Abstract
The 2019 novel coronavirus (2019-nCoV/SARS-CoV-2) originally arose as part of a major outbreak of respiratory disease centered on Hubei province, China. It is now a global pandemic and is a major public health concern. Taxonomically, SARS-CoV-2 was shown to be a Betacoronavirus (lineage B) closely related to SARS-CoV and SARS-related bat coronaviruses, and it has been reported to share a common receptor with SARS-CoV (ACE-2). Subsequently, betacoronaviruses from pangolins were identified as close relatives to SARS-CoV-2. Here, we perform structural modeling of the SARS-CoV-2 spike glycoprotein. Our data provide support for the similar receptor utilization between SARS-CoV-2 and SARS-CoV, despite a relatively low amino acid similarity in the receptor binding module. Compared to SARS-CoV and all other coronaviruses in Betacoronavirus lineage B, we identify an extended structural loop containing basic amino acids at the interface of the receptor binding (S1) and fusion (S2) domains. We suggest this loop confers fusion activation and entry properties more in line with betacoronaviruses in lineages A and C, and be a key component in the evolution of SARS-CoV-2 with this structural loop affecting virus stability and transmission.
Keywords: SARS-CoV-2, COVID-19, coronavirus, spike protein, cleavage sites
Abbreviations: SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; ACE2, angiotensin-converting enzyme 2; NTD, N-terminal domain; RBD, receptor binding domain; RBM, receptor binding motif; MHV, murine hepatitis virus; IBV, infectious bronchitis virus
Introduction
Coronaviruses are zoonotic pathogens that are well known to evolve environmentally and infect many mammalian and avian species [1]. These diverse viruses often have effective transmission and immune evasion strategies, especially when outbreaks occur within dense human populations. In the past two decades, coronavirus outbreaks have arisen in human populations around the world, each with unique features but also sharing several similarities. Severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2002 in Guangdong province, China, causing an outbreak that spread to 26 countries, with more than 8000 infections and 774 deaths and a case fatality rate of 9.5% [2]. More recently, the ongoing Middle East respiratory syndrome coronavirus (MERS-CoV) outbreak that originated in 2012 in Saudi Arabia has spread to 27 countries with 2494 infections and 858 deaths, with a case fatality rate of 34.4% [3]. The recent surfacing of the novel coronavirus SARS-CoV-2 (first identified on December 12th, 2019) was initially detected in Wuhan, Hubei Province, China, and has now spread globally via travelers and breached the boundaries of 182 countries/regions [4,5]. On March 11th, 2020, the World Health Organization officially declared a global pandemic. The rapidly evolving situation has prompted most affected countries to impose tight restrictions on border movements and unprecedented statewide lockdown measures. At the time of writing (April 06th, 2020), over 1.2 million cases and approximately 70,000 fatalities have been reported globally [5]. Similar to SARS-CoV and MERS-CoV, SARS-CoV-2 infections in the early stages of the outbreak were observed in family clusters and hospital personnel [4,[6], [7], [8]]. The outbreak occurring during the winter is another commonality between SARS-CoV and SARS-CoV-2 [9]. Currently, sustained community-based spread is occurring, which would make SARS-CoV-2 a community-acquired respiratory coronavirus, along with the other less pathogenic human community-acquired respiratory coronaviruses 229E, OC43, HKU-1, and NL63 [10].
Clinical signs associated with SARS-CoV-2 include pneumonia, fever, dry cough, headache, and dyspnea, which may progress to respiratory failure and death [7,9,11]. The incubation period for SARS-CoV-2 seems to be longer than for SARS-CoV and MERS-CoV, which have a mean incubation time of 5 to 7 days leading to challenges in contact tracing [12]. Preexisting conditions and comorbidities such as hypertension, diabetes, cardiovascular disease, or kidney disease affect the severity of pathogenesis attributed to SARS-CoV and MERS-CoV, and thus far, similar patterns seem to exist with SARS-CoV-2 [7,11]. SARS-CoV and MERS-CoV seem to exhibit deleterious morbidity and mortality on the elderly population (> 60 years of age), with most deaths occurring in this age group, and SARS-CoV-2 is currently portraying a comparable trend [7].
The coronaviruses belong to the Coronaviridae family and the Orthocoronaviridae subfamily, which is divided in four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. SARS-CoV, MERS-CoV, and SARS-CoV-2 are all betacoronaviruses, a genus that includes many viruses that infect humans, bats, and other domesticated and wild animals [13]. Betacoronaviruses have many similarities within the ORF1ab polyprotein and most structural proteins; however, the spike protein and accessory proteins portray significant diversity [14]. MERS-CoV has maintained a stable genome since its emergence in 2012, unlike other coronaviruses that readily evolve and can undergo notable recombination events [15].
Alphacoronaviruses and betacoronaviruses including SARS-CoV, MERS-CoV, and SARS-CoV-2, and other human coronaviruses like HCoV-NL63 are thought to have originated in bats [[14], [15], [16]]. Gammacoronaviruses and deltacoronaviruses are reported to have an avian origin but are known to infect both mammals and avian species [17]. Human infections of bat-origin viruses typically occur through intermediate hosts. For SARS-CoV, these hosts are palm civets (Paguma larvata) and racoon dogs (Nyctereutes procyonoides). For MERS-CoV, the known host is the dromedary camel (Camelus dromedarius) [14,18]. SARS-CoV antibodies were first detected in palm civets and the animal handlers in wet markets [14]. MERS-CoV is thought to have been circulating for at least 30 years within the dromedary camel population based on retrospective antibody testing of serum from 1983 [14]. The source of the SARS-CoV-2 outbreak has been linked to the Huanan seafood wholesale market in Wuhan, where it was initially confirmed that the majority of cases with onset prior to January 1st, 2020, were linked to the market [19]. The market sells many species including seafood, birds, snakes, marmots, and bats [7]. The market was closed on January 1st, 2020, and sampling and decontamination have occurred in order to find the source of the infection. Origination of SARS-CoV-2 from bats has been strongly supported, but the presumed intermediate host remains to be identified. Recent reports have suggested the Malayan pangolins (Manis javanica) as a possible intermediate host for SARS-CoV-2, with putative recombination signals found between pangolin, bat, and human coronavirus sequences [7,9,[20], [21], [22]]. However, the direct precursor sequence to the currently circulating human SARS-CoV-2 and putative intermediate host remain to be identified.
The coronavirus spike protein (S) is the primary determinant of viral tropism and is responsible for receptor binding and membrane fusion. It is a large (approx. 180 kDa) glycoprotein that is present on the viral surface as a prominent trimer, and it is composed of two domains, S1 and S2 [23]. The S1 domain mediates receptor binding and is divided into two sub-domains, with the N-terminal subdomain (NTD) often binding sialic acid and the C-terminal subdomain (also known as C-domain) binding a specific proteinaceous receptor [24]. The receptor for SARS-CoV has been identified as angiotensin-converting enzyme 2 (ACE2), similar to what has been recently identified with SARS-CoV-2 [9,14,25]. HeLa cells transfected to express or not the ACE2 receptor from either human, Chinese horseshoe bat, mouse, civet, and pig were infected with SARS-CoV-2, and it was reported that the virus was able to use all receptors except mouse ACE2 [9]. SARS-CoV-2 was not found to use dipeptidyl peptidase 4 (DPP4), the receptor for MERS-CoV [9,14,25]. Following receptor binding, the S2 domain mediates viral–membrane fusion through the exposure of a highly conserved fusion peptide [26,27]. The fusion peptide is activated through proteolytic cleavage at a site found immediately upstream (S2′), which is common to all coronaviruses. In many (but not all) coronaviruses, additional proteolytic priming occurs at a second site located at the interface of the S1 and S2 domains (S1/S2) [28]. The use of proteases in priming and activation, combined with receptor binding and ionic interactions (e.g. H+ and Ca2+) together control viral stability and transmission, and modulate the conformational changes in the S protein that dictate the viral entry process into host cells [23,26,29]. Specifically, SARS-CoV and MERS-CoV both infect type II pneumocytes in vivo; however, they individually infect ciliated bronchial epithelial cells and non-ciliated bronchial epithelial cells, respectively [14]. SARS-CoV-2 can infect ex vivo with the same range of cell culture lines as SARS-CoV and MERS-CoV, e.g. Vero E6, Huh-7 cells, though primary human airway epithelial cells have been reported to be its preferential cell type [15,25,30]. Overall, how cell tropism of SARS-CoV-2 reflects a balance of receptor binding, endosomal environment, and protease activation, and the specifics of these mechanisms remain to be determined.
The rapid dissemination and sharing of information during the SARS-CoV-2 pandemic has surpassed that of both MERS-CoV and SARS-CoV, where the latter virus was only identified after several months and with a genome available a month later [7]. The SARS-CoV-2 was identified and a genome sequence was available within a month from the initial surfacing of the agent in patients [7]. Initial reports identified that SARS-CoV-2 contains six major open-reading frames in the viral genome and various accessory proteins [9]. The SARS-like (SL) virus BatCoV-RaTG13 (also named Bat-SL-RaTG13) was observed to have a remarkably high degree of genomic sequence identity with that of SARS-CoV-2 at over 96% overall sequence identity, and with two other bat SARS-like viruses (Bat-SL-CoV-ZC45 and Bat-SL-CoV-ZXC21), both having around 88% sequence identity compared with SARS-CoV-2 on a genome-wide level [9]. When SARS-CoV-2 is compared to the clinically relevant human coronaviruses SARS-CoV and MERS-CoV, pairwise percent identities fall to around 79.6% and 50% on a genomic level, respectively [4,9]. The S protein of SARS-CoV-2 was found to be approximately 75% homologous to the SARS-CoV spike [7,9].
In this study, we perform phylogenetic, bioinformatic, and homology structural modeling analyses of SARS-CoV-2 S, in comparison with closely related viruses. We identify a distinct four residue insert (featuring two arginine residues) that maps to the S1/S2 priming loop of SARS-CoV-2, which is missing from all other SARS-CoV-related viruses but present in MERS-CoV S and in many other coronaviruses. We discuss the importance of this extended basic loop for S protein-mediated membrane fusion and its implications for virus transmission.
Results
Comparison of amino acid identity of the spike (S) protein of SARS-CoV-2 with human SARS-CoV
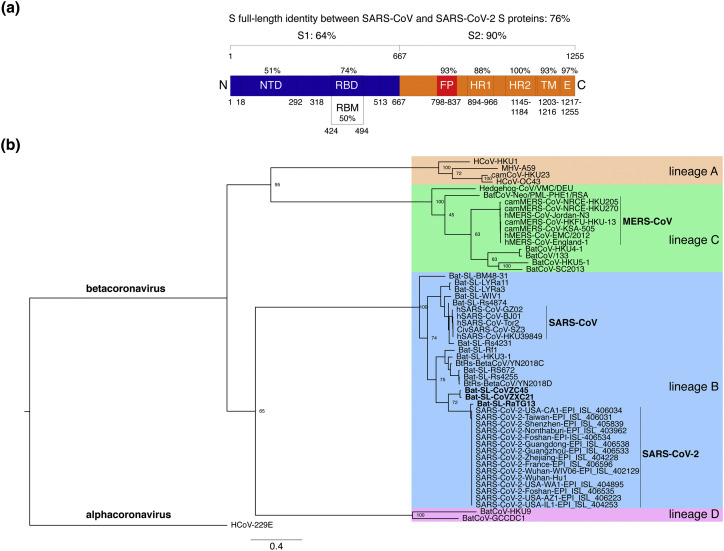
To obtain an initial assessment of shared and/or specific features of the SARS-CoV-2 spike (S) envelope glycoprotein, a protein sequence alignment was performed to compare the sequence of the Wuhan-Hu-1 strain of the novel coronavirus with that of the closely related human SARS-CoV S strain Tor2 sequence (Supplementary Figure 1). The overall percent protein sequence identity found by the alignment was 76% (Figure 1(a)). A breakdown of the functional domains of the S protein, based on the SARS-CoV S sequence, reveals that the S1 receptor-binding domain was less conserved (64% identity) than the S2 fusion domain (90% identity). Within S1, the NTD was found to be less conserved (51% identity) compared to the receptor binding domain (RBD; 74% identity), which is part of the C-terminal subdomain (Figure 1(a)). The relatively high degree of sequence identity for the RBD is consistent with the view that SARS-CoV-2, like SARS-CoV, may use ACE2 as its host cell receptor [9,25,31]. Interestingly, when the more defined receptor binding motif (RBM) was analyzed (i.e. the region of SARS-CoV S containing residues that were shown to directly contact the ACE2 receptor) the identity between the two sequences drops to 50% (Figure 1(a)), in this case hinting at possible differences in binding residues involved in the interaction with the receptor and/or binding affinities [[31], [32], [33]]. As expected, within the well-conserved S2 domain, subdomain identities were high for the fusion peptide region (FP, 93% identity), high for the heptad-repeat 1 region (HR1, 88% identity), identical for HR2 (100% identity) and high for both the transmembrane and the C-terminal endodomain (TM, 93% identity; E, 97% identity) (Figure 1(a)).
Figure 1.
Comparative analyses of SARS-CoV-2 S protein sequence. (a) Protein sequence identities between SARS-CoV-2 S with SARS-CoV S. The S protein sequences were aligned using MAFFT and the sequence identities obtained for the full-length and domains/subdomains are shown on the S protein diagram. Amino acid numbering and delineations of domains and subdomains are based on the SARS-CoV S protein. (b) Phylogenetic analysis of SARS-CoV-2 S protein. The S protein sequence of 15 isolates of SARS-CoV-2 was aligned using MAFFT with representatives of all four Betacoronavirus lineages. A Maximum-Likelihood tree was generated based on the alignment. The tree was rooted using the alphacoronavirus HCoV-229E S sequence. Highly pathogenic betacoronaviruses SARS-CoV and MERS-CoV are highlighted (bold font) along with bat SARS-like coronaviruses closely related to SARS-CoV-2 (Bat-SL-CoVZC45, Bat-SL-CoVZXC21, and Bat-SL-RaTG13). Number at nodes indicates bootstrap support (100 replicates), and the scale bar indicates the estimated number of substitutions per site. Accession numbers of sequences used in the analyses are found in the Materials and Methods section.
Phylogenetic analysis of SARS-CoV-2 S with other betacoronaviruses
Early phylogenetic studies on SARS-CoV-2 genomic sequences revealed that it clustered closely with sequences originating from SARS-like sequences from bats, within lineage B of the Betacoronavirus genus. Lineage A groups prototypical coronaviruses such as murine hepatitis virus (MHV) and human coronaviruses HCoV-HKU1 and HCoV-OC43. The other highly pathogenic coronavirus, MERS-CoV is found within lineage C, along with related camel-derived MERS-CoV. Lineage C also groups viruses from bats and other mammals such as hedgehogs. Lineage D contains viral species infecting bats. To gain a better understanding of both shared and specific features of SARS-CoV-2 S protein, a phylogenetic analysis centered on S protein sequences of representatives of the four Betacoronavirus lineages was carried out (Figure 1(b)). Fifteen sequences of SARS-CoV-2 S sequences obtained from NCBI and GISAID from China and various export locations worldwide were analyzed along with representative members of lineages A–D betacoronaviruses. The analysis confirmed that all SARS-CoV-2 S sequences clustered very closely with bat SARS-like sequences, with the closest matching sequence corresponding to a bat coronavirus (bat-CoV) strain named Bat-SL-RaTG13. Other closely related sequences found were from Bat-SL-CoVZC45 and Bat-SL-CoVZXC21. The sub-clade that groups SARS-CoV-2, Bat-SL-RaTG13, Bat-SL-CoVZC45, and Bat-SL-CoVZXC21 is distinct from the one grouping human and civet SARS-CoV along with other related bat SARS-like viruses, such as Bat-SL-LYRa3.
While bat coronaviruses are the established reservoir for SARS-CoV-2, the presumed intermediate host remains to be determined. Based on the high degree of sequence identity in the RBD of sequences found in Malayan pangolins (M. javanica), it was proposed that these mammals could be intermediate hosts [[20], [21], [22]]. Subsequent whole-genome analysis revealed 85.5%–92.% identity to SARS-CoV-2, which is less than what is observed for Bat-SL-RaTG13 (96.3%) [34]. To explore the relationship of pangolin CoVs to SARS-CoV-2, we performed a phylogenetic analysis of the spike gene of pangolin CoVs isolated from animals smuggled into China in 2017 (Guangxi Province, GX) and 2019 (Guangdong Province, GD), compared to SARS-CoV-2, BatCoV-RaTG13, and other betacoronaviruses (Figure 2 ). Pairwise comparison between SARS-CoV-2 S protein and that of BatCoV-RaTG13 and representative sequences from Guangxi pangolin (2017, abbreviated GX here) and Guangdong pangolin (2019, abbreviated GD) confirm that overall BatCoV-RaTG13 had the highest identity: 97% overall, 96% and 100% for S1 and S2, respectively (Figure 2(a)). The analysis reveals that pangolin S protein sequences are more divergent overall (92% identity for GX and 89% identity for GD), with most of the divergence concentrating in the S1 domain. Notably, while the NTD domain of both GX and GD pangolin sequences fell to 88% and 67% identity, respectively, the RBD domain of the GD domain was confirmed to be remarkably well conserved compared to SARS-CoV-2 (97% identity compared to 87% identity for GX pangolin and 89% for BatCoV-RaTG13). These observations are in line with previously reported putative recombination events occurring between pangolin, bat, and human betacoronavirus sequences [22]. Phylogenetically, pangolin spike sequences from 2017 (GX) form a subclade that groups with RaTG13 (Figure 2(b)). Interestingly, the 2019 pangolin (GD) sequence appears to branch off early (i.e. it could represent an S protein sequence that diverged earlier) and forms its own subclade, distinct from the RaTG13/2017pangolin and SARS-CoV-2/environmental subclades.
Figure 2.
SARS-CoV-2 S protein sequence relatedness with betacoronavirus lineage B sequences from bats, pangolins, and environmental samples. (a) S protein sequences were aligned using MAFFT and a pairwise analysis of the protein sequence identities comparing the S protein and its subdomains of SARS-CoV-2 with either BatCoV-RaTG13 (blue % numbers), 2017 pangolin coronavirus from Guangxi (GX) Province (red % numbers, BetaCoV/pangolin/Guangxi/P4L/2017 | EPI_ISL_410538), or 2019 pangolin coronavirus from Guangdong (GD) Province (orange % numbers, BetaCoV/pangolin/Guandong/1/2019 | EPI_ISL_410721) are shown. Amino acid numbering based on SARS-CoV-2 sequence. (b) A Maximum-Likelihood tree was generated based on the above-mentioned alignment. The tree was rooted using the lineage B betacoronavirus BatCoV-HKU9 S sequence. Number at nodes indicates bootstrap support (100 replicates), and the scale bar indicates the estimated number of substitutions per site. Accession numbers of sequences used in the analyses are found in the Materials and Methods section. Blue asterisk highlights Bat-SL-RaTG13, and white triangles highlight SARS-CoV-2 from environmental samples.
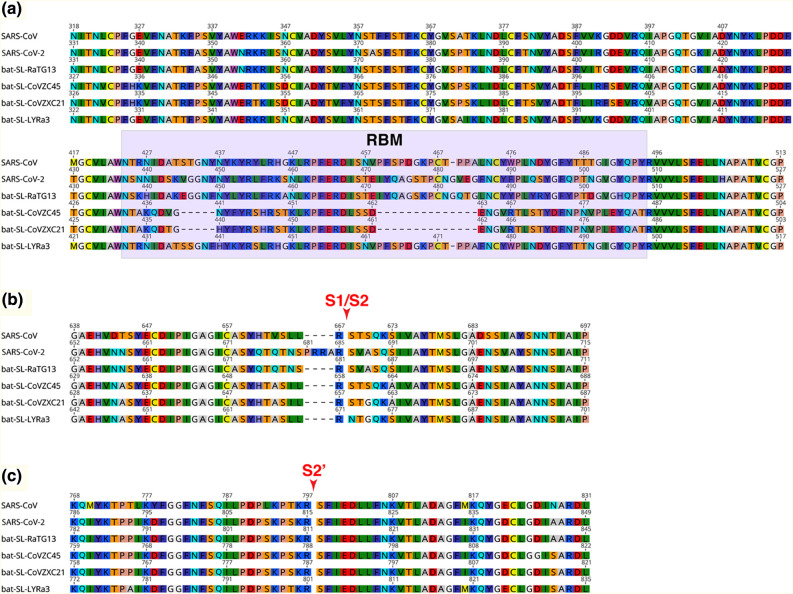
Alignments of RBD and cleavage sites of SARS-CoV-2 and other bat-CoVs
An S protein sequence alignment focusing on the RBD region of SARS-CoV-2, SARS-CoV, and bat-SARS-related viruses reveals that the N-terminal half of the RBD is relatively well conserved, whereas the C-terminal half, which contains the RBM, exhibits more variation (Figure 3(a)). Notably, Bat-SL-CoV-ZC45 and Bat-SL-CoV-ZXC21 both have two deletions of 5 and 14 residues within the RBD. The composition of residues found at the two known coronavirus S cleavage sites was performed using alignment data (Figure 3(b) and (c)). The region around arginine 667 (R667) of SARS-CoV S, the S1/S2 cleavage site, aligned well with SARS-CoV-2 and the bat SARS-related sequences [56]. Notably, an arginine at the position corresponding to SARS-CoV R667 is conserved for the other five sequences analyzed. The alignment shows that SARS-CoV-2 contains a four amino acid insertion 681PRRA684 that is not found in any other sequences analyzed, including the closely related bat-SL-RaTG13 (Figure 3(b)). Together with the conserved R685 amino acid found in SARS-CoV-2 at the putative S1/S2 cleavage site, the insertion introduces a stretch of three basic arginine residues that could potentially be recognized by members of the pro-protein convertase family of proteases [35,36]. This insertion was conserved for all 15 SARS-CoV-2 sequences analyzed (Supplementary Figure 2). Within the Betacoronavirus genus, the presence of a basic stretch of residues at the S1/S2 site is found for a number of species from lineages A (HCoV-HKU1, MHV, HCoV-OC43) and C (MERS-CoV, BatCoV-HKU5). The four-amino-acid insertion feature appears unique among lineage B viruses, as all other species analyzed in the extended alignment, none contained the stretch of basic residues identified in SARS-CoV-2 S (Supplementary Figure 2). As expected from previous analyses, the S2′ cleavage site, located immediately upstream of the fusion peptide and corresponding to the residue position R797 in the case of SARS-CoV, was strictly conserved for SARS-CoV-2 and closely related bat SARS-related sequences (Figure 3(c)). Of note, the leucine (L) residue found at position 792 of the SARS-CoV sequence is substituted to serine (S) residue for SARS-CoV-2 S as well as the bat SARS-related sequences. The fusion peptide sequence was found to be well conserved for all sequences analyzed.
Figure 3.
Sequence alignments of S protein regions of SARS-CoV-2 with closely related species. S protein sequences from SARS-CoV-2, SARS-CoV, and closely related bat coronaviruses, bat-SL-RaTG13, bat-SL-CoVZC45, bat-SL-CoVZXC21, and bat-SL-LYRa3 were aligned. The regions corresponding to the RBD (a), the S1/S2 cleavage site (red arrow (b)), and S2′ cleavage site (red arrow (c)) are shown. Accession numbers of sequences used in the analyses are found in the Materials and Methods section.
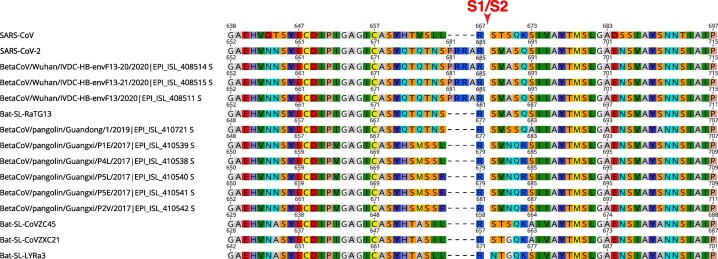
Notably, protein alignment analyses show that pangolin sequences (as for other viruses in betacoronaviruses lineage B) do not show the presence of the predicted proteolytically sensitive S1/S2 fusion activation site present in SARS-CoV-2 (Figure 4 ). Only environmental samples taken from the Huanan Seafood Market appear to also harbor the S1/S2 insert. Interestingly, the 2019 pangolin sequence shares the same motif immediately upstream of the S1/S2 insert found in SARS-CoV-2 S, 675QTQTNS680 (SARS-CoV-2 numbering). This is also shared with BatCoV-RaTG13, but not with the other pangolin sequences from 2017, which harbor a distinct motif, 673HSMSSL/F678 (BetaCoV/pangolin/Guangxi/P4L/2017 | EPI_ISL_410538 numbering).
Figure 4.
Amino acid composition of the S1/S2 region of SARS-CoV-2 and related sequences from bats, pangolins, and environmental samples. S protein sequences from SARS-CoV-2, SARS-CoV, and closely related environmental, pangolin, and bat coronaviruses were aligned using MAFFT. The region corresponding to the S1/S2 cleavage site (red arrow) is shown. Accession numbers of sequences used in the analyses are found in the Materials and Methods section.
SARS-CoV-2 S protein homology structure modeling
To gain a deeper understanding of common and possibly distinguishing structural features found in SARS-CoV-2 S protein, homology modeling was undertaken. The analysis of modeled proteins provides a powerful tool to identify predicted structural characteristics, which can translate into structure–function changes in the studied protein. Our laboratory has previously taken advantage of these tools, for structure–function studies of other CoVs S proteins [23,37,38]. To perform the modeling, it is first necessary to identify a suitable protein structure to be used as template, which will determine the accuracy of the predicted model. The S protein structure of several CoVs including the following have been reported previously: Alphacoronavirus: HCoV-NL63 and feline coronavirus UU4 (FCoV-UU4); Betacoronavirus: HCoV-HKU1, MHV, SARS-CoV, and MERS-CoV; Gammacoronavirus: infectious bronchitis virus (IBV); and Deltacoronavirus: porcine deltacoronavirus (PDCoV) [[39], [40], [41], [42], [43], [44], [45]]. Considering that genome and S protein alignments have showed that the SARS-CoV-2 belongs to the Betacoronavirus genus, we focused our analysis on the S structures from viruses belonging to this genus. To select the template structure, the S protein amino acid sequences from four representative betacoronaviruses (HCoV-HKU1, MHV, SARS-CoV, and MERS-CoV) were aligned and the solved S protein structures were compared to determine their amino acid identity and the overall structural organization similarities among these proteins (Supplementary Figure 3A). We observed an average of ~ 30% identity among the four viral S proteins at the amino acid level, with the exception of HCoV-HKU1 and MHV, which share an amino acid identity of 59% at the S protein (Supplementary Figure 3A). Despite the differences at the amino acid level, the overall structure of the four Betacoronavirus S proteins showed a similar folding pattern (Supplementary Figure 3B), and major differences can only be spotted at specific sections of the functional domains where flexible loops are abundant (e.g. RBD and cleavage sites). Considering this, we used Modeller (v. 9.23, University of California) to build a first set of models for the SARS-CoV-2 S protein based on each of the above-mentioned structures (Supplementary Figure 4). Interestingly, we found no major differences at the secondary structures among the SARS-CoV-2 S protein-predicted models depending on the S structure that was used as template for the modeling construction. However, extended flexible loops at the RBD and/or clashes between S monomers at the S2 domain level were observed in the SARS-CoV-2 S models based on HCoV-HKU1, MHV, and MERS-CoV (Supplementary Figure 4, first three panels). In contrast, the predicted SARS-CoV-2 S model based on the SARS-CoV S structure displayed a much better organized folding and no major clashes were observed between the S monomers (Supplementary Figure 4, last panel).
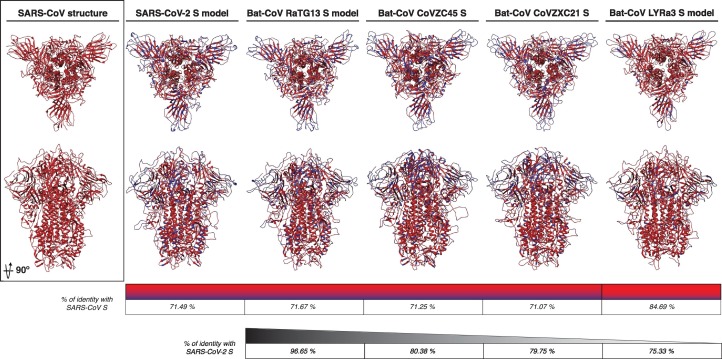
As we described previously, the identity between SARS-CoV-2 and SARS-CoV at the S protein amino acid level was 76%, and phylogenetic analyses grouped SARS-CoV-2 in the lineage B of the Betacoronavirus genus, closely related to SARS-CoV, as well as to other CoVs originating in bats (Figure 1(b)). These two considerations, in addition to our preliminary modeling results, suggested SARS-CoV S as the most suitable template for modeling the SARS-CoV-2 S protein. Taking an alternative approach, the S protein sequence of SARS-CoV-2 was submitted to two structure homology modeling servers Phyre 2 and RaptorX [46,47]. For both cases, the structural models with highest homology scores were based on the SARS-CoV S template structure (PDB ID 5X58, data not shown), confirming the choice of using SARS-CoV S as template for generating structural models of SARS-CoV-2.
To better compare the predicted structural characteristics of the SARS-CoV-2, we also performed homology modeling of four S proteins from Bat-CoVs belonging to lineage B in our phylogenetic analysis, which are closely related to SARS-CoV-2. The modeled S proteins from the Bat-CoVs RaTG13, CoVZC45, CoVZXC21, and LYRa3 were compared to the predicted structure of SARS-CoV-2 S and to the template structure of SARS-CoV (Figure 5 ). The amino acid homology of the modeled S proteins in comparison to the template SARS-CoV S was ~ 71% for all the Bat-CoV S, with the exception of the LYRa3 S, which shares a homology of 84.7% with the template S. Overall, all the modeled S proteins shared a similar folding pattern in comparison to SARS-CoV S and both S1 and S2 domains showed a uniform organization (Figure 5). As expected, differences were mostly observed at the flexible loops forming the “head” of the S1 domain, especially at the NTD region (RBD region), where most of the amino acid variation was observed (Figures 3(a) and 5). The S protein amino acid identity among the Bat-CoV (including SARS-CoV-2) ranged between 75.3% and 96.7%, with LYRa3 and RaTG13 S proteins having the lowest and highest identity to SARS-CoV-2, respectively. Despite amino acid variability, no major changes in the secondary structures and the overall folding of the proteins were observed among the modeled S structures of these viruses, suggesting a conserved organization for all the S proteins of the lineage B including SARS-CoV-2. Nevertheless, differences at the flexible loops in both domains were observed, and their impact in the SARS-CoV-2 S protein function must be further studied.
Figure 5.
SARS-CoV-2 and bat-CoVs S protein models. The modeled S protein of SARS-CoV-2, RaTG13, CoVZC45, CoVCZXC21, and LYRa3 is compared to SARS-CoV S structure. The amino acid homology between the modeled proteins and SARS-CoV S is noted in red and amino acid differences in blue in both models and identity scale. The S amino acid identity between SARS-CoV-2 and bat-CoVs is also noted (black identity scale).
Structural modeling of the predicted RBM SARS-CoV-2
It was recently reported that the SARS-CoV-2 binds ACE2 as a receptor to infect target cells [9,25,33,48]. This finding appears to agree with previous reports describing the ability of bat-CoVs to successfully bind and use ACE2 as a cellular receptor for infection [[49], [50], [51]]. This conservation in the receptor usage among SARS-CoV and SARS-like bat-CoVs, contrasts with the high variability that are observed at the amino acid sequence of the RBM (Figure 3(a)). Considering this high variability, we compared the predicted RBM structure of the SARS-CoV-2 and bat-CoVs to the one of SARS-CoV. Interestingly, despite the variability, the modeled SARS-CoV-2-predicted RBM displayed a similar organization to SARS-CoV (Figure 6 , top panel). This was also observed in the RaTG3 and LYRa3-predicted RBM structures (Figure 6, middle left and bottom right panels), suggesting that the RBM organization is well conserved among these viruses. In contrast, the predicted RBM of the CoVZC45 and CoVZXC21 viruses showed a different folding at this region in comparison to SARS-CoV (Figure 6, middle right and bottom left panels). These two last viruses showed a 5- and a 14-amino-acid deletion, respectively, in the RBM sequence (Figure 3(a)), which can explain the differential folding in the modeled proteins.
Figure 6.
SARS-CoV-2 and bat-CoVs modeled RBM. Surface view of SARS-CoV S structure and SARS-CoV-2, RaTG13, CoVZC45, CoVCZXC21, and LYRa3 S models. SARS-CoV RBM (red) and flanking residues (yellow) are noted. RBM in the modeled structures is also noted according to their amino acid homology (red) and differences (blue) to SARS-CoV.
Structural modeling of SARS-CoV-2 S reveals a proteolytically sensitive loop
The alignment in Figure 3(b) shows a four-amino-acid insertion 681PRRA684, as well as a conserved arginine corresponding to R685 at the S1/S2 site of the SARS-CoV-2. This insertion, which appears to be unique among lineage B betacoronaviruses, suggests a differential mechanism of activation for the SARS-CoV-2 compared to other SARS-CoV and SARS-like BatCoV. At the structural level, the S1/S2 site has been shown to be difficult to solve for most CoVs structures, resulting in either incomplete structures (missing the complete S1/S2 site) or structures with an altered (i.e. mutated) S1/S2 site [42,44,45]. Solving the structure of the S1/S2 site was also found to be an issue in the SARS-CoV S structure we used for our modeling analyses. We have previously shown that the S1/S2 site can be modeled in other CoV S proteins, and it appears to be organized as a flexible exposed loop that extends from the S structure and suggest it could be easily accessible for proteolytic activation [37].
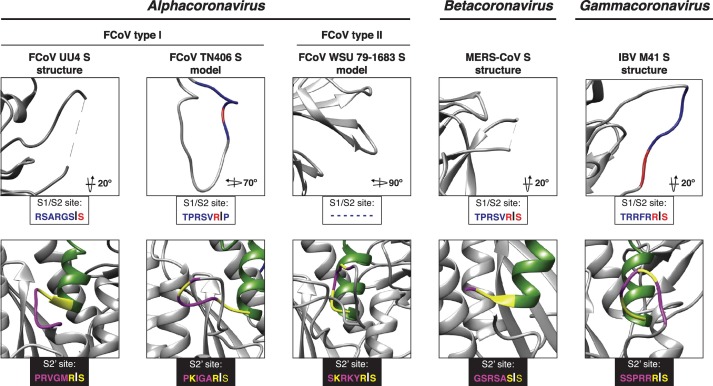
To better study the S1/S2 site structural organization, we modeled the SARS-CoV S protein based on the S structure of MHV (S1/S2 site mutated in the structure), and MERS-CoV and SARS-CoV (S1/S2 site missing in the structure) to see if the predicted structure of the S1/S2 site was similar despite the template structure. We observed no differences in the modeled SARS-CoV S protein at the S1/S2 site, predicting an exposed flexible loop in all the three models (data not shown). Based on this, we proceeded to compare the S1/S2 site, as well as other major functional elements of the S2 domain (i.e. S2′ site and fusion peptide), in the predicted structure in our SARS-CoV, SARS-CoV-2, and Bat-CoV S models (Figure 7 ). Remarkably, two features appear to exhibit distinctive characteristics in the SARS-CoV-2 S model: the fusion peptide, which is predicted to be organized in a more compact conformation for SARS-CoV-2 S than in SARS-CoV S (Figure 7, surface models), and the region corresponding to the S1/S2 cleavage site, which contains R667 in the case of SARS-CoV (Figure 7, S1/S2 alignment box and ribbon models). For SARS-CoV and the bat-CoV proteins, the S1/S2 site forms a short loop that appears flanking closely to the side of the trimeric structure. In the case of SARS-CoV-2 S, the S1/S2 site is predicted to form an extended loop that protrudes to the exterior of the trimer (Figure 7). This feature suggests that the S1/S2 loop in SARS-CoV-2 S could be more exposed for proteolytic processing by host cell proteases. As mentioned before, solving the structure of the S1/S2 site appears to present difficulties for most of the reported CoV S structures (Figure 8 , top panel). However, the exposed loop feature has been demonstrated in both modeled and cryo-EM CoV S structures with similar amino acid sequences at the S1/S2 site (i.e. FCoV and IBV, respectively) (Figure 8, top panel). Interestingly, FCoV viruses do not always display a S1/S2 site (Figure 8, top panel), which results in distinct cell entry mechanisms. We also performed an analysis of the S2′ site of the SARS-CoV-2 in comparison to SARS-CoV and bat-CoV S proteins. As expected, differences in the modeled S2′ site structure were not predicted in any of the studied spikes (Figure 7, S2′ ribbon models). This agrees with the fact that the S2′ site appears to be conserved in the studied sequences (Figure 7, S2′ alignment box) and as we described previously, the SARS-CoV functional R797 residue at the cleavage position 1 (P1) as well as the serine at position 798 (cleavage position P1′) are conserved in the SARS-CoV-2 and among the compared bat-CoVs. This feature also appears to be conserved in other CoVs (Figure 8, bottom panel); however, mutations in the residues immediately upstream of the SARS-CoV R797 residue (or equivalent in each virus) have been shown to result in changes in the proteolytical requirements in other CoVs [28]. We observed mutations L to S and T to S, which are located upstream of the P1 arginine at positions P3 and P6 in the S2′ site of the SARS-CoV-2 in comparison to SARS-CoV. These mutations are not predicted to alter the structure of the S2′ in the SARS-CoV-2 (Figure 7, S2′ ribbon models).
Figure 7.
SARS-CoV-2 S1/S2 and S2′ activation sites. The S1/S2 and S2′ activation sites of SARS-CoV and SARSCoV-2 S models are shown in surface and ribbon views. S1/S2 and S2′ sites of bat-CoVs are shown in ribbon view. Amino acid homology to SARS-CoV is noted as follows: S1/S2 site: homology (red) and differences (blue); S2′ site: homology (yellow) and differences (magenta). Amino acid alignments of the S1/S2 and S2′ sites are shown, and homology is also noted.
Figure 8.
CoVs S1/S2 and S2′ site. The S1/S2 and S2′ activation sites of FCoV, MERS-CoV and IBV. S models are shown ribbon views. Amino acid homology to SARS-CoV is noted as follows: S1/S2 site: homology (red) and differences (blue); S2′ site: homology (yellow) and differences (magenta). Amino acid sequences of the S1/S2 and S2′ sites are shown.
Discussion
The current COVID-19 pandemic caused by SARS-CoV-2 is evidence of the potential of coronaviruses to continuously evolve in wild reservoirs and jump to new species. Our study aims to contribute to our understanding of the SARS-CoV-2 from a phylogenetic and structural point of view, focusing on the functional and the proteolytically sensitive sites of the S protein. Using phylogenetic analysis, we showed that the SARS-CoV-2 S protein is closely related to other SARS-like viruses originating in bats (Figure 1(b)), which agrees with early similar reports [34]. The identity of the S protein of BatCoV-RaTG13 strain of bat coronavirus was shown to be 96.65%, suggesting this virus as the closest relative to SARS-CoV-2. While the origin of the novel coronavirus appears to be in bat reservoirs, there is still no definitive evidence of the possible intermediate host that could transmit the virus to humans. Recent reports have suggested the Malayan pangolins as an intermediate host for the SARS-CoV-2. In our analysis, we found that the pangolin spike sequences grouped in a subclade branching from RaTG13. We additionally observed that the 2019 (Guangdong) pangolin sequence appeared to branch off early in a distinct subclade from RaTG13. Based on these findings, we hypothesize that despite of having a common origin in bats, the phylogenetic relationship between pangolin CoVs and SARS-CoV-2 is not sufficient to support the claim that pangolins harbor the direct precursor to the currently circulating human SARS-CoV-2. In fact, our analysis suggests that both humans and pangolins could be considered final hosts of their respective coronavirus.
In this study, we show the presence of a distinct insert that maps to the S1/S2 priming loop of the SARS-CoV-2 spike protein and is not shared with SARS-CoV or any SARS-related viruses in Betacoronavirus lineage B. During the preparation of this manuscript, the cryo-electron microscopy structures of SARS-CoV-2 S have recently been determined [33,52,53]. These studies have revealed in detail the structure of the SARS-CoV-2 spike RBD and how it contacts the ACE2 host cell receptor with notable differences compared to SARS-CoV [54]. It has been described that the SARS-CoV RBM “down” conformation packs more closely to the NTD of the S protein [33]. In the same report, the SARS-CoV-2 RBD was shown to be angled to the center of the trimmer in its down conformation, which differs to the SARS-CoV RBM structure. Interestingly, we observed in our models that the RBD is predicted to pack similarly to SARS-CoV and the RBM is also predicted to organize as a flexible loop with similar structure despite the lower amino acid identity between these two proteins (Figure 6). In a more recent report, it has been shown that ACE2-binding mode of both SARS-CoV and SARS-CoV-2 RBDs is nearly identical, which supports the claim that the flexibility in the RBM is key to compensate the amino acid differences between the two CoVs proteins and agrees with our predicted models [54].
One of the common difficulties of the CoVs S protein cryo-EM studies is the difficulty to solve proteolytically sensitive regions in the protein [39,55]. Since the S1/S2 region is proteolytically sensitive, it is common to introduce mutations in this site to prevent proteolytic priming and to allow efficient heterologous expression and purification [42]. This has resulted in a difficulty to solve the S1/S2 region in most of the available CoV S protein structures, with a few exceptions for the S proteins of viruses belonging to the Alphacoronavirus and Gammacoronavirus genera [40,44]. Considering that this region is proteolytically sensitive and has been shown to play a major role in the S protein function in other CoVs, the use of in silico modeling tools has become a useful alternative to study this region in the context of the structural organization of the protein [37,38]. The SARS-CoV-2 S protein is not an exception to this issue and the recently reported structures do not allow resolution of the S1/S2 priming loop and/or have the loop mutated. We used the structural modeling approach to better understand the organization of this region, which is not only suggested to be functionally active, but has been reported as one of the major differences between the SARS-CoV-2 and its closest relative RaTG13 [25,33]. We observed that the SARS-CoV-2 S1/S2 site is predicted to be organized as an exposed flexible loop, suggesting that the site is easily available for protease cleavage and suggesting a major role in SARS-CoV-2 S function (Figure 7).
The significance of the SARS-CoV-2 spike protein S1/S2 priming loop is yet to be explored experimentally, but we consider it may fundamentally change the entry pathway of this virus compared to other known viruses in Betacoronavirus lineage B. The presence of the extended S1/S2 priming loop containing paired basic residues predicts that SARS-CoV-2 S would most likely be cleaved by Golgi-resident proprotein convertases such as furin during virus assembly and delivery to the cell surface. Indeed, analysis of Western blots of VSV-pseudoparticles containing SARS-CoV-2 S have shown the presence of cleaved S, in contrast to pseudoparticles containing SARS-CoV S [25]. In the case of MERS-CoV, but not SARS-CoV, it is known that priming of S by “pre-cleavage” occurs at the S1/S2 site, giving SARS-CoV-2 cleavage activation properties more in line with MERS-CoV than SARS-CoV [25,[56], [57], [58]]. The extended structural loop may also allow enhanced priming by trypsin-like proteases (TTSPs) or even cathepsins. SARS-CoV-2 is currently believed to be highly SARS-CoV-like with respect to its receptor binding, and the modeling studies reported here are broadly in line with this finding despite the relatively low amino acid identity in the RBM. However, it is important to remember that changes in protease usage may allow coronaviruses to undergo receptor-independent entry (virus–cell fusion) as well as affect syncytia formation (cell–cell fusion) and tissue pathology [[59], [60], [61]].
Our study provides a structural context to the S1/S2 insert, which has also been reported by others [52,62]. The presence of a distinct insert containing paired basic residues in the S1/S2 priming loop is common in many coronaviruses in Betacoronavirus lineage C (e.g. MERS-CoV), as well as in lineage A (e.g. mouse hepatitis virus, MHV) and lineage D, and is universally found in Gammacoronavirus S (e.g. IBV) [40]. It is noticeably absent in most Alphacoronaviruses, with the clear exception of type I canine and feline coronaviruses [28,37]. One feature of the distinct insert for of SARS-CoV-2 that warrants attention relates to potential changes as the virus evolves. An equivalent loop is present in influenza HA (in this case adjacent to the fusion peptide), and insertions of basic residues into the loop are a primary marker of conversion from low pathogenicity to highly pathogenic avian influenza virus (e.g. H5N1) [63]. In coronaviruses, such loop modifications are known to affect MHV pathogenesis and to modulate neurovirulence and neuroinvasiveness of HCoV-OC43 [64,65]. The FCoV is another example where S1/S2 loop modifications appear to lead directly to changes in viral pathogenesis [37,66,67]. In the case of FCoV, the equivalent proteolytically sensitive structural loop is within a hypervariable region of the spike gene, suggesting that this region of spike is a significant driver of virus evolution [67].
At present, SARS-CoV-2 is behaving in a distinct manner compared to SARS-CoV. We believe our findings are of special importance considering that the available data indicates ACE2 as a suitable cellular receptor for SARS-CoV-2 entry [48,68]. In our modeling analysis, we observed that the RBM of the SARS-CoV-2 predicted a similar organization as SARS-CoV and that deletions at this RBM region in other bat-CoVs are reported to not impact its ability to bind ACE2 [[49], [50], [51]]. This suggests that instead of receptor binding, the S1/S2 loop is a distinctive feature relevant to SARS-CoV-2 pathogenesis and marks a unique similarity to MERS-CoV. We would predict that the distinct insert in SARS-CoV-2 S would give the virus biological properties more in line with MERS-CoV and not SARS-CoV, especially with regard to its cell entry pathway. However, it may also impact virus spread and transmission. While many epidemiological features of SARS-CoV-2 still need to be resolved, there are many features of transmission that appear to align more with MERS-CoV than SARS-CoV. One component of transmission is the reproductive number (R0), which is currently thought to be approximately 2.0–3.0 for SARS-CoV-2, broadly in line with than for SARS-CoV, and while MERS-CoV has a low R0 in humans (< 1), it is high in camels and in outbreak situations (> 3) [[69], [70], [71], [72], [73]]. Another study has reported that the serial interval (time from the disease onset in a patient to the onset of the disease in secondary case) can be estimated to be below 4 days, suggesting that transmission can occur before the onset of clinical signs [74]. These two parameters highlight the high transmissibility of the SARS-CoV-2. One notable feature of the S protein S1/S2 cleavage site was first observed during the purification of the MHV S protein for structural analysis [42]. MHV with an intact cleavage loop was unstable when expressed, and so we consider that the S1/S2 loop controls virus stability, likely via access to the down-stream S2′ site that regulates fusion peptide exposure and activity. As such, it will interesting to monitor the effects of S1/S2 loop insertions and proteolytic cleavability in the context of virus transmission, in addition to virus entry, pathogenesis, and evolution.
Materials and Methods
Sequences
Amino acid sequences of the S protein used in the phylogenetic analysis were obtained from GISAID and NCBI GenBank. GISAID accession numbers (in parenthesis) from which whole-genome sequences were obtained were as follows: SARS-CoV-2-Foshan-EPI_ISL_406535 (EPI_ISL_406535), SARS-CoV-2-Foshan-EPI-ISL-406534 (EPI-ISL-406534), SARS-CoV-2-France-EPI_ISL_406596 (EPI_ISL_406596), SARS-CoV-2-Guangdong-EPI_ISL_406538 (EPI_ISL_406538), SARS-CoV-2-Guangzhou-EPI_ISL_406533 (EPI_ISL_406533), SARS-CoV-2-Nonthaburi-EPI_ISL_403962 (EPI_ISL_403962), SARS-CoV-2-Shenzhen-EPI_ISL_405839 (EPI_ISL_405839), SARS-CoV-2-Taiwan-EPI_ISL_406031 (EPI_ISL_406031), SARS-CoV-2-USA-AZ1-ISL_406223 (EPI_ISL_406223), SARS-CoV-2-USA-CA1-ISL_406034 (EPI_ISL_406034), SARS-CoV-2-USA-IL1-EPI_ISL_404253 (EPI_ISL_404253), SARS-CoV-2-USA-WA1-EPI_ISL_404895 (EPI_ISL_404895), SARS-CoV-2-Wuhan-WIV06-EPI_ISL_402129 (EPI_ISL_402129), SARS-CoV-2-Zhejiang-EPI_ISL_404228 (EPI_ISL_404228), Bat-SL-RaTG13 (EPI_ISL_402131), BetaCoV/pangolin/Guandong/1/2019 | EPI_ISL_410721 (EPI_ISL_410721), BetaCoV/pangolin/Guangxi/P5E/2017 | EPI_ISL_410541 (EPI_ISL_410541), BetaCoV/pangolin/Guangxi/P2V/2017 | EPI_ISL_410542 (EPI_ISL_410542), BetaCoV/pangolin/Guangxi/P5L/2017 | EPI_ISL_410540 (EPI_ISL_410540), BetaCoV/pangolin/Guangxi/P4L/2017 | EPI_ISL_410538 (EPI_ISL_410538), BetaCoV/pangolin/Guangxi/P1E/2017 | EPI_ISL_410539 (EPI_ISL_410539), BetaCoV/Wuhan/IVDC-HB-envF13–20/2020 | EPI_ISL_408514 (EPI_ISL_408514), BetaCoV/Wuhan/IVDC-HB-envF13/2020 | EPI_ISL_408511 (EPI_ISL_408511), and BetaCoV/Wuhan/IVDC-HB-envF13–21/2020 | EPI_ISL_408515 (EPI_ISL_408515). GenBank accession numbers (in parenthesis) from which whole-genome or S gene sequences were obtained were as follows: SARS-CoV-2-Wuhan-Hu1 (MN908947.3), Bat-SL-CoVZC45 (MG772933.1), Bat-SL-CoVZXC21 (MG772934.1), Bat-SL-LYRa3 (KF569997.1), BatCoV/133 (DQ648794.1), BatCoV-GCCDC1 (NC_030886.1), BatCoV-HKU4–1 (EF065505.1), BatCoV-HKU5–1 (EF065509.1), BatCoV-HKU9 (NC_009021.1), BatCoV-Neo/PML-PHE1/RSA (KC869678.4), BatCoV-SC2013 (KC869678.4), Bat-CoV-BM48–31 (NC_014470.1), Bat-SL-HKU3–1 (DQ022305.2), Bat-SL-LYRa11 (KF569996.1), Bat-SL-Rf1 (DQ412042.1), Bat-SL-Rs4231 (KY417146.1), Bat-SL-Rs4255 (KY417149.1), Bat-SL-Rs4874 (KY417150.1), Bat-SL-RS672 (FJ588686.1), Bat-SL-WIV1 (KC881007.1), BtRs-BetaCoV/YN2018C (MK211377.1), BtRs-BetaCoV/YN2018D (MK211378.1), camMERS-CoV-HKFU-HKU-13 (KJ650295.1), camMERS-CoV-HKU23 (KF906251.1), camMERS-CoV-KSA-505 (KJ713295.1), camMERS-CoV-NRCE-HKU205 (KJ477102.1), camMERS-CoV-NRCE-HKU270 (KJ477103.2), CivSARS-CoV-SZ3 (P59594.1), HCoV-229E (NC_002645.1), HCoV-HKU1 (AY597011.2), HCoV-OC43 (KF963244.1), Hedgehog-CoV/VMC/DEU (KC545383.1), hMERS-CoV-EMC/2012 (JX869059.2), hMERS-CoV-England-1 (KC164505.2), hMERS-CoV-Jordan-N3 (KC776174.1), hSARS-CoV-BJ01 (AY278488.2), hSARS-CoV-GZ02 (AY390556.1), hSARS-CoV-HKU39849 (JN854286.1), hSARS-CoV-Tor2 (NC_004718.3), MHV-A59 (M18379.1), and Racoon dog SL-SZ13 (AY304487.1).
For S protein modeling, amino acid sequences of SARS-CoV Urbani (AAP13441.1), SARS-CoV-2-Wuhan-Hu1 (MN908947.3), Bat-SL-CoVZC45 (MG772933.1), Bat-SL-CoVZXC21 (MG772934.1), Bat-SL-LYRa3 (KF569997.1), and FCoV WSU-79-1683 (JN634064.1) were obtained from the NCBI GenBank, and Bat-SL-RaTG13 (EPI_ISL_402131) was obtained from GISAID database. Amino acid sequence of the FCoV-TN406 S was provided by Prof. Susan Baker (Loyola University Chicago).
Amino acid alignments and phylogenetic trees
Sequences alignments were performed on coronavirus S protein sequences using MAFFT v7.388 [75,76]. Maximum-Likelihood phylogenetic trees were based on the S protein alignments and were generated using PhyML [77]. Numbers at nodes indicate bootstrap support (100 bootstraps). Sequence alignment display and formatting was performed using Geneious R10 (Biomatters), and phylogenetic tree display and formatting was performed using FigTree 1.4.3 (http://tree.bio.ed.ac.uk/).
S protein modeling
Beta-CoV S protein structures and amino acid sequences were obtained from the RCSB Protein Data Bank: HCoV-HKU1 (PDB No. 5I08), MHV (PDB No. 3JCL), MERS-CoV (PDB No. 6Q05), SARS-CoV (PDB No. 5X58), FCoV-UU4 (PDB No. 6JX7), IBV-M41 (PDB No. 6CV0), and HCoV-NL63 (PDB No. 5SZS). Pairwise amino acid alignments between each of the Beta-CoV and the SARS-CoV-2 were performed using Geneious Prime® (v.2019.2.3. Biomatters Ltd.) and exported as *.FASTA file extension for further application. S protein models were built using UCSF Chimera (v.1.14, University of California) through modeler homology tool of the Modeller extension (v.9.23, University of California) and edited using PyMOL (v.2.0.7, Schrodinger LLC.). SARS-CoV-2 S models built was based on HCoV-HKU1, MHV, MERS-CoV, and SARS-CoV S structures. Models for the Bat-CoV RaTG13, Bat-CoV CoVZC45, Bat-CoV CoVZXC21, Bat-CoV LYRa3, and SARS-CoV were built based on SARS-CoV S structure. Finally, FCoV-TN406 and FCoV WSU-78-1683 S models were built based on HCoV-NL63 S structure. Additional SARS-CoV S models based on MHV and MERS-CoV S structures were built to validate our modeling approach (data not shown).
Acknowledgments
Acknowledgments
We thank all members of the Whittaker and Daniel labs at Cornell University for comments and discussion.
Funding
Work in the author's laboratory is supported by the National Institutes of Health (research grant R01AI35270).
Edited by Eric O. Freed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2020.04.009.
Appendix A. Supplementary data
Supplementary Figures
References
- 1.Menachery V.D., Graham R.L., Baric R.S. Jumping species-a mechanism for coronavirus persistence and survival. Curr. Opin. Virol. 2017;23:1–7. doi: 10.1016/j.coviro.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. 2004.
- 3.World Health Organization. Middle East respiratory syndrome coronavirus (MERS-CoV). 2019.
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johns Hopkins University CfSSaE . 2019. Coronavirus COVID-19 Global Cases. [Google Scholar]
- 6.Chan J.F., Yuan S., Kok K.H., To KK, Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12 doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phan L.T., Nguyen T.V., Luong Q.C., Nguyen T.V., Nguyen H.T., Le H.Q. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N. Engl. J. Med. 2020;382:872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McIntosh K., Perlman S. In: Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases (Ninth Edition) Bennett J.E., Dolin R., Blaser M.J., editors. Elsevier; Philadelphia: 2020. 155—Coronaviruses, including severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) pp. 2072–2080. e3. [Google Scholar]
- 11.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu J.T., Leung K., Leung G.M. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International Committee on Taxonomy of Viruses (ICTV) 2018. Virus Taxonomy: 2018b Release. [Google Scholar]
- 14.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perlman S. Another decade, another coronavirus. N. Engl. J. Med. 2020;382:760–762. doi: 10.1056/NEJMe2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huynh J., Li S., Yount B., Smith A., Sturges L., Olsen J.C. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu L., Zhang Y., Liu Y., Chen Z., Deng H., Ma Z. Angiotensin-converting enzyme 2 (ACE2) from raccoon dog can serve as an efficient receptor for the spike protein of severe acute respiratory syndrome coronavirus. J. Gen. Virol. 2009;90:2695–2703. doi: 10.1099/vir.0.013490-0. [DOI] [PubMed] [Google Scholar]
- 19.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong M.C., Javornik Cregeen S.J., Ajami N.J., Petrosino J.F. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. bioRxiv. 2020 doi: 10.1101/2020.02.07.939207. [DOI] [Google Scholar]
- 21.Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.-J. Isolation and characterization of 2019-nCoV-like coronavirus from Malayan pangolins. bioRxiv. 2020 doi: 10.1101/2020.02.17.951335. [DOI] [Google Scholar]
- 22.Lam T.T.-Y., Shum M.H.-H., Zhu H.-C., Tong Y.-G., Ni X.-B., Liao Y.-S. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 23.Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai A.L., Millet J.K., Daniel S., Freed J.H., Whittaker G.R. The SARS-CoV fusion peptide forms an extended bipartite fusion platform that perturbs membrane order in a calcium-dependent manner. J. Mol. Biol. 2017;429:3875–3892. doi: 10.1016/j.jmb.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J. Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millet J., Whittaker G. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heald-Sargent T., Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science (New York, N.Y.) 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 33.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.) 2020:eabb2507. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seidah N.G., Sadr M.S., Chretien M., Mbikay M. The multifaceted proprotein convertases: their unique, redundant, complementary, and opposite functions. J. Biol. Chem. 2013;288:21473–21481. doi: 10.1074/jbc.R113.481549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seidah N.G. The proprotein convertases, 20 years later. Methods Mol. Biol. 2011;768:23–57. doi: 10.1007/978-1-61779-204-5_3. [DOI] [PubMed] [Google Scholar]
- 37.Jaimes J.A., Millet J.K., Stout A.E., Andre N.M., Whittaker G.R. A tale of two viruses: the distinct spike glycoproteins of feline coronaviruses. Viruses. 2020;12 doi: 10.3390/v12010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaimes J.A., Whittaker G.R. Feline coronavirus: insights into viral pathogenesis based on the spike protein structure and function. Virology. 2018;517:108–121. doi: 10.1016/j.virol.2017.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang J., Zheng Y., Yang Y., Liu C., Geng Q., Luo C. Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang J., Zheng Y., Yang Y., Liu C., Geng Q., Tai W. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state. J. Virol. 2018;92 doi: 10.1128/JVI.01556-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J., DiMaio F. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walls A.C., Tortorici M.A., Frenz B., Snijder J., Li W., Rey F.A. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang T.J., Chang Y.C., Ko T.P., Draczkowski P., Chien Y.C., Chang Y.C. Cryo-EM analysis of a feline coronavirus spike protein reveals a unique structure and camouflaging glycans. Proc. Natl. Acad. Sci. U. S. A. 2020;117:1438–1446. doi: 10.1073/pnas.1908898117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S., Li W., Liu S., Xu J. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res. 2016;44:W430–W435. doi: 10.1093/nar/gkw306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menachery V.D., Yount B.L., Jr., Sims A.C., Debbink K., Agnihothram S.S., Gralinski L.E. SARS-like WIV1-CoV poised for human emergence. Proc. Natl. Acad. Sci. U. S. A. 2016;113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X.L., Hu B., Wang B., Wang M.N., Zhang Q., Zhang W. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J. Virol. 2015;90:3253–3256. doi: 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. 281-92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific analysis of the SARS-CoV-2 glycan shield. bioRxiv. 2020 doi: 10.1101/2020.03.26.010322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 55.Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kleine-Weber H., Elzayat M.T., Hoffmann M., Pohlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018;8:16597. doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gallagher T.M., Buchmeier M.J., Perlman S. Cell receptor-independent infection by a neurotropic murine coronavirus. Virology. 1992;191:517–522. doi: 10.1016/0042-6822(92)90223-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Menachery V.D., Dinnon K.H., 3rd, Yount B.L., Jr., McAnarney E.T., Gralinski L.E., Hale A. Trypsin treatment unlocks barrier for zoonotic bat coronavirus infection. J. Virol. 2020;94 doi: 10.1128/JVI.01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phillips J.M., Gallagher T., Weiss S.R. Neurovirulent murine coronavirus JHM.SD uses cellular zinc metalloproteases for virus entry and cell–cell fusion. J. Virol. 2017;91 doi: 10.1128/JVI.01564-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou H., Chen X., Hu T., Li J., Song H., Liu Y. A novel bat coronavirus reveals natural insertions at the S1/S2 cleavage site of the Spike protein and a possible recombinant origin of HCoV-19. bioRxiv. 2020 doi: 10.1101/2020.03.02.974139. [DOI] [Google Scholar]
- 63.Chen J., Lee K.H., Steinhauer D.A., Stevens D.J., Skehel J.J., Wiley D.C. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 64.Frana M.F., Behnke J.N., Sturman L.S., Holmes K.V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 1985;56:912–920. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Le Coupanec A., Desforges M., Meessen-Pinard M., Dube M., Day R., Seidah N.G. Cleavage of a neuroinvasive human respiratory virus spike glycoprotein by proprotein convertases modulates neurovirulence and virus spread within the central nervous system. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.André N.M., Cossic B., Davies E., Miller A.D., Whittaker G.R. Distinct mutation in the feline coronavirus spike protein cleavage activation site in a cat with feline infectious peritonitis-associated meningoencephalomyelitis. J. Feline Med. Surg. Open Reports. 2019;5 doi: 10.1177/2055116919856103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Licitra B.N., Millet J.K., Regan A.D., Hamilton B.S., Rinaldi V.D., Duhamel G.E. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg. Infect. Dis. 2013;19:1066–1073. doi: 10.3201/eid1907.121094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Q., Herrmann A. Fast assessment of human receptor-binding capability of 2019 novel coronavirus (2019-nCoV) bioRxiv. 2020 doi: 10.1101/2020.02.01.930537. [DOI] [Google Scholar]
- 69.Choi S., Jung E., Choi B.Y., Hur Y.J., Ki M. High reproduction number of Middle East respiratory syndrome coronavirus in nosocomial outbreaks: mathematical modelling in Saudi Arabia and South Korea. J. Hosp. Infect. 2018;99:162–168. doi: 10.1016/j.jhin.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dighe A., Jombart T., van Kerkhove M., Ferguson N. A mathematical model of the transmission of Middle East respiratory syndrome coronavirus in dromedary camels (Camelus dromedarius) Int. J. Infect. Dis. 2019;79:1. [Google Scholar]
- 71.Lipsitch M., Cohen T., Cooper B., Robins J.M., Ma S., James L. Transmission dynamics and control of severe acute respiratory syndrome. Science (New York, N.Y.) 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu T., Hu J., Kang M., Lin L., Zhong H., Xiao J. Transmission dynamics of 2019 novel coronavirus (2019-nCoV) bioRxiv. 2020 doi: 10.1101/2020.01.25.919787. [DOI] [Google Scholar]
- 73.Zhou T., Liu Q., Yang Z., Liao J., Yang K., Lü X. Preliminary prediction of the basic reproduction number of the Wuhan novel coronavirus 2019-nCoV. arXiv Preprint arXiv. 2020;200110530 doi: 10.1111/jebm.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishiura H., Linton N.M., Akhmetzhanov A.R. Serial interval of novel coronavirus (COVID-19) infections. Int. J. Infect. Dis. 2020;93:284–286. doi: 10.1016/j.ijid.2020.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katoh K., Misawa K., Kuma K., Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Re. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures