptosis.
This classification of the ferroptosis inducers has provided a useful framework for studying and understanding ferroptosis. Classical ferroptosis inducers inhibit either of the two antioxidant systems during lipid peroxidation. One is the antiporter SLC7A11/system xc− (solute carrier family 7 member 11), which imports cysteine (the oxidized form) into cells with a 1:1 counter-transport of glutamate (the most abundant neurotransmitter, an excess of which can cause neurotoxicity and oxidative injury) and then induces the production of glutathione (GSH), a major free radical scavenger. A number of small molecules or drugs such as erastin, sulfasalazine, and sorafenib induce ferroptosis by the inhibition of SLC7A11/system xc− (Dixon et al., 2012). Erastin, as the first-used ferroptosis inducer, has other direct binding targets, such as voltage dependent anion channel 2 (VDAC2) and voltage dependent anion channel 3 (VDAC3), which also contribute to ferroptosis (Yagoda et al., 2007). The second antioxidant system is the selenium-containing glutathione peroxidase 4 (GPX4), a phospholipid hydroperoxidase that can directly reduce phospholipid hydroperoxide production. The antioxidant activity of GPX4 requires GSH, the pharmacological inhibition of GPX4 by RSL3 or FIN56, causes ferroptotic cell death (Yang et al., 2014). Notably, the conditional depletion of GPX4 in tissues or cells can cause oxidative injury or cell death in both ferroptosis-dependent and -independent manners (Canli et al., 2016; Cole-Ezea et al., 2012; Friedmann Angeli et al., 2014; Kang et al., 2018a; Matsushita et al., 2015; Ran et al., 2006; Ran et al., 2004; Ran et al., 2003; Seiler et al., 2008), indicating broad roles for GPX4 in different RCD scenarios. In contrast, antioxidants, including N-acetyl cysteine and vitamin E, can protect against ferroptosis caused by the classical ferroptosis inducers (e.g., erastin or RSL3) or GPX4 depletion in vivo and in vitro. In addition to GPX4, apoptosis inducing factor mitochondria associated 2 (AIFM2) (Bersuker et al., 2019; Doll et al., 2019) or endosomal sorting complexes required for transport (ESCRT)-III (Dai et al., 2020b; Dai et al., 2020c) also limits membrane damage through increased CoQ10 production or plasma membrane repair during ferroptosis.
Other components of an integrated stress response, such as transcription factors (e.g., nuclear factor, erythroid 2 like 2 [NFE2L2/NRF2] (Chen et al., 2017; Shin et al., 2018; Sun et al., 2016; Wu et al., 2011) and tumor protein p53 [TP53] (Gnanapradeepan et al., 2018; Jiang et al., 2015; Kang et al., 2019; Xie et al., 2017)), heat shock response (Sun et al., 2015), the UPR (Sauzay et al., 2018; Zhu et al., 2017), cell adhesion (Wu et al., 2019a; Yang et al., 2019b), and autophagy (discussed below) also play a context-dependent role in the control of oxidative injury-mediated ferroptosis.
Role of autophagy in ferroptosis
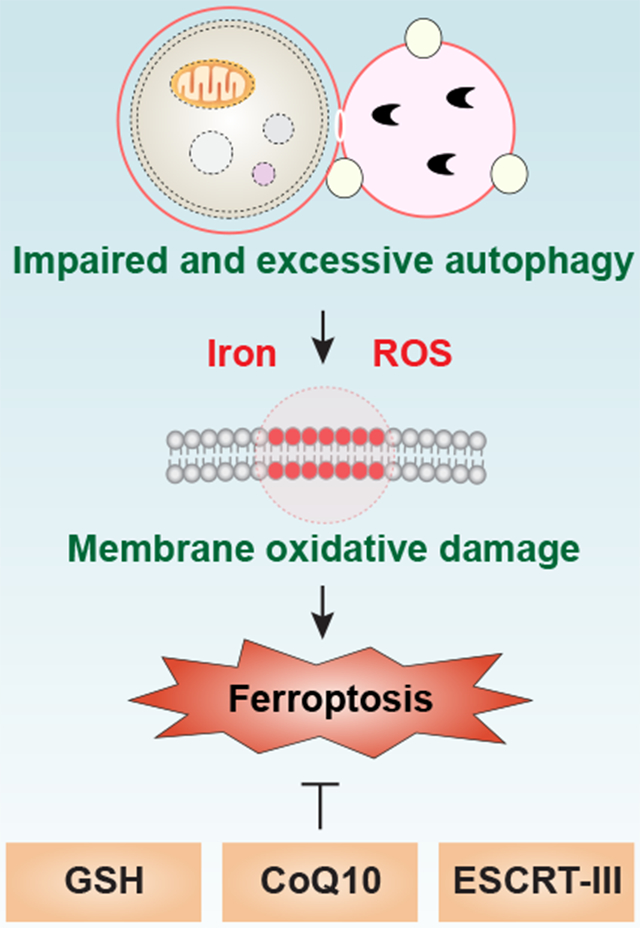
Increased autophagy flux is observed in various cells in response to classical ferroptosis activators, such as erastin and RSL3. While appropriate autophagy has likely evolved as a pro-survival response, excessive autophagy, especially selective autophagy, and impaired lysosomal activity may promote ferroptotic cell death. Here, we outline the key types of selective autophagy or regulators of autophagic machinery in driving ferroptosis (Fig. 5).
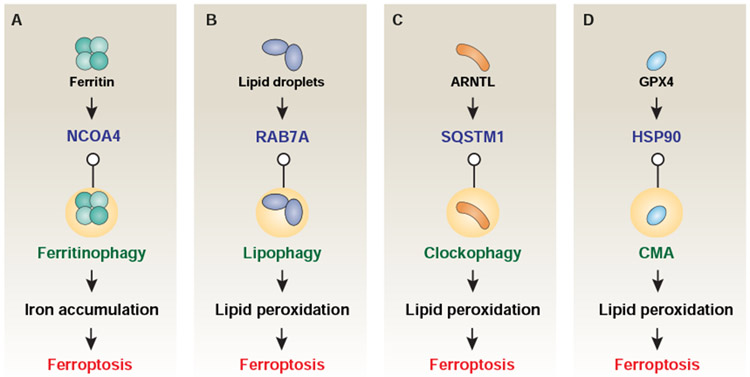
Figure 5. Role of selective autophagy in ferroptosis.
(A) NCOA4-mediated ferritinophagy promotes iron accumulation in ferroptosis. (B) RAB7A-mediated lipophagy, and (C) SQSTM1-mediated clockophagy promote lipid peroxidation in ferroptosis, and (D) HSP90-mediated chaperone-mediated autophagy (CMA) promote lipid peroxidation in ferroptosis.
NCOA4-dependent ferritinophagy
Ferritinophagy is the process of autophagic degradation of the iron-storage protein ferritin, which is critical for the regulation of cellular iron levels. Ferritin is composed of 24 subunits of FTH1 and FTL, and can store up to 4500 atoms of iron. The combination of autophagosome isolation and quantitative proteomics has identified nuclear receptor coactivator 4 (NCOA4) as a cargo receptor responsible for autophagy-dependent ferritin degradation, likely because the C terminus of NCOA4 binds a conserved surface arginine (R23) on FTH1 in phagophores, and subsequently in autophagosomes and autolysosomes (Mancias et al., 2014). NCOA4-dependent ferritinophagy promotes ferroptosis through releasing free iron from ferritin. The depletion or inhibition of NCOA4 or ATG protein (e.g., ATG3, ATG5, ATG7, and ATG13) inhibits ferritin degradation and therefore reduce free iron levels and thus limit subsequent oxidative injury during ferroptosis (Gao et al., 2016; Hou et al., 2016). As a feedback mechanism, the level of intracellular NCOA4 is regulated by cellular iron (Mancias et al., 2014). When cellular iron levels are high, NCOA4 can be degraded subsequent to its ubiquitination by HECT and RLD domain containing E3 ubiquitin protein ligase 2 (HERC2, a ubiquitin E3 ligase) through the ubiquitin-proteasome pathway (Mancias et al., 2015), suggesting a negative correlation of HERC2 with ferroptosis induction. Moreover, deficient ferritinophagy may increase the activity of iron responsive element binding protein 2 (IREB2/IRP2), a central posttranscriptional regulator of iron metabolism, and subsequently upregulate TF as a feedback mechanism (Dixon et al., 2012). Mitochondrial ferritin and iron uptake also limit or amplify the iron toxicity and ferroptosis (Wang et al., 2016; Yuan et al., 2016a). Other than NCOA4, it remains unknown whether additional mitochondrial factors are required for the selective autophagic degradation of ferritin in mitochondria. Ferritinophagy-mediated ferroptosis is implicated in hepatic fibrosis and neurodegenerative diseases (Kong et al., 2019; Zhang et al., 2018) and can be used to kill cancer under specific circumstances (Du et al., 2018; Lin et al., 2016). A further understanding of different pathways leading to ferritin degradation, including autophagy-independent lysosomal degradation of ferritin (Goodwin et al., 2017), and their contribution to intracellular and systemic iron homeostasis will be important for the therapeutic manipulation of ferroptosis.
RAB7A-dependent lipophagy
Free fatty acids can be esterified into triglycerides and cholesterol esters in cells to reduce their toxicity. This anabolic reaction occurs in the ER, causing the deposition of neutral lipids in a lenticular microdomain between the bilayer membranes of the ER, ultimately leading the outer leaves of the layer to expand to form a unique spherical organelle, the lipid droplet (Liu and Czaja, 2013). Lipid droplets are complex organelles that were previously thought to be inert sites for fat fixation and are now known to have a unique, dynamic proteome. The lipid droplet proteome fluctuates with changes in cellular metabolic status, thus influencing the composition of neutral lipids in organelles. Lipophagy, the autophagic digestion of lipid droplets can release free fatty acids, which then serve as a fuel for mitochondrial beta-type oxidation. The process by which intracellular lipid droplets are selectively transported by autophagosomes for lysosomal decomposition, lipophagy, provides another potential pathway for regulating cellular lipid levels, and therefore the propensity to cell death. The level of lipid droplets is negatively related to oxidative stress-induced ferroptosis (Bai et al., 2018). Increased lipid droplet formation by the upregulation of tumor protein D52 (TPD52) suppresses RSL3-induced ferroptosis in hepatocytes (Bai et al., 2018). The main function of RAB7A (a member of the RAS oncogene family) is to mediate the recruitment of lipid droplets by multivesicular bodies and lysosomes during lipid phagocytosis. In contrast, increased RAB7A-dependent lipophagy promotes lipid droplet degradation and therefore increases lipid peroxidation-mediated ferroptosis (Bai et al., 2018). It remains unknown whether other regulators of lipophagy, such as glycine N-methyltransferase (GNMT), transcription factor EB (TFEB), and forkhead box O1 (FOXO1), contribute to lipophagy-mediated ferroptosis in diseases caused by fat accumulation.
SQSTM1-dependent clockophagy
Circadian clocks are endogenous, entrainable rhythmic oscillations with a periodicity of approximately 24 hours that play a fundamental role in internal cycles of behavior, physiology, and metabolism. The core oscillation mechanism is a transcriptional-translational negative feedback loop, in which ARNTL and clock circadian regulator (CLOCK) are core nuclear factors driving circadian clocks through regulating the expression of a number of genes via the binding of E-box motifs in their promoters. Clockophagy is a recently identified type of selective autophagy for the degradation of ARNTL during ferroptosis induced by type-2 ferroptosis inducers (e.g., RSL3 and FIN56), but not type-1 ferroptosis inducers (e.g., erastin, sulfasalazine, and sorafenib) (Liu et al., 2019; Yang et al., 2019a). Mass spectrometric mechanistic investigation of ARNTL-binding protein indicates that the cargo receptor SQSTM1 (but not NBR1, OPTN, CALCOCO2, or NCOA4) makes a major contribution to autophagic ARNTL degradation (Yang et al., 2019a). Although clockophagy requires several ATG proteins, such as ATG5 and ATG7, it is mechanistically distinct from classical autophagy, in thus far that clockophagy does not appear to require ATG9A (Yang et al., 2019a). Autophagic ARNTL degradation promotes ferroptotic cell death through blocking hypoxia inducible factor 1 subunit alpha (HIF1A)-dependent fatty acid uptake and lipid storage. HIF1A, one of the major transcription factors regulating hypoxic response, plays a pro-survival role in ferroptosis through the upregulation of fatty acid binding protein 3 (FABP3, muscle and heart) and fatty acid binding protein 7 (FABP7, brain) for lipid storage (Yang et al., 2019a). The level of HIF1A is further regulated by egl-9 family hypoxia inducible factor 2 (EGLN2/PHD1), an ARNTL target gene in RSL3-induced ferroptosis (Yang et al., 2019a). In addition to HIF1A, endothelial PAS domain protein 1 (EPAS1/HIF2A) regulates ferroptosis in kidney cancer (Zou et al., 2019). It also remains unclear how the disruption of circadian rhythms might affect ferroptosis sensitivity.
Chaperone-mediated autophagy
Chaperone-mediated autophagy (CMA) is a type of selective autophagy that uses molecular chaperones to deliver certain cytosolic proteins to lysosomes for degradation based on the recognition of specific amino acid sequences. Heat shock protein family A (Hsp70) member 8 (HSPA8/HSC70), the constitutively expressed member of the Hsp70 family, is a major molecular chaperone responsible for the recognition of cytosolic proteins that contain a KFERQ-like motif within the sequence of a protein. GPX4 protein degradation by CMA seems to be a universal event in response to various ferroptosis activators (Muller et al., 2017; Shimada et al., 2016; Zhu et al., 2017). HSPA5, an ER stress-associated molecular chaperone, can limit erastin-induced GPX4 degradation and therefore protects against ferroptotic cell death in pancreatic cancer cells (Zhu et al., 2017). HSPA8-dependent CMA also contributes to erastin-induced GPX4 degradation in breast cancer cells (Wu et al., 2019b). Moreover, HSP90 can increase the protein stability of lysosomal associated membrane protein 2A (LAMP2A), a CMA receptor, to enhance GPX4 degradation during ferroptosis (Wu et al., 2019b). These findings establish a model of interaction between CMA and autophagy to determine GPX4 protein stability in ferroptosis. However, the structural basis of GPX4 degradation remains poorly understood.
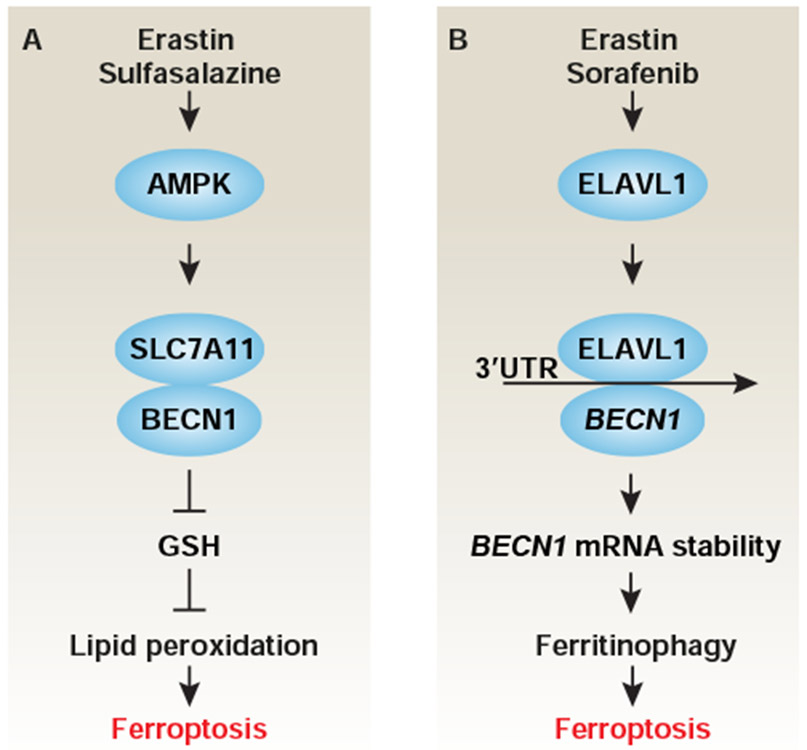
BECN1-mediated oxidative injury
BECN1 was originally identified as a BCL2 (an anti-apoptotic protein)-binding protein through yeast two-hybrid assays (Liang et al., 1998). Later, BECN1 was reported as the mammalian homolog of yeast Vps30/Atg6, and is involved in autophagy induction and tumor suppression (Liang et al., 1999). Autophagy-independent functions of BECN1 were also demonstrated. The multifunctional roles of BECN1 in cell survival and death appear to rely on its binding partners. In the case of ferroptosis, BECN1 was identified as a novel SLC7A11/system xc−-binding protein to inhibit transport activity in cancer cells in response to type 1 ferroptosis inducers (e.g., erastin, sulfasalazine, and sorafenib) (Kang et al., 2018b; Song et al., 2018) (Fig. 6A). This process is controlled by AMPK-mediated BECN1 phosphorylation at S90 and S93. Thus, the mutation of these BECN1 phosphorylation sites or inhibtion of AMPK activity limits the formation of a BECN1-SLC7A11 / system xc− complex and blocks ferroptosis (Song et al., 2018). In contrast, the administration of the BECN1 activator peptide Tatbeclin 1 enhances the anticancer activity of the SLC7A11/system xc− inhibitor through the induction of lipid peroxidation-dependent ferroptosis (Song et al., 2018). In addition to protein-protein interactions, BECN1-mediated ferroptosis is regulated by BECN1 mRNA stability. Consistent with this notion, the RNA-binding protein ELAV like RNA binding protein 1 (ELAVL1) can increase BECN1 mRNA stability to trigger subsequent ferritinophagy in hepatic stellate cells (Zhang et al., 2018) (Fig. 6B). These findings indicate that BECN1 regulates ferroptosis through both autophagy-dependent and -independent mechanisms. becn1 knockout mice exhibit late-onset lung carcinomas, hepatocellular carcinomas, and lymphomas (Liang et al., 1999), and hence may constitute a convenient model for studying the impact of autophagy-dependent ferroptosis on tumorigenesis.
Figure 6. Role of BECN1 in ferroptosis.
(A) AMPK-mediated BECN1 phosphorylation at S90 and S93 is required for the formation of a BECN1-SLC7A11/system xc− complex, which leads to lipid peroxidation-dependent ferroptosis via the inhibition of SLC7A11/system xc− activity. (B) ELAVL1-mediated BECN1 mRNA stability increases autophagosome formation and subsequent ferritinophagy-mediated ferroptosis.
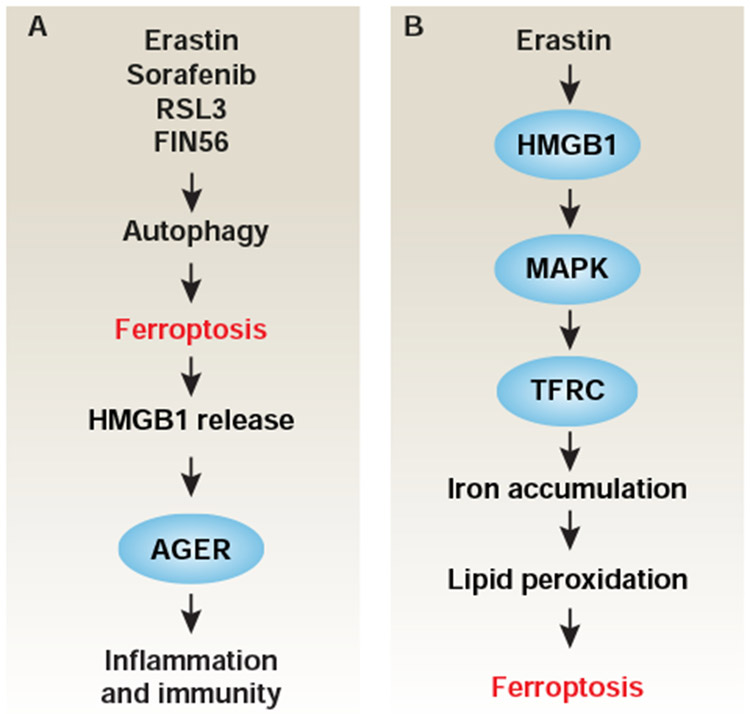
HMGB1-mediated danger signals
HMGB1 plays a dual role in the regulation of stress responses. Most intracellular HMGB1 localizes in the nucleus and functions as a DNA chaperone to regulate gene transcription, replication, and recombination. Moreover, HMGB1 is a damage-associated molecular pattern (DAMP) and can be secreted by immune cells or passively released from dead or dying cells. Extracellular HMGB1 acts as a danger signal to trigger immune responses during various types of cell death, including ferroptosis (Wen et al., 2019). Various ferroptosis activators (e.g., erastin, sorafenib, RSL3, and FIN56) can induce HMGB1 release, and this process is inhibited by pharmacological (using ferrostatin-1, liproxstatin-1, baicalein, bafilomycin A1, or chloroquine) or genetic (using Acs14 shRNA or atg5−/− or atg7−/− cells) inhibition of autophagy or ferroptosis (Wen et al., 2019). Moreover, HMGB1 release mediates ferroptotic cell death-induced inflammatory responses in macrophages through the activation of advanced glycosylation end-product specific receptor (AGER/RAGE), but not toll like receptor 4 (TLR4) (Wen et al., 2019) (Fig. 7A). HMGB1 also plays a location-dependent role in promoting autophagy. Cytosolic HMGB1 is a BECN1-binding protein for the induction of autophagosome formation (Tang et al., 2010b). Nuclear HMGB1 regulates heat shock protein family B (small) member 1 (HSPB1) expression for actin filament assembly that is involved in the regulation of autophagy membrane dynamics (Tang et al., 2011). Extracellular HMGB1 induces autophagy through the activation of the PIK3C3 complex (Tang et al., 2010a). Interestingly, intracellular HMGB1 positively regulates erastin-induced ferroptosis in leukemia cells through the upregulation of TFRC expression downstream of the activation of mitogen-activated protein kinase (MAPK, e.g., MAPK/JNK and MAPK/p38) (Ye et al., 2019) (Fig. 7B). The broader interconnectivity of HMGB1 in autophagy, ferroptosis, and immune response requires further elucidation.
Figure 7. Role of HMGB1 in ferroptosis.
(A) Autophagy-dependent ferroptotic cell death promotes HMGB1 release, which favors inflammation and immunity response through binding its receptor AGER. (B) HMGB1 promotes ferroptosis through upregulation of MAPK-dependent TFRC expression.
Conclusions and perspectives
In the past 5 years, studies of ferroptosis have been vastly expanded and the knowledge of the mechanism for ferroptosis has been ever increasing, revealing a surprising degree of complexity. Ferroptosis was originally recognized as a novel cell death modality that is morphologically, biochemically, and genetically distinct from other forms of cell death including apoptosis, necrosis, and autophagy. However, recent studies indicate that ferroptosis may occur while sharing common signals or regulators with other types of RCD such as apoptosis (Hong et al., 2017; Huang et al., 2018), necroptosis (Muller et al., 2017), autophagy (Gao et al., 2016; Hou et al., 2016), and lysosomal cell death (Gao et al., 2018; Torii et al., 2016). Indeed, mixed types of cell death appear to be more prevalent in human disease than “pure” types, although one type of cell death may dominate over others at a particular stage. In this review, we highlighted the regulation of autophagy-dependent ferroptosis and the involvement of a diverse array of molecular factors in this process. While certain types of selective autophagy (e.g., ferritinophagy, lipophagy, clockophagy, and CMA) play a significant role in promoting ferroptosis, the molecular, structural, and metabolic basis of autophagy-dependent ferroptosis still remains largely unknown. Although mitochondrial dysfunction and ER stress are related to ferroptosis in some cases (Dixon et al., 2014; Gao et al., 2019; Lee et al., 2018; Zhu et al., 2017), the functional role of mitophagy, reticulophagy, or the associated autophagy receptor in ferroptosis remains poorly understood. It is also a challenge to define the threshold or checkpoints associated with pro-survival and pro-death autophagy in ferroptosis. More recently, autophagy-dependent ferroptosis has been related to pancreatic tumor growth and progression through the released KRAS protein-mediated tumor-associated macrophage polarization (Dai et al., 2020a), indicating a dual role of ferroptosis in tumorigenesis and cancer therapy. More connections between autophagy-dependent ferroptosis and human disease will likely be demonstrated in the near future.
Supplementary Material
Highlights.
Autophagy is a lysosome-dependent cellular degradation mechanism.
Ferroptosis is an iron-dependent form of oxidative cell death.
Ferroptosis requires the autophagy machinery for its induction and execution.
Autophagy-dependent ferroptosis is implicated in diseases and pathological conditions.
Significance.
Autophagy plays a fundamental role in controlling of cellular homeostasis, thus determining the cellular fate. In many cases, autophagy is presumably activated by dying cells as a part of the programmed cell survival mechanism in response to stress. However, impaired and excessive autophagy promotes cell death, namely autophagy-dependent cell death. Of note, ferroptosis has been recognized as a form of autophagy-dependent cell death in some conditions. The ties between autophagy and ferroptosis could potentially allude to a complex interplay between metabolism dysfunction and oxidative stress. As a result, it is important to monitor autophagic activity or flux during ferroptosis. There are multiple compounds or drugs that inhibit the different types of autophagy-dependent cell death. A deeper understanding of the process and function of autophagy-dependent ferroptosis is critical for creating innovative therapeutic strategies for oxidative stress-related diseases, including neurodegenerative disorders and cancer.
Acknowledgments
We apologize to the researchers who were not referenced due to space limitations. We thank Dave Primm (Department of Surgery, University of Texas Southwestern Medical Center) for his critical reading of the manuscript. This work was supported by grants from the US National Institutes of Health (GM131919 [D.J.K.]) and the American Cancer Society (Research Scholar Grant RSG-16-014-01-CDD [D.T.]). G.K. is supported by the Ligue contre le Cancer (équipe labellisée); Agence National de la Recherche (ANR) – Projets blancs; ANR under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Chancelerie des universités de Paris (Legs Poix), Fondation pour la Recherche Médicale (FRM); a donation by Elior; European Research Area Network on Cardiovascular Diseases (ERA-CVD, MINOTAUR); the European Union Horizon 2020 Project Oncobiome; Fondation Carrefour; Institut National du Cancer (INCa); Inserm (HTE); Institut Universitaire de France; LeDucq Foundation; the LabEx Immuno-Oncology; the RHU Torino Lumiere; the Seerave Foundation; the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); and the SIRIC Cancer Research and Personalized Medicine (CARPEM).
Footnotes
Declaration of interests
The authors declare no conflicts of interest or financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aits S, Kricker J, Liu B, Ellegaard AM, Hamalisto S, Tvingsholm S, Corcelle-Termeau E, Hogh S, Farkas T, Holm Jonassen A, et al. (2015). Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 11, 1408–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, and Harper JW (2018). Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol 20, 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Meng L, Han L, Jia Y, Zhao Y, Gao H, Kang R, Wang X, Tang D, and Dai E (2018). Lipid storage and lipophagy regulates ferroptosis. Biochem Biophys Res Commun. [DOI] [PubMed] [Google Scholar]
- Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, Roberts MA, Tong B, Maimone TJ, Zoncu R, et al. (2019). The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhujabal Z, Birgisdottir AB, Sjottem E, Brenne HB, Overvatn A, Habisov S, Kirkin V, Lamark T, and Johansen T (2017). FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep 18, 947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialik S, Dasari SK, and Kimchi A (2018). Autophagy-dependent cell death - where, how and why a cell eats itself to death. J Cell Sci 131. [DOI] [PubMed] [Google Scholar]
- Bogdan AR, Miyazawa M, Hashimoto K, and Tsuji Y (2016). Regulators of Iron Homeostasis: New Players in Metabolism, Cell Death, and Disease. Trends Biochem Sci 41, 274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, and Parker R (2013). Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canli O, Alankus YB, Grootjans S, Vegi N, Hultner L, Hoppe PS, Schroeder T, Vandenabeele P, Bornkamm GW, and Greten FR (2016). Glutathione peroxidase 4 prevents necroptosis in mouse erythroid precursors. Blood 127, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Tavana O, Chu B, Erber L, Chen Y, Baer R, and Gu W (2017). NRF2 Is a Major Target of ARF in p53-Independent Tumor Suppression. Mol Cell 68, 224–232 e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Xiao Y, Chai P, Zheng P, Teng J, and Chen J (2019). ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy. Curr Biol 29, 846–855 e846. [DOI] [PubMed] [Google Scholar]
- Cherra SJ 3rd, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, and Chu CT (2010). Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol 190, 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chino H, Hatta T, Natsume T, and Mizushima N (2019). Intrinsically Disordered Protein TEX264 Mediates ER-phagy. Mol Cell 74, 909–921 e906. [DOI] [PubMed] [Google Scholar]
- Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, Tyurin VA, Yanamala N, Shrivastava IH, Mohammadyani D, et al. (2013). Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol 15, 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole-Ezea P, Swan D, Shanley D, and Hesketh J (2012). Glutathione peroxidase 4 has a major role in protecting mitochondria from oxidative damage and maintaining oxidative phosphorylation complexes in gut epithelial cells. Free Radic Biol Med 53, 488–497. [DOI] [PubMed] [Google Scholar]
- Cui Y, Parashar S, Zahoor M, Needham PG, Mari M, Zhu M, Chen S, Ho HC, Reggiori F, Farhan H, et al. (2019). A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science 365, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Cherra SJ 3rd, Kulich SM, Tandon A, Park D, and Chu CT (2009). Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem 284, 13843–13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai E, Han L, Liu J, Xie Y, Kroemer G, Klionsky DJ, Zeh HJ, Kang R, Wang J, and Tang D (2020a). Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai E, Meng L, Kang R, Wang X, and Tang D (2020b). ESCRT-III-dependent membrane repair blocks ferroptosis. Biochem Biophys Res Commun 522, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai E, Zhang W, Cong D, Kang R, Wang J, and Tang D (2020c). AIFM2 Blocks Ferroptosis Independent of Ubiquinol Metabolism. Biochem Biophys Res Commun. [DOI] [PubMed]
- Denton D, and Kumar S (2019). Autophagy-dependent cell death. Cell Death Differ 26, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deosaran E, Larsen KB, Hua R, Sargent G, Wang Y, Kim S, Lamark T, Jauregui M, Law K, Lippincott-Schwartz J, et al. (2013). NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci 126, 939–952. [DOI] [PubMed] [Google Scholar]
- Dikic I, and Elazar Z (2018). Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol 19, 349–364. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. (2012). Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Patel DN, Welsch M, Skouta R, Lee ED, Hayano M, Thomas AG, Gleason CE, Tatonetti NP, Slusher BS, et al. (2014). Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife 3, e02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon SJ, Winter GE, Musavi LS, Lee ED, Snijder B, Rebsamen M, Superti-Furga G, and Stockwell BR (2015). Human Haploid Cell Genetics Reveals Roles for Lipid Metabolism Genes in Nonapoptotic Cell Death. ACS Chem Biol 10, 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll S, Freitas FP, Shah R, Aldrovandi M, da Silva MC, Ingold I, Grocin AG, Xavier da Silva TN, Panzilius E, Scheel CH, et al. (2019). FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698. [DOI] [PubMed] [Google Scholar]
- Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A, et al. (2017). ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol 13, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al. (2015). Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Wang T, Li Y, Zhou Y, Wang X, Yu X, Ren X, An Y, Wu Y, Sun W, et al. (2018). DHA inhibits proliferation and induces ferroptosis of leukemia cells through autophagy dependent degradation of ferritin. Free Radic Biol Med. [DOI] [PubMed]
- Fan W, Nassiri A, and Zhong Q (2011). Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc Natl Acad Sci U S A 108, 7769–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch A, Eggenhofer E, et al. (2014). Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 16, 1180–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, St-Pierre P, Shankar J, Wang PT, Joshi B, and Nabi IR (2013). Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Mol Biol Cell 24, 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Hase K, Wada K, and Kabuta T (2015). An RNautophagy/DNautophagy receptor, LAMP2C, possesses an arginine-rich motif that mediates RNA/DNA-binding. Biochem Biophys Res Commun 460, 281–286. [DOI] [PubMed] [Google Scholar]
- Fumagalli F, Noack J, Bergmann TJ, Cebollero E, Pisoni GB, Fasana E, Fregno I, Galli C, Loi M, Solda T, et al. (2016). Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol 18, 1173–1184. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, et al. (2018). Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell death and differentiation 25, 486–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Chen D, Si J, Hu Q, Qin Z, Fang M, and Wang G (2015). The mitochondrial protein BNIP3L is the substrate of PARK2 and mediates mitophagy in PINK1/PARK2 pathway. Hum Mol Genet 24, 2528–2538. [DOI] [PubMed] [Google Scholar]
- Gao H, Bai Y, Jia Y, Zhao Y, Kang R, Tang D, and Dai E (2018). Ferroptosis is a lysosomal cell death process. Biochem Biophys Res Commun 503, 1550–1556. [DOI] [PubMed] [Google Scholar]
- Gao M, Monian P, Pan Q, Zhang W, Xiang J, and Jiang X (2016). Ferroptosis is an autophagic cell death process. Cell Res 26, 1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Yi J, Zhu J, Minikes AM, Monian P, Thompson CB, and Jiang X (2019). Role of Mitochondria in Ferroptosis. Mol Cell 73, 354–363 e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica D, Lahiri V, and Klionsky DJ (2018). Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, and Springer W (2010). PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12, 119–131. [DOI] [PubMed] [Google Scholar]
- Geng N, Shi BJ, Li SL, Zhong ZY, Li YC, Xua WL, Zhou H, and Cai JH (2018). Knockdown of ferroportin accelerates erastin-induced ferroptosis in neuroblastoma cells. Eur Rev Med Pharmacol Sci 22, 3826–3836. [DOI] [PubMed] [Google Scholar]
- Gnanapradeepan K, Basu S, Barnoud T, Budina-Kolomets A, Kung CP, and Murphy ME (2018). The p53 Tumor Suppressor in the Control of Metabolism and Ferroptosis. Front Endocrinol (Lausanne) 9, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Sanchez JA, Carty L, Iruarrizaga-Lejarreta M, Palomo-Irigoyen M, Varela-Rey M, Griffith M, Hantke J, Macias-Camara N, Azkargorta M, Aurrekoetxea I, et al. (2015). Schwann cell autophagy, myelinophagy, initiates myelin clearance from injured nerves. J Cell Biol 210, 153–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin JM, Dowdle WE, DeJesus R, Wang Z, Bergman P, Kobylarz M, Lindeman A, Xavier RJ, McAllister G, Nyfeler B, et al. (2017). Autophagy-Independent Lysosomal Targeting Regulated by ULK1/2-FIP200 and ATG9. Cell Rep 20, 2341–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso D, Ropolo A, Lo Re A, Boggio V, Molejon MI, Iovanna JL, Gonzalez CD, Urrutia R, and Vaccaro MI (2011). Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem 286, 8308–8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P, Morozzi G, Holper S, Mari M, Harwardt MI, Yan R, Muller S, Reggiori F, Heilemann M, and Dikic I (2017). Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, and Ohsumi Y (2007). The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 282, 37298–37302. [DOI] [PubMed] [Google Scholar]
- Heo JM, Ordureau A, Paulo JA, Rinehart J, and Harper JW (2015). The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell 60, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, Ordureau A, Swarup S, Paulo JA, Shen K, Sabatini DM, and Harper JW (2018). RAB7A phosphorylation by TBK1 promotes mitophagy via the PINK-PARKIN pathway. Sci Adv 4, eaav0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman A, Holst CR, Latham JC, Bruegger JJ, Ulas G, McCusker KP, Amagata A, Davis D, Hoff KG, Kahn-Kirby AH, et al. (2018). Vitamin E hydroquinone is an endogenous regulator of ferroptosis via redox control of 15-lipoxygenase. PLoS One 13, e0201369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Lee DH, Lee YS, Jo MJ, Jeong YA, Kwon WT, Choudry HA, Bartlett DL, and Lee YJ (2017). Molecular crosstalk between ferroptosis and apoptosis: emerging role of ER stress-induced p53-independent PUMA expression. Oncotarget 8, 115164–115178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W, Xie Y, Song X, Sun X, Lotze MT, Zeh HJ 3rd, Kang R, and Tang D (2016). Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 12, 1425–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Yang M, Deng J, Li P, Su W, and Jiang R (2018). Upregulation and activation of p53 by erastininduced reactive oxygen species contribute to cytotoxic and cytostatic effects in A549 lung cancer cells. Oncol Rep 40, 2363–2370. [DOI] [PubMed] [Google Scholar]
- Hung YH, Chen LM, Yang JY, and Yang WY (2013). Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun 4, 2111. [DOI] [PubMed] [Google Scholar]
- Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, Baer R, and Gu W (2015). Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachim J, Jefferies HB, Razi M, Frith D, Snijders AP, Chakravarty P, Judith D, and Tooze SA (2015). Activation of ULK Kinase and Autophagy by GABARAP Trafficking from the Centrosome Is Regulated by WAC and GM130. Mol Cell 60, 899–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, and Lamark T (2020). Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J Mol Biol 432, 80–103. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, and Yoshimori T (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19, 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Mao G, Qu F, Angeli JP, Doll S, Croix CS, Dar HH, Liu B, Tyurin VA, Ritov VB, et al. (2017). Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol 13, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Hoshijima M, Seo K, Bedja D, Sysa-Shah P, Andrabi SA, Chen W, Hoke A, Dawson VL, Dawson TM, et al. (2014). Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J 33, 2798–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Kroemer G, and Tang D (2019). The tumor suppressor protein p53 and the ferroptosis network. Free Radic Biol Med 133, 162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, and Tang D (2017). Autophagy and Ferroptosis - What's the Connection? Curr Pathobiol Rep 5, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Zeh HJ, Lotze MT, and Tang D (2011). The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 18, 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Zeng L, Xie Y, Yan Z, Zhou B, Cao L, Klionsky DJ, Tracey KJ, Li J, Wang H, et al. (2016). A novel PINK1- and PARK2-dependent protective neuroimmune pathway in lethal sepsis. Autophagy 12, 2374–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Zeng L, Zhu S, Xie Y, Liu J, Wen Q, Cao L, Xie M, Ran Q, Kroemer G, et al. (2018a). Lipid Peroxidation Drives Gasdermin D-Mediated Pyroptosis in Lethal Polymicrobial Sepsis. Cell Host Microbe 24, 97–108 e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Zhu S, Zeh HJ, Klionsky DJ, and Tang D (2018b). BECN1 is a new driver of ferroptosis. Autophagy 14, 2173–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, et al. (2015). Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358. [DOI] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, and Guan KL (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, and Emr SD (2000). Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerver L, Papadopoulos C, Liu B, Kravic B, Rota G, Brecht L, Veenendaal T, Polajnar M, Bluemke A, Ehrmann M, et al. (2019). The ubiquitin-conjugating enzyme UBE2QL1 coordinates lysophagy in response to endolysosomal damage. EMBO Rep, e48014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Z, Liu R, and Cheng Y (2019). Artesunate alleviates liver fibrosis by regulating ferroptosis signaling pathway. Biomed Pharmacother 109, 2043–2053. [DOI] [PubMed] [Google Scholar]
- Kroemer G, and Levine B (2008). Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol 9, 1004–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, and Levine B (2010). Autophagy and the integrated stress response. Mol Cell 40, 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, and Youle RJ (2015). The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, and Tournier C (2011). The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy 7, 689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MY, Sumpter R Jr., Zou Z, Sirasanagandla S, Wei Y, Mishra P, Rosewich H, Crane DI, and Levine B (2017). Peroxisomal protein PEX13 functions in selective autophagy. EMBO Rep 18, 48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Lee DH, Choudry HA, Bartlett DL, and Lee YJ (2018). Ferroptosis-Induced Endoplasmic Reticulum Stress: Cross-talk between Ferroptosis and Apoptosis. Mol Cancer Res 16, 1073–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, and Kroemer G (2019). Biological Functions of Autophagy Genes: A Disease Perspective. Cell 176, 11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang Y, Cheng X, Yuan H, Zhu S, Liu J, Wen Q, Xie Y, Liu J, Kroemer G, et al. (2018). PINK1 and PARK2 Suppress Pancreatic Tumorigenesis through Control of Mitochondrial Iron-Mediated Immunometabolism. Dev Cell 46, 441–455 e448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Han H, Zhou MT, Yang B, Ta AP, Li N, Chen J, and Wang W (2017). Proteomic Analysis of the Human Tankyrase Protein Interaction Network Reveals Its Role in Pexophagy. Cell Rep 20, 737–749. [DOI] [PubMed] [Google Scholar]
- Li Y, Jiang X, Zhang Y, Gao Z, Liu Y, Hu J, Hu X, Li L, Shi J, and Gao N (2019). Nuclear accumulation of UBC9 contributes to SUMOylation of lamin A/C and nucleophagy in response to DNA damage. J Exp Clin Cancer Res 38, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, and Jung JU (2006). Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 8, 688–699. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, and Levine B (1999). Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676. [DOI] [PubMed] [Google Scholar]
- Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, and Levine B (1998). Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 72, 8586–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Zhang Z, Chen L, Zhou Y, Zou P, Feng C, Wang L, and Liang G (2016). Dihydroartemisinin (DHA) induces ferroptosis and causes cell cycle arrest in head and neck carcinoma cells. Cancer Lett 381, 165–175. [DOI] [PubMed] [Google Scholar]
- Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang SM, Lian G, Liu Q, Ruan K, et al. (2012). GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336, 477–481. [DOI] [PubMed] [Google Scholar]
- Liu J, Yang M, Kang R, Klionsky DJ, and Tang D (2019). Autophagic degradation of the circadian clock regulator promotes ferroptosis. Autophagy 15, 2033–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, and Czaja MJ (2013). Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ 20, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al. (2012). Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol 14, 177–185. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang W, Li Y, Xiao Y, Cheng J, and Jia J (2015). The 5-Lipoxygenase Inhibitor Zileuton Confers Neuroprotection against Glutamate Oxidative Damage by Inhibiting Ferroptosis. Biol Pharm Bull 38, 1234–1239. [DOI] [PubMed] [Google Scholar]
- Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, et al. (2013). Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J 32, 2336–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias JD, Pontano Vaites L, Nissim S, Biancur DE, Kim AJ, Wang X, Liu Y, Goessling W, Kimmelman AC, and Harper JW (2015). Ferritinophagy via NCOA4 is required for erythropoiesis and is regulated by iron dependent HERC2-mediated proteolysis. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancias JD, Wang X, Gygi SP, Harper JW, and Kimmelman AC (2014). Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, Li F, Gemperline DC, Book AJ, and Vierstra RD (2015). Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol Cell 58, 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M, Freigang S, Schneider C, Conrad M, Bornkamm GW, and Kopf M (2015). T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. J Exp Med 212, 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer CA, Kaliappan A, and Dennis PB (2009). A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5, 649–662. [DOI] [PubMed] [Google Scholar]
- Michiorri S, Gelmetti V, Giarda E, Lombardi F, Romano F, Marongiu R, Nerini-Molteni S, Sale P, Vago R, Arena G, et al. (2010). The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ 17, 962–974. [DOI] [PubMed] [Google Scholar]
- Mizushima N (2019). The ubiquitin E2 enzyme UBE2QL1 mediates lysophagy. EMBO Rep, e49104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, and Yoshimori T (2007). How to interpret LC3 immunoblotting. Autophagy 3, 542–545. [DOI] [PubMed] [Google Scholar]
- Molejon MI, Ropolo A, Re AL, Boggio V, and Vaccaro MI (2013). The VMP1-Beclin 1 interaction regulates autophagy induction. Sci Rep 3, 1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AS, and Holzbaur EL (2016). Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc Natl Acad Sci U S A 113, E3349–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller T, Dewitz C, Schmitz J, Schroder AS, Brasen JH, Stockwell BR, Murphy JM, Kunzendorf U, and Krautwald S (2017). Necroptosis and ferroptosis are alternative cell death pathways that operate in acute kidney failure. Cell Mol Life Sci 74, 3631–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakawa T, Yamaguchi O, Hashimoto A, Hikoso S, Takeda T, Oka T, Yasui H, Ueda H, Akazawa Y, Nakayama H, et al. (2015). Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun 6, 7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrow L, and Debnath J (2013). Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol 8, 105–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, and Yoshimori T (2017). New insights into autophagosome-lysosome fusion. J Cell Sci 130, 1209–1216. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ishii J, Asai E, and Ohsumi Y (2012). Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy 8, 177–186. [DOI] [PubMed] [Google Scholar]
- NaveenKumar SK, SharathBabu BN, Hemshekhar M, Kemparaju K, Girish KS, and Mugesh G (2018). The Role of Reactive Oxygen Species and Ferroptosis in Heme-Mediated Activation of Human Platelets. ACS Chem Biol 13, 1996–2002. [DOI] [PubMed] [Google Scholar]
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. (2013). mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 15, 406–416. [DOI] [PubMed] [Google Scholar]
- Niu TK, Cheng Y, Ren X, and Yang JM (2010). Interaction of Beclin 1 with survivin regulates sensitivity of human glioma cells to TRAIL-induced apoptosis. FEBS Lett 584, 3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan TE, Johnson LR, Kang HH, and Sun JC (2015). BNIP3- and BNIP3L-Mediated Mitophagy Promotes the Generation of Natural Killer Cell Memory. Immunity 43, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstein A, Jeffrey PD, and Shi Y (2007). Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem 282, 13123–13132. [DOI] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R Jr., Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, et al. (2011). Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbye A, Fengsrud M, and Seglen PO (2007). Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy 3, 300–322. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, and Johansen T (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282, 24131–24145. [DOI] [PubMed] [Google Scholar]
- Papadopoulos C, and Meyer H (2017). Detection and Clearance of Damaged Lysosomes by the Endo-Lysosomal Damage Response and Lysophagy. Curr Biol 27, R1330–R1341. [DOI] [PubMed] [Google Scholar]
- Papandreou ME, and Tavernarakis N (2019). Nucleophagy: from homeostasis to disease. Cell Death Differ 26, 630–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Seo M, Jung CH, Grunwald D, Stone M, Otto NM, Toso E, Ahn Y, Kyba M, Griffin TJ, et al. (2018). ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy 14, 584–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, and Levine B (2005). Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939. [DOI] [PubMed] [Google Scholar]
- Princely Abudu Y, Pankiv S, Mathai BJ, Hakon Lystad A, Bindesboll C, Brenne HB, Yoke Wui Ng M, Thiede B, Yamamoto A, Mutugi Nthiga T, et al. (2019). NIPSNAP1 and NIPSNAP2 Act as "Eat Me" Signals for Mitophagy. Dev Cell 49, 509–525 e512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Q, Gu M, Van Remmen H, Strong R, Roberts JL, and Richardson A (2006). Glutathione peroxidase 4 protects cortical neurons from oxidative injury and amyloid toxicity. J Neurosci Res 84, 202–208. [DOI] [PubMed] [Google Scholar]
- Ran Q, Liang H, Gu M, Qi W, Walter CA, Roberts LJ 2nd, Herman B, Richardson A, and Van Remmen H (2004). Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J Biol Chem 279, 55137–55146. [DOI] [PubMed] [Google Scholar]
- Ran Q, Van Remmen H, Gu M, Qi W, Roberts LJ 2nd, Prolla T, and Richardson A (2003). Embryonic fibroblasts from Gpx4+/− mice: a novel model for studying the role of membrane peroxidation in biological processes. Free Radic Biol Med 35, 1101–1109. [DOI] [PubMed] [Google Scholar]
- Reichelt ME, Mellor KM, Curl CL, Stapleton D, and Delbridge LM (2013). Myocardial glycophagy - a specific glycogen handling response to metabolic stress is accentuated in the female heart. J Mol Cell Cardiol 65, 67–75. [DOI] [PubMed] [Google Scholar]
- Rello-Varona S, Lissa D, Shen S, Niso-Santano M, Senovilla L, Marino G, Vitale I, Jemaa M, Harper F, Pierron G, et al. (2012). Autophagic removal of micronuclei. Cell Cycle 11, 170–176. [DOI] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, and Guan KL (2013). ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15, 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, and Wang J (2008). Essential role for Nix in autophagic maturation of erythroid cells. Nature 454, 232–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent G, van Zutphen T, Shatseva T, Zhang L, Di Giovanni V, Bandsma R, and Kim PK (2016). PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J Cell Biol 214, 677–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauzay C, Louandre C, Bodeau S, Anglade F, Godin C, Saidak Z, Fontaine JX, Usureau C, Martin N, Molinie R, et al. (2018). Protein biosynthesis, a target of sorafenib, interferes with the unfolded protein response (UPR) and ferroptosis in hepatocellular carcinoma cells. Oncotarget 9, 8400–8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf MB, Keulers TG, Vooijs MA, and Rouschop KM (2016). LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J 30, 3961–3978. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, and Elazar Z (2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26, 1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler A, Schneider M, Forster H, Roth S, Wirth EK, Culmsee C, Plesnila N, Kremmer E, Radmark O, Wurst W, et al. (2008). Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell metabolism 8, 237–248. [DOI] [PubMed] [Google Scholar]
- Sharma V, Verma S, Seranova E, Sarkar S, and Kumar D (2018). Selective Autophagy and Xenophagy in Infection and Disease. Front Cell Dev Biol 6, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Skouta R, Kaplan A, Yang WS, Hayano M, Dixon SJ, Brown LM, Valenzuela CA, Wolpaw AJ, and Stockwell BR (2016). Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat Chem Biol 12, 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Kim EH, Lee J, and Roh JL (2018). Nrf2 inhibition reverses resistance to GPX4 inhibitor-induced ferroptosis in head and neck cancer. Free Radic Biol Med 129, 454–462. [DOI] [PubMed] [Google Scholar]
- Shintoku R, Takigawa Y, Yamada K, Kubota C, Yoshimoto Y, Takeuchi T, Koshiishi I, and Torii S (2017). Lipoxygenase-mediated generation of lipid peroxides enhances ferroptosis induced by erastin and RSL3. Cancer Sci 108, 2187–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, and Czaja MJ (2009). Autophagy regulates lipid metabolism. Nature 458, 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, Burman JL, Li Y, Zhang Z, Narendra DP, et al. (2018). Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Smith M, and Wilkinson S (2017). ER homeostasis and autophagy. Essays Biochem 61, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, von Kriegsheim A, Behrends C, and Wilkinson S (2018). CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev Cell 44, 217–232 e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhu S, Chen P, Hou W, Wen Q, Liu J, Xie Y, Liu J, Klionsky DJ, Kroemer G, et al. (2018). AMPK-Mediated BECN1 Phosphorylation Promotes Ferroptosis by Directly Blocking System Xc(−) Activity. Curr Biol 28, 2388–2399 e2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascon S, Hatzios SK, Kagan VE, et al. (2017). Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 171, 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, and Tang D (2016). Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 63, 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Ou Z, Xie M, Kang R, Fan Y, Niu X, Wang H, Cao L, and Tang D (2015). HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene 34, 5617–5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szargel R, Shani V, Abd Elghani F, Mekies LN, Liani E, Rott R, and Engelender S (2016). The PINK1, synphilin-1 and SIAH-1 complex constitutes a novel mitophagy pathway. Hum Mol Genet 25, 3476–3490. [DOI] [PubMed] [Google Scholar]
- Tabata K, Matsunaga K, Sakane A, Sasaki T, Noda T, and Yoshimori T (2010). Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain. Mol Biol Cell 21, 4162–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Berghe TV, Vandenabeele P, and Kroemer G (2019). The molecular machinery of regulated cell death. Cell Res 29, 347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, Benschop R, Sparvero LJ, Amoscato AA, Tracey KJ, et al. (2010a). HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene 29, 5299–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ 3rd, et al. (2010b). Endogenous HMGB1 regulates autophagy. J Cell Biol 190, 881–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, Van Houten B, Zeh HJ 3rd, and Lotze MT (2011). High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab 13, 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, and Yoshimori T (2010). The origin of the autophagosomal membrane. Nat Cell Biol 12, 831–835. [DOI] [PubMed] [Google Scholar]
- Torii S, Shintoku R, Kubota C, Yaegashi M, Torii R, Sasaki M, Suzuki T, Mori M, Yoshimoto Y, Takeuchi T, et al. (2016). An essential role for functional lysosomes in ferroptosis of cancer cells. Biochem J 473, 769–777. [DOI] [PubMed] [Google Scholar]
- Turco E, Fracchiolla D, and Martens S (2020). Recruitment and Activation of the ULK1/Atg1 Kinase Complex in Selective Autophagy. J Mol Biol 432, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L, Joza N, Vitale I, Morselli E, Tailler M, et al. (2009). The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ 16, 1006–1017. [DOI] [PubMed] [Google Scholar]
- Wang W, and Subramani S (2017). Role of PEX5 ubiquitination in maintaining peroxisome dynamics and homeostasis. Cell Cycle 16, 2037–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Serricchio M, Jauregui M, Shanbhag R, Stoltz T, Di Paolo CT, Kim PK, and McQuibban GA (2015). Deubiquitinating enzymes regulate PARK2-mediated mitophagy. Autophagy 11, 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Chang SY, Wu Q, Gou YJ, Jia L, Cui YM, Yu P, Shi ZH, Wu WS, Gao G, et al. (2016). The Protective Role of Mitochondrial Ferritin on Erastin-Induced Ferroptosis. Front Aging Neurosci 8, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Chiang WC, Sumpter R Jr., Mishra P, and Levine B (2017). Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 168, 224–238 e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, et al. (2013). EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 154, 1269–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Q, Liu J, Kang R, Zhou B, and Tang D (2019). The release and activity of HMGB1 in ferroptosis. Biochem Biophys Res Commun 510, 278–283. [DOI] [PubMed] [Google Scholar]
- Wenzel SE, Tyurina YY, Zhao J, St Croix CM, Dar HH, Mao G, Tyurin VA, Anthonymuthu TS, Kapralov AA, Amoscato AA, et al. (2017). PEBP1 Wardens Ferroptosis by Enabling Lipoxygenase Generation of Lipid Death Signals. Cell 171, 628–641 e626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YC, and Holzbaur EL (2014). Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A 111, E4439–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Minikes AM, Gao M, Bian H, Li Y, Stockwell BR, Chen ZN, and Jiang X (2019a). Intercellular interaction dictates cancer cell ferroptosis via NF2-YAP signalling. Nature 572, 402–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KC, Cui JY, and Klaassen CD (2011). Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci 123, 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Geng Y, Lu X, Shi Y, Wu G, Zhang M, Shan B, Pan H, and Yuan J (2019b). Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc Natl Acad Sci U S A 116, 2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyant GA, Abu-Remaileh M, Frenkel EM, Laqtom NN, Dharamdasani V, Lewis CA, Chan SH, Heinze I, Ori A, and Sabatini DM (2018). NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 360, 751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Hou W, Song X, Yu Y, Huang J, Sun X, Kang R, and Tang D (2016). Ferroptosis: process and function. Cell Death Differ 23, 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, and Tang D (2015). Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 11, 28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Zhu S, Song X, Sun X, Fan Y, Liu J, Zhong M, Yuan H, Zhang L, Billiar TR, et al. (2017). The Tumor Suppressor p53 Limits Ferroptosis by Blocking DPP4 Activity. Cell Rep 20, 1692–1704. [DOI] [PubMed] [Google Scholar]
- Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, Wolpaw AJ, Smukste I, Peltier JM, Boniface JJ, et al. (2007). RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 447, 864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Abe K, Tatemichi Y, and Fujiki Y (2014). The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy 10, 1549–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Li Y, Yan G, Liu T, Feng C, Gong R, Yuan Y, Ding F, Zhang L, Idiiatullina E, et al. (2017). Inhibition of iron overload-induced apoptosis and necrosis of bone marrow mesenchymal stem cells by melatonin. Oncotarget 8, 31626–31637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Chen P, Liu J, Zhu S, Kroemer G, Klionsky DJ, Lotze MT, Zeh HJ, Kang R, and Tang D (2019a). Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci Adv 5, eaaw2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WH, Ding CC, Sun T, Rupprecht G, Lin CC, Hsu D, and Chi JT (2019b). The Hippo Pathway Effector TAZ Regulates Ferroptosis in Renal Cell Carcinoma. Cell Rep 28, 2501–2508 e2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, Kim KJ, Gaschler MM, Patel M, Shchepinov MS, and Stockwell BR (2016). Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci U S A 113, E4966–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. (2014). Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, and Stockwell BR (2008). Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chemistry & biology 15, 234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, and Klionsky DJ (2010). Eaten alive: a history of macroautophagy. Nat Cell Biol 12, 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chai W, Xie M, Yang M, Yu Y, Cao L, and Yang L (2019). HMGB1 regulates erastin-induced ferroptosis via RAS-JNK/p38 signaling in HL-60/NRAS(Q61L) cells. Am J Cancer Res 9, 730–739. [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Yasuda S, Fujita T, Hamasaki M, Murakami A, Kawawaki J, Iwai K, Saeki Y, Yoshimori T, Matsuda N, et al. (2017). Ubiquitination of exposed glycoproteins by SCF(FBXO27) directs damaged lysosomes for autophagy. Proc Natl Acad Sci U S A 114, 8574–8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii SR, and Mizushima N (2017). Monitoring and Measuring Autophagy. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Li X, Zhang X, Kang R, and Tang D (2016a). CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation. Biochem Biophys Res Commun 478, 838–844. [DOI] [PubMed] [Google Scholar]
- Yuan H, Li X, Zhang X, Kang R, and Tang D (2016b). Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun 478, 1338–1343. [DOI] [PubMed] [Google Scholar]
- Yun J, Puri R, Yang H, Lizzio MA, Wu C, Sheng ZH, and Guo M (2014). MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. Elife 3, e01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tripathi DN, Jing J, Alexander A, Kim J, Powell RT, Dere R, Tait-Mulder J, Lee JH, Paull TT, et al. (2015). ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol 17, 1259–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yao Z, Wang L, Ding H, Shao J, Chen A, Zhang F, and Zheng S (2018). Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy 14, 2083–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Liu J, Kang R, Klionsky DJ, Kroemer G, and Tang D (2019). Ferroptosis is a type of autophagy-dependent cell death. Semin Cancer Biol. [DOI] [PubMed] [Google Scholar]
- Zhou B, Zhang JY, Liu XS, Chen HZ, Ai YL, Cheng K, Sun RY, Zhou D, Han J, and Wu Q (2018). Tom20 senses iron-activated ROS signaling to promote melanoma cell pyroptosis. Cell Res 28, 1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Ma K, Gao R, Mu C, Chen L, Liu Q, Luo Q, Feng D, Zhu Y, and Chen Q (2017). Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res 27, 184–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Zhang Q, Sun X, Zeh HJ 3rd, Lotze MT, Kang R, and Tang D (2017). HSPA5 Regulates Ferroptotic Cell Death in Cancer Cells. Cancer Res 77, 2064–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Palte MJ, Deik AA, Li H, Eaton JK, Wang W, Tseng YY, Deasy R, Kost-Alimova M, Dancik V, et al. (2019). A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun 10, 1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zutphen T, Veenhuis M, and van der Klei IJ (2008). Pex14 is the sole component of the peroxisomal translocon that is required for pexophagy. Autophagy 4, 63–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.