Abstract
Background
The pandemic of coronavirus disease 2019 (COVID-19) has become a global threat to public health. The lipid pathophysiology in COVID-19 is unknown.
Methods
In this retrospective longitudinal study, we monitored the serum lipids in 17 surviving and 4 non-surviving COVID-19 cases prior to their viral infections and duration the entire disease courses.
Results
In surviving cases, the low-density lipoprotein (LDL) levels decreased significantly on admission as compared with the levels before infection; the LDL levels remained constantly low during the disease progression and resumed to the original levels when patients recovered (pre-infection: 3.5 (3.0–4.4); on admission: 2.8 (2.3–3.1), p < 0.01; progression: 2.5 (2.3–3.0); discharge: 3.6 (2.7–4.1); median (IQR), in mmol/L). In non-surviving patients, LDL levels showed an irreversible and continuous decrease until death (1.1 (0.9–1.2), p = 0.02 versus the levels on admission). The ratio changes of LDL levels inversely correlated with ratio changes of high-sensitivity C-reactive protein levels. Logistic regression analysis showed increasing odds of lowered LDL levels associated with disease progression (odds ratio: 4.48, 95% IC: 1.55–12.92, p = 0.006) and in-hospital death (odds ratio: 21.72, 95% IC: 1.40–337.54, p = 0.028).
Conclusions
LDL levels inversely correlated to disease severities, which could be a predictor for disease progress and poor prognosis.
Abbreviation: ALP, alkaline phosphatase; ALT, alanine aminotransferase; AST, aspartate aminotransferase; COVID-19, coronavirus disease 2019; CRP, C-reactive protein; hsCRP, high-sensitivity C-reactive protein; GGT, gamma-glutamyl transferase; HDL, high-density lipoprotein; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TC, total cholesterol
Keywords: COVID-19, LDL, Lipid, Odds ratio
1. Introduction
Dear Sir,
The pandemic of coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a global threat to public health [1]. SARS-COV-2 is a single-strand RNA virus with 96.3% sequence identity the bat coronavirus RaTG13 [[2], [3], [4]]. The spike protein of SARS-COV-2 is responsible for entry of the host cells via binding to surface angiotensin converting enzyme 2 [2,5]. The estimated mortality rate for COVID-19 is about 2.3%, with a range of 6 to 41 days from the onset of symptoms to death [6,7]. The prominent pathological changes in lungs include edema, proteinaceous exudate, and multinucleated pneumocytes [8,9]. We here first report the lipid pathophysiology in COVID-19.
2. Methods
We performed a retrospective longitudinal analysis of COVID-19 patients (n = 21) who were admitted between January 18 and February 8, 2020, into the department of infectious diseases at Zhongnan Hospital of Wuhan University in Wuhan, China. The study was approved by the Institutional Review Board at the hospital (No. 2020011). The patients who had a routine laboratory test including lipids in our hospital between January 9 and 17, 2020 prior to their SARS-CoV2 infection and tested positive for SARS-CoV-2 during the COVID-19 epidemic were included in this study. All patients were admitted to our hospital between January 18 and February 8, 2020, and were either discharged or died by March 15, 2020. Healthy subjects (n = 31) and patients with chronic obstructive pulmonary disease (COPD, n = 21) who had lipid tests in our hospital between September 2019 to October 2019 were included as normal and non-COVID-19 controls. Electronic medical records including demographic, clinical treatment, and laboratory data, were extracted. Detailed methods were described in the supplementary materials. Data were presented as median (interquartile range, IQR) or mean (standard deviation, s.d.) and a Mann-Whitney U test was used to compare differences between groups.
3. Results
The average age of the patients was 62.5 ± 12.6 years; 52% were male and 48% were female. 62% had morbidities such as diabetes, hypertension, cardiovascular disorders, hyperlipidemia, or kidney diseases. The onset symptoms of the patients were: fever (86%), cough (95%), fatigue (71%), shortness of breath (43%) and diarrhea (10%) (Table 1a ). The severities of the admitted patients were 62% of mild, 19% of severe, and 19% of critical, a classification according to Chinese Center for Disease Control (CDC) guidelines [1]. Patients received standard treatments based on the guidelines of Chinese CDC, including antiviral remedies (arbidol, lopinavir and ribonavir, interferon α inhalation), anti-inflammatory treatments (corticosteroid), and immune-modulator (thymalfasin, immunoglobulin) (Table 1a). Severe and critical cases were given high-flow oxygen, non-invasive or invasive ventilation, depending on their morbidities. Seventeen patients recovered and were discharged, while four patients did not survive.
Table 1a.
Clinical characteristics of COVID-19 patients.
| No. (%) |
|||
|---|---|---|---|
| Total (n = 21) | Survival (n = 17) | Non-survival (n = 4) | |
| Age, mean (s.d.) | 62.5 (12.6) | 61.5 (9.5) | 79.7 (14.3)⁎ |
| Male | 11 (52.3) | 9 (53.9) | 2 (50.0) |
| Comorbidities | 13 (61.9) | 9 (53.9) | 4 (100) |
| Type 2 diabetes | 3 (14.3) | 3 (17.6) | 0 (0) |
| Hypertension | 10 (47.6) | 8 (35.3) | 2 (50.0) |
| Cardiovascular disease | 5 (23.8) | 3 (17.6) | 2 (50.0) |
| Kidney diseases | 1 (4.8) | 1 (5.8) | 0 (0) |
| Onset symptoms | |||
| Fever | 18 (85.7) | 14 (82.3) | 4 (100) |
| Cough | 20 (95.2) | 16 (94.1) | 4 (100) |
| Fatigue | 15 (71.4) | 11 (64.7) | 4 (100) |
| Shortness of breath | 9 (42.8) | 5 (29.4) | 4 (100) |
| Diarrhea | 2 (9.5) | 2 (11.7) | 0 (0) |
| Treatment | |||
| Arbidol | 15 (71.4) | 12 (70.6) | 3 (75.0) |
| Lopinavir/ritonavir | 7 (33.3) | 4 (23.5) | 3 (75.0) |
| Interferon inhalation | 7 (33.3) | 4 (23.5) | 3 (75.0) |
| Corticosteroid | 3 (14.3) | 3 (17.6) | 0 (0) |
| Thymalfasin | 6 (28.6) | 6 (35.3) | 0 (0) |
| Oxygen | 8 (38.1) | 4 (23.5) | 4 (100) |
| Mechanical ventilation | 4 (19.0) | 0 (0) | 4 (100) |
p < 0.05, as compared to the patient group that survived.
In the stage of disease progression, there were significantly higher levels of C-reactive protein (CRP) (78 (28–134) versus 15 (7–36), in mg/L, p = 0.042) and interleukin-6 (IL-6) (195 (127–280) versus 12 (4–18), in pg/ml, p < 0.001), but lower levels of CD8+ subpopulation T cells (33 (23–135) versus 273 (122–377), cells/μl, p = 0.016) in non-surviving patients as compared with the patients survived (Table 1b ). Furthermore, in surviving patients, increases in levels of high-sensitivity CRP (hsCRP) were significantly associated with the disease severity (pre-infection: 1.2 (0.7–5.9); on admission: 11 (5.9–26), p < 0.02; progression: 9.8 (5.3–45); discharge: 2.2 (1.2–7.0), in mg/L) (Supplementary Table S1). The number of lymphocytes (LY) decreased significantly at the time of admission as compared with before infection; it did not show a full recovery at the time of discharge (pre-infection: 1.6 (1.4–2.2); on admission: 1.0 (0.8–1.5), p < 0.02; progression: 0.9 (0.5–1.4); discharge: 1.2 (0.8–1.9), in x109/L) (Supplementary Table S1). In patients did not survive, there was a continuous increase in hsCRP level or lymphopenia until death (Supplementary Table S1).
Table 1b.
Laboratory tests for COVID-19 patients during the stage of disease progression.
| Laboratory tests | Reference value | No. (%) |
|||
|---|---|---|---|---|---|
| Total (n = 21) | Survival (n = 17) | Non-survival(n = 4) | p | ||
| IL-6 (pg/ml) | 0.1–2.9 | 15 (8–79, 19) | 12 (4–18, 15) | 195 (127–280, 4) | <0.001 |
| CRP (mg/L) | <4 | 20 (7–45, 21) | 15 (7–36, 17) | 78 (28–134, 4) | 0.042 |
| CD3+ (cells/μl) | 805 to 4459 | 562 (203–939, 19) | 680 (303–945, 15) | 232 (129–340, 4) | 0.062 |
| CD4+ (cells/μl) | 345 to 2350 | 297 (135–557, 19) | 416 (172–557, 15) | 168 (108–250, 4) | 0.121 |
| CD8+ (cells/μl) | 345 to 2350 | 166 (50–336, 19) | 273 (122–377, 15) | 33 (23–135, 4) | 0.016 |
| CD4+/CD8+ ratio | 0.96–2.05 | 1.7 (1.2–3.1, 19) | 1.6 (1.3–2.4, 15) | 5.0 (2.0–6.2, 4) | 0.079 |
| CD19+ (cells/μl) | 240 to 1317 | 104 (49–174, 19) | 104 (49–236, 15) | 84 (34–134, 4) | 0.650 |
| CD16+/CD56+ (cells/μl) | 210 to 1514 | 164 (49–252, 19) | 168 (106–252, 15) | 33 (10–223, 4) | 0.124 |
P value is compared the group that survived with the group that did not survive by a Mann-Whitney U test. Data is presented as median (IQR, n).
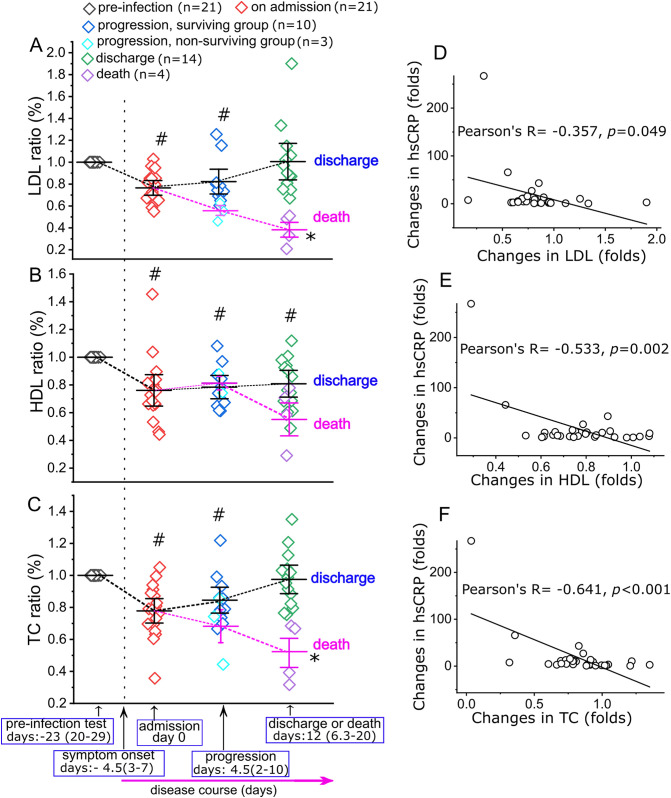
We next analyzed the serum lipid levels (in mmol/L) of the patients before they were infected by SARS-CoV-2 and during their entire courses of the disease. The average timeline of disease course was shown in Fig. 1 . The low-density lipoprotein (LDL) levels in all patients showed significant decreases at the time on admission as compared to the levels prior to infection (pre-infection: 3.5 (3.0–4.4); admission: 2.8 (2.3–3.1), p < 0.01); the LDL levels remained relatively low during the treatment (2.5 (2.3–3.0)) and returned to the levels prior to infection in patients that survived by the time of discharge (3.6 (2.7–4.1)) (Fig. 1, Supplementary Table S1). The high-density lipoprotein (HDL) levels also showed significant decreases at the time on admission as compared to levels prior to infection (pre-infection: 1.4 (1.0–1.8); admission: 1.1 (0.8–1.4), p = 0.03). Unlike LDL, HDL levels remained relatively low during the treatment stage and after recovery (progression: 1.2 (1.0–1.4); discharge: 1.0 (1.0–1.4)) (Fig. 1, Supplementary Table S1). Total cholesterol (TC) levels showed a pattern similar to LDL during the disease course (pre-infection: 5.2 (4.6–6.5); admission: 3.5 (3.3–4.1), p < 0.01; progression: 4.5 (4.0–4.8); discharge: 5.2 (4.4–6.1); Fig. 1, Supplementary Table S1).
Fig. 1.
Ratio changes for LDL (A), HDL (B) and TC (C) in COVID-19 patients during the course of disease. For each data point, the ratio is normalized to the levels of LDL, HDL and TC prior to infection for the same patient. The days listed are the duration (median (IQR)) for each period during the disease course. The date on admission is set as “day 0”. Data is presented as “mean ± 95% confidence interval”. # indicates p < 0.05 as compared to the levels of pre-infection stage, and * indicates p < 0.05 as compared to the levels on admission, by a Mann-Whitney U test. The sample actual values at each stage are listed in Supplementary Table S1. The ratios of hsCRP at each time point are normalized to the levels prior to infection in the same patients. Pearson correlation coefficient analysis show the ratios of hsCRP significantly inversely correlated with the ratios of LDL (D), HDL (E) and TC (F) during the disease course; n = 33 data pairs for each analysis.
The LDL, HDL and TC levels in the patients that did not survive (n = 4) decreased continuously until death (Fig. 1, Supplementary Table S1). Particularly, LDL levels showed an irreversible decrease by the most percentage (~60%) until death (1.1 (0.9–1.2), p = 0.02 versus the levels on admission) (Fig. 1, Supplementary Table S1). Supplementary table S2 showed a timeline of disease course for a non-surviving patient, from hospital admission to death, with continuous and irreversible decreases in LDL, HDL, and TC levels. LDL, HDL and TC levels in normal subjects and non-COVID-19 patients (COPD) control groups did not show significant difference as compared with the levels in COVID-19 patients before their viral infections (Supplementary Table S3). Logistic regression analysis showed increasing odds of lowered LDL levels associated with disease progression (Chi-Square = 7.49; p = 0.006; odds ratio: 4.48, 95% IC: 1.55–12.92, p = 0.006) and in-hospital mortality (Chi-Square = 10.87; p = 0.001; odds ratio: 21.72, 95% IC: 1.40–337.54, p = 0.028). The ratio changes of LDL, HDL and TC inversely correlated with the ratio changes of hsCRP during the disease course (Fig. 1).
4. Discussion
In this study, we report the dyslipidemia in COVID-19 patients and demonstrate that the degrees of decreased LDL levels have high odds associated with severity and mortality of the disease. LDL, together with other risk factors [10] such as older age, high Sequential Organ Failure Assessment, and d-dimer, may aid physicians in identifying patients with poor prognosis at an early phase.
There are several possible explanations for aberrant lipid levels in COVID-19 patients. First, it may result from a liver injury. Although liver functions only exhibit mild abnormalities in protein synthesis (Supplementary Table 1), whether lipid biogenesis has been impaired is yet to be determined. Second, viral infection induced pro-inflammatory cytokines modulate lipid metabolism including oxidation of LDL by reactive oxygen species singling to facilitate LDL clearance [[11], [12], [13]]. A measurement of oxidized LDL in patient's serum will aid in determining this mechanism. Third, COVID-19 patients may have an increased vascular permeability caused by viral-induced inflammation. Exudates have been found evidently in the early phase of COVID-19 lung pathology [8]. Exudative fluids, containing high levels of protein and cholesterol, are caused by inflammation-related vascular permeability [14,15], which may be one possible mechanism underlying our data. The major limitations of this study include a small size of patients and lack of information of specific lipoproteins and oxidized LDLs. Another limitation is lack of a data set with lipids monitoring on a hospitalized non-COVID-19 disease group to further address the specificity between LDL levels and COVID-19 severity. We also posit that the dyslipidemia plays an important role in pathological development of COVID-19, which mechanism needs an urgent investigation. In conclusion, our results demonstrate that LDL decrease is associated with pathological course of COVID-19, which can serve a factor to access the disease progression and mortality.
Declaration of competing interest
The authors do not have any professional and financial affiliations that may be perceived to have biased the presentation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.metabol.2020.154243.
Contributor Information
Wenbin Tan, Email: wenbin.tan@uscmed.sc.edu.
Yongxi Zhang, Email: znact1936@126.com.
Appendix A. Supplementary data
Supplementary methods and data
References
- 1.Xu X., Chen P., Wang J. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The species severe acute respiratory syndrome-related coronavirus: classifying 2019-NCOV and naming it SARS-COV-2. Nat Microbiol. 2020 doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., Yang X.L., Wang X.G. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wan Y., Shang J., Graham R. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol. 2020 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020:2762130. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 7.Wang W., Tang J., Wei F. Updated understanding of the outbreak of 2019 novel coronavirus (2019-NCOV) in Wuhan, China. J Med Virol. 2020;92:441–447. doi: 10.1002/jmv.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian S., Hu W., Niu L. Pulmonary pathology of early phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J Thorac Oncol. 2020 doi: 10.1016/j.jtho.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Z., Shi L., Wang Y. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020 doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou F., Yu T., Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funderburg N.T., Mehta N.N. Lipid abnormalities and inflammation in HIV inflection. Curr HIV/AIDS Rep. 2016;13:218–225. doi: 10.1007/s11904-016-0321-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsoukas M.A., Farr O.M., Mantzoros C.S. Leptin in congenital and HIV-associated lipodystrophy. Metabolism. 2015;64:47–59. doi: 10.1016/j.metabol.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 13.Vu C.N., Ruiz-Esponda R., Yang E. Altered relationship of plasma triglycerides to HDL cholesterol in patients with HIV/HAART-associated dyslipidemia: further evidence for a unique form of metabolic syndrome in HIV patients. Metabolism. 2013;62:1014–1020. doi: 10.1016/j.metabol.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Light R.W., Macgregor M.I., Luchsinger P.C. Pleural effusions: the diagnostic separation of transudates and exudates. Ann Intern Med. 1972;77:507–513. doi: 10.7326/0003-4819-77-4-507. [DOI] [PubMed] [Google Scholar]
- 15.Heffner J.E., Sahn S.A., Brown L.K. Multilevel likelihood ratios for identifying exudative pleural effusions. Chest. 2002;121:1916–1920. doi: 10.1378/chest.121.6.1916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods and data