Abstract
The role of human adenovirus (HAdV) infection in different acute diseases, such as febrile exudative tonsillitis, conjunctivitis, and pharyngoconjunctival fever is well established. However, the relationships, if any, of HAdV persistence and reactivation in the development of the chronic adenotonsillar disease is not fully understood. The present paper reports a 3‐year cross‐sectional hospital‐based study aimed at detecting and quantifying HAdV DNA and mRNA of the HAdV hexon gene in adenoid and palatine tonsil tissues and nasopharyngeal secretions (NPS) from patients with adenotonsillar hypertrophy or recurrent adenotonsillitis. HAdV C, B, and E were detectable in nearly 50% of the patients, with no association with the severity of airway obstruction, nor with the presence of recurrent tonsillitis, sleep apnea or otitis media with effusion (OME). Despite the higher rates of respiratory viral coinfections in patients with HAdV, the presence of other viruses, including DNA and RNA viruses, had no association with HAdV replication or shedding in secretions. Higher HAdV loads in adenoids showed a significant positive correlation with the presence of sleep apnea and the absence of OME. Although this study indicates that a significant proportion (~85%) of individuals with chronic adenotonsillar diseases have persistent nonproductive HAdV infection, including those by HAdV C, B, and E, epithelial and subepithelial cells in tonsils seem to be critical for HAdV C production and shedding in NPS in some patients, since viral antigen was detected in these regions by immunohistochemistry in four patients, all of which were also positive for HAdV mRNA detection.
Keywords: chronic adenotonsillar disease, hexon mRNA, human adenovirus (HAdV)
Highlights
• Adenoids and palatine tonsils from patients with chronic adenotonsilar disease are frequently infected with HAdV‐B, HAdV‐C and HAdV‐E.
• Only a small proportion of patients with the chronic adenotonsilar disease has a productive adenoviral infection, characterized by the detection of viral gene expression.
• There are no obvious association between adenovirus replication and viral coinfection.
• HAdV infection was not associated with the severity of nasal obstruction or the presence of recurrent tonsillitis.
1. INTRODUCTION
Human adenovirus (HAdV) is a nonenveloped icosahedral DNA virus that is highly prevalent in human populations.1 Since its discovery in the early 1950s,2, 3 more than 84 HAdV genotypes, including all the 50 previously characterized serotypes were described. Currently, seven HAdV species (A‐G) have been identified and are classified in the genus Mastadenovirus of the family Adenoviridae.4 HAdV can infect a large variety of cell types and tissues in humans, leading to a broad array of diseases, including acute respiratory infections (ARI),5 febrile exudative tonsillitis,6 acute conjunctivitis,7 cystitis, gastroenteritis,4 and rare cases of encephalitis,8 myocarditis,9 and hepatitis.10 Although HAdV infections are generally asymptomatic in immunocompetent individuals, acute HAdV diseases have a significant impact on children (especially under 4 years of age), elderly, immunosuppressed individuals, and military recruits.4
While HAdV can replicate in several cells types in vitro and is associated with productive infections in different tissues in humans, several HAdV species present varied tissue specificities. For instance, HAdV C (serotypes 1, 2, 5, and 6) are commonly associated with acute tonsillitis and respiratory diseases, whereas HAdV‐F (serotypes 40 and 41) and HAdV D (serotypes 8, 19, and 37) are typically associated with gastrointestinal infections and a relatively severe and highly contagious form of epidemic keratoconjunctivitis, respectively.1
Following the HAdV replication cycle, the viral genome can persist in the nucleus.11, 12 Such a fact is best exemplified by the persistence of HAdV C after primary infections of the respiratory tract, with intermittent viral excretion in nasopharyngeal secretions (NPS) and feces.13, 14, 15
Numerous studies have shown that lymphocytes of tonsils and adenoids are essential sites of HAdV persistence, namely of the species C.16 Indeed, seminal studies indicated the ability of HAdV to persist in tonsils and adenoids, since it was possible to recover HAdV from these tissues weeks to months after the establishment of explant cultures.2, 17 More recent studies using tissue cell separation and sorting have revealed that HAdV DNA is present in T lymphocytes of tonsils and adenoids.18 In addition, several established human lymphocyte cell lines, including a lymphoblastoid cell line derived from a bone marrow transplant recipient with adenovirus pneumonia, may sustain prolonged and noncytopathic adenovirus infection.19, 20
Although substantial knowledge has been obtained regarding mechanisms associated with viral persistence in human cell lines in vitro,21 the strategies of viral persistence and reactivation in human lymphoid tissues in vivo have been poorly elucidated. In fact, the cells types involved in the process of viral reactivation in vivo and the possible roles that HAdV replication may play in the development of chronic diseases, such as adenotonsillar hypertrophy and recurrent tonsillitis, is not entirely understood. The present cross‐sectional study of HAdV replication in adenotonsillar hypertrophy was conducted to help comprehend the association between viral replication in those tissues and shedding in secretions and the development of adenotonsillar hypertrophy and recurrent tonsillitis. Quantification of the HadV genome and the detection of mRNA of the HAdV hexon gene was performed in human adenoids and tonsils, NPS, and peripheral blood (PB) from patients with tonsillar hypertrophy and were compared to those obtained in samples from control patients.
2. PATIENTS AND METHODS
2.1. Ethics
The present study was conducted according to the principles expressed in the Helsinki Declaration and was approved by the local Research Ethics Committee (#10466/2008). All patients and caregivers signed informed consent and voluntarily agreed to participate in the survey.
2.2. Study design
This was a cross‐sectional study that evaluated the presence of HAdV in different samples of tissues and secretions from the upper respiratory tract of children with obstructive sleep apnea (OSA) or recurrent tonsillitis, comparing the results with control patients.
2.3. Patients and samples
Fragments of surgically removed adenoids and palatine tonsils (PTs), as well as samples of NPS and PB, were obtained from 180 patients (93 males) aged 1 to 18 years (median 5.0 years) who underwent adenotonsillectomy due to OSA or recurrent tonsillitis. Small punch biopsies from tonsillar tissues, NPS, and PB were also obtained from 12 control patients (7 males, median 3.0 years) undergoing cochlear implantation in the absence of chronic adenotonsillitis, without ARI symptoms and with normal nasofibroscopy. All patients enrolled in the study were undergoing treatment at the Otorhinolaryngology Division of the Clinical Hospital of the University of São Paulo Medical School, in the city of Ribeirão Preto, Brazil, from May 2010 to July 2012. Exclusion criteria for both the patient and control groups comprised the presence of ARI symptoms at the time of the surgical procedure and the use of antibiotics within one month before surgery. OSA was diagnosed by clinical evaluation, and recurrent tonsillitis using Paradise criteria.22 A detailed description of the criteria used for disease classification and the methods used in clinical sample processing was previously published by our research group.23
2.4. DNA and RNA extraction
Tissue samples, including those from sick and healthy individuals, were maintained in a preservative solution (RNA later; Invitrogen, Carlsbad, CA) at −86oC until nucleic acid extraction. DNA and RNA were extracted from approximately 30.0 mg of adenotonsillar tissue samples using the AllPrep DNA and RNA mini kits (Qiagen, Hilden, Germany), respectively. Total nucleic acids were extracted from 200 μL of NPS and 1.0 mL of PB using the QIAamp MinElute Virus Spin Kit and the QIAamp RNA and DNA blood mini kit, respectively, both from Qiagen GmbH. All nucleic acid extraction procedures were performed according to the manufacturer's instructions.
2.5. Detection and quantification of HAdV genomes
HAdV detection was performed by TaqMan real‐time PCR (real‐time PCR) following a previously published protocol.23 Briefly, the final reaction volume (10.0 μL), which contained 50.0 ng of DNA, 10 mM of forward and reverse primers (HAdV‐F: 5′‐GCCACGGTGGGGTTTCTAAACTT‐3′; HAdV‐R: 5′‐ GCCCCAGTGGTCTTACATGCACAT‐3′), 5 mM of the probe (HAdV‐P: 5′‐FAM‐TGCACCAGACCCGGGCTCAGGTACTCCGA‐TAMRA), and 5.0 μL of TaqMan master mix (Applied Biosystems, Foster City, CA), underwent the following cycling parameters: 95°C for 10 minutes, followed by 45 cycles of 95°C for 15 seconds, and 60°C for 1 minute. All real‐time PCR assays were done on a StepOne Plus thermocycler (Applied Biosystems), and qPCRs for the β‐actin and RNAseP reference genes were conducted simultaneously in all tissues or secretion samples, respectively.23 Applicable measures to prevent cross‐contamination of the real‐time PCR reactions were taken, including sample handling and mix preparations done in separate rooms. In addition, all the real‐time PCR plates included appropriate blanks.
The quantitative PCR (qPCR) for HAdV was targeted to the same region used for viral detection (hexon gene). To quantify the viral genomes, all qPCR assays included a standard curve produced using serial decimal dilutions of a plasmid in which the target DNA sequence of the HAdV hexon gene had been cloned, and the detection limit of the assay was approximately one copy of the HAdV hexon gene. A qPCR for HAdV was considered positive when the threshold was reached before the 40th cycle. All HAdV qPCR assays were performed in triplicate, and the results were normalized by amplification of the β‐actin or RNAseP gene included in duplicate in all tested batches. With this approach, viral loads were determined as the number of copies of HAdV DNA per gram of tissue of mL of NPS or blood
2.6. Detection of hexon gene mRNA
Hexon gene mRNA detection was performed by real‐time reverse transcription PCR (real‐time RT‐PCR) in tissue and NPS samples to ascertain the presence of HAdV replication using the same strategy used in our previously published human bocavirus (HBoV) study.24 Briefly, reverse transcription (RT) was performed on 1.0 mg of RNA using 10 pmol of oligo (dT) primer, according to the manufacturer's protocol. Real‐time PCR was then carried out using 150 ng of cDNA, with 10 mM of each primer (HAdV‐F and HAdV‐R), 5 mM of the probe (HAdV‐P), and 5.0 mL of TaqMan universal PCR master mix (Applied Biosystems), following the conditions described above. The total RNA extraction product was treated with DNAse I (Invitrogen) for 2 hours before the PCR to ensure target‐specific amplification. As a negative control, the same RNA extraction product was used for real‐time PCR without previous RT. Samples were considered PCR‐positive for HAdV hexon mRNA only when they were also simultaneously negative for the same target using the extracted RNA without previous RT. All samples, including all cDNAs and the RNAs pretreated with DNAse, were tested by real‐time PCR for β‐actin mRNA, following the previously described protocol.24
2.7. HAdV molecular typing
A molecular typing assay based on conventional nested‐PCR amplification and sequencing of a hypervariable region contained in the hexon gene was performed to determine which species of HAdV were present in the patients included in this study, following a previously published protocol.25 Briefly, the first PCR reaction was conducted using a final volume of 50.0 μL containing 100 ng of DNA, 0.2 μM of forward and reverse primers (AdhexF1: 5′‐TICTTTGACATICGIGGIGTICTIGA‐3′ and AdhexR1: 5′‐CTGTCIACIGCCTGRTTCCACA‐3′), 10 mM Tris‐HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 200 μM of each deoxynucleotide triphosphate, and 1 U of Taq DNA polymerase (Invitrogen). The cycling conditions were: 94°C for 2 minutes, followed by 35 cycles of 94°C for 1 minute, 45°C for 1 minute, and 72°C for 2 minutes, and a final extension at 72°C for 5 minutes. For the second PCR (nested reaction), 0.5 μL of the first PCR product was amplified using the same parameters described above, with the following forward and reverse primers, respectively: AdhexF2: 5′‐GGYCCYAGYTTYAARCCCTAYTC‐3′ and AdhexR2: 5′‐GGTTCTGTCICCCAGAGARTCIAGCA‐3′. The amplified products were separated on 1% agarose gels, and the nested‐PCR products were purified using the QIAquick PCR Purification Kit (Qiagen, Chatsworth, CA, USA). Sanger sequencing was performed in both directions using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit Ver. 3.1 and the AdhexF2 and AdhexR2 primers on an ABI 3100 DNA Sequencer (Applied Biosystems).
2.8. Phylogenetic analysis
A maximum likelihood (ML) phylogenetic tree was inferred using nucleotide sequences from strains of adenoviruses described in this study and representative members of the Adenoviridae family. Multiple sequence alignment was generated using MAFFT v.726 with manual adjustments. The ML tree was constructed using the IQ‐TREE version 1.6.8 software with 1000 ultrafast bootstraps and the best‐fit nucleotides model determined by Bayesian Information Criterion, which considered 88 reversible DNA substitution models.27, 28 Statistical support for individual nodes was estimated using the bootstrap value, and the phylogenetic tree was visualized with the FigTree (v.1.4.2) program.
2.9. Detection of other respiratory viruses
In this study, the association between the replication of HAdV and the presence of other respiratory viruses in adenotonsillar tissue were analyzed. All samples were tested for the presence of the following respiratory viruses by real‐time PCR, according to previously described procedures23: human enterovirus, human rhinovirus, human respiratory syncytial virus, human metapneumovirus, influenza A and B, human parainfluenza, human coronavirus 229E and OC43, and HBoV.
2.10. Immunohistochemistry for HAdV in adenotonsillar tissue
Positive and negative tissues for HAdV by real‐time PCR were tested regarding the presence of HAdV antigen by immunohistochemistry. Fragments of adenoid and PT tissues were fixed for 12 hours in formaldehyde (10%), dehydrated, embedded in paraffin, and subsequently sectioned and placed on microscope slides. Tissue sections were deparaffinized in xylene and rehydrated in decreasing concentrations of ethanol. For antigen retrieval, the sections were treated with trypsin (0.05% in distilled water with 0.1% calcium chloride; pH 7.8) at 37°C for 15 minutes. To detect HAdV antigen, the tissue sections were washed in phosphate‐buffered saline (PBS), incubated for 1 hour in PBS with bovine serum albumin (BSA) and 3% horse serum, and incubated for 2 hours with anti‐HAdV mouse monoclonal antibody (MAB8052; Millipore, Billerica, MA) diluted 1:1000 in PBS/BSA (pH 7.4) with 0.1% of Triton X‐100 (Sigma‐Aldrich, St. Louis, MO) at room temperature. The sections were then incubated with biotinylated horse antimouse IgG (Vector, Burlingame, CA) diluted 1:2000 in PBS (pH 7.4) for 30 minutes at room temperature. Detection of the biotinylated antibody was carried out with 1:300 Streptavidin‐peroxidase Ultrasensitive Polymer (Sigma‐Aldrich), and color development was obtained using NovaRED (Vector). The slides were counterstained with hematoxylin and eosin and mounted with Permount (Thermo Fisher Scientific, Waltham, MA). For the positive controls, HAdV‐infected Hep‐2 cells (HAdV 7 [ATCC VR‐7]) were suspended in a small volume of human plasma, clotted by treatment with thrombin, then fixed and paraffin‐embedded. Equally treated noninfected Hep‐2 cells were used as negative controls.
2.11. Statistical analysis
The patient groups were compared using the Chi‐square and Fisher's Exact tests; viral loads among patient groups were assessed using the Mann‐Whitney or unpaired t test. Comparisons between three or more groups were conducted with one‐way analysis of variance and the Bonferroni test. All assays were carried out using the GraphPad Prism software version 5.00 for Mac (GraphPad Software, San Diego, CA), and a P value of less than or equal to 0.05 was adopted for significance.
3. RESULTS
3.1. Frequency of HAdV
Of the 180 patients with chronic adenotonsillar diseases, 95 (52.8%) had HAdV detected by real‐time PCR in adenoids and/or tonsils. In 43 of the 95 patients (45.3%), HAdV was found simultaneously in the tissues and NPS, suggesting that some patients could have productive HAdV infection in the adenoid and/or PTs. The virus was not identified by real‐time PCR in PB from any of the enrolled patients, indicating the lack of viremia, in spite of HAdV detection in the upper airways.
HAdV was detected significantly (P < 0.05) more often in adenoids (48.9%) than PTs (27.2%) (Tables 1 and 2), and the frequency of HAdV detection in tonsillar tissues from patients with chronic adenotonsillar disease was not significantly different from that observed in the tissues from the control patients (Tables 1 and 2).
Table 1.
Clinical and demographic data of patients with HAdV detected in adenoids
| Disease patients group | Control patients group | |||
|---|---|---|---|---|
| HAdV + | HAdV − | HAdV + | HAdV − | |
| Patients | 85 (48.9%) | 89 (51.1%) | 3 (25.0%) | 9 (75.0%) |
| Males | 40 (47.1%) | 51 (57.3%) | 1 (33.3%) | 6 (66.6%) |
| Age (median of years) | 5.0 | 5.0 | 3.0 | 3.0 |
| Viral coinfection* | 62 (72.9%) | 45 (50.6%) | 2 (66.6%) | 4 (44.4%) |
| 0%‐50% nasal obstruction | 8 (9.4%) | 9 (10.1%) | – | – |
| 50%‐75% nasal obstruction | 36 (42.3%) | 41 (46.1%) | – | – |
| 75%‐100% nasal obstruction | 41 (48.3%) | 39 (43.8%) | – | – |
| Sleep apnea | 50 (58.8%) | 54 (60.6%) | – | – |
| Otitis media with effusion | 12 (14.1%) | 18 (20.2%) | – | – |
| Allergy | 27 (31.8%) | 21 (23.6%) | – | – |
Abbreviation: HAdV, human adenovirus.
P < 0.05.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Table 2.
Clinical and demographic data of patients with HAdV detected in palatine tonsils
| Disease patients proup | Control patients group | |||
|---|---|---|---|---|
| HAdV + | HAdV − | HAdV + | HAdV − | |
| Patients | 49 (27.2%) | 131 (72.8%) | 2 (16.6%) | 10 (83.3%) |
| Males | 23 (46.9%) | 70 (53.4%) | 1 (50.0%) | 6 (60.0%) |
| Age (median of years) | 5.0 | 5.0 | 3.0 | 3.0 |
| Viral coinfection* | 33 (67.3%) | 15 (11.4%) | 0 (0.0%) | 0 (0.0%) |
| Recurrent Tonsillitis | 29 (59.2%) | 84 (46.6%) | – | – |
| Tonsillar hypertrophy | 41 (83.7%) | 107 (81.7%) | – | – |
| Sleep apnea | 29 (59.2%) | 75 (57.3%) | – | – |
| Otitis media with effusion | 8 (16.3%) | 22 (16.8%) | – | – |
| Allergy | 14 (28.6%) | 34 (25.9%) | – | – |
Abbreviation: HAdV, human adenovirus.
P < 0.05.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Detection of HAdV in adenoids or PTs was not significantly associated with any of the specific clinical features, including a degree of nasal obstruction, sleep apnea, otitis media with effusion (OME), and allergy (Tables 1 and 2). Among all the analyzed parameters, the only fact worth mentioning was that the viral codetections were significantly more frequent (P = 0.002) in tissues positive for HAdV than in HAdV‐negative ones (Table 2).
3.2. HAdV viral load
The median HAdV load determined by qPCR in adenoids from patients with chronic adenotonsillar disease was 1.6 × 105 copies of genome/g (mean 9.4 × 106 ± 5.1 × 107 copies/g), while in the control patients, the median HAdV load was 2.6 × 106 copies/g (mean 3.9 × 107 ± 6.5 × 107 copies/g). In the PTs from patients with chronic adenotonsillar disease, the median HAdV load was 5.5 × 104 copies/g (mean 7.0 × 105 ± 1.8 × 106 copies/g), whereas, in the control group, the median was 1.8 × 105 copies/g (mean 1.8 × 105 ± 3.2 × 104 copies/g). Regarding the NPS samples, the median HAdV load was 1.4 × 104 copies/mL (mean 1.2 × 106 ± 7.2 × 106 copies/mL) and 3.4 × 104 copies/mL (mean 3.4 × 104 ± 4.7 × 104 copies/mL) in the patients with and without chronic adenotonsillar disease, respectively. The median HAdV load was almost three times higher in the adenoids than the other infection sites, although the difference was not significant (Figure 1A). However, HAdV loads in the adenoids were not uniformly high when compared to the other sampling sites of the same patients (Figure 1B). Differences in HAdV viral loads between patients with chronic adenotonsillar disease and the controls were not significant (Figure 1C‐E).
Figure 1.
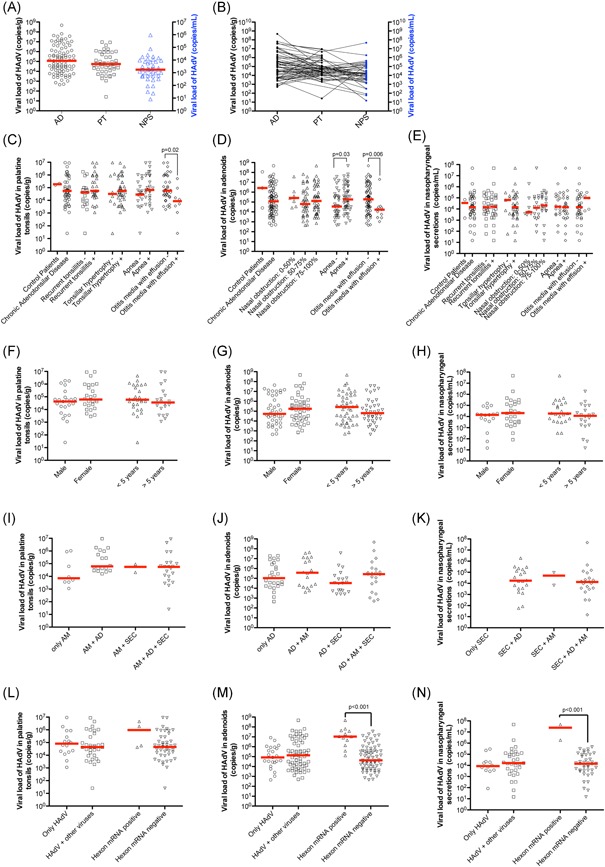
Viral load of HAdV in adenoids, palatine tonsils, and nasopharyngeal secretions by qPCR. A, HAdV viral loads in ADs, PTs, and NPS from patients with adenotonsillar chronic diseases. B, Patterns of HAdV viral loads in patients with simultaneous detection in several sites. The viral loads in the same patient were connected by a straight line. C, HAdV viral loads in palatine tonsils in the different clinical conditions. D, HAdV viral loads in adenoids in the different clinical conditions. E, HAdV viral loads in NPS in the different clinical conditions. F, HAdV viral loads in palatine tonsils according to sex and age. G, HAdV viral loads in adenoids according to sex and age. H, HAdV viral load in NPS according to sex and age. I, HAdV viral loads in palatine tonsils from patients with this virus detectable only in this tissue or when the agent was also detectable in other sites, including as adenoids and NPS. J, HAdV viral loads in adenoids from patients with this virus detectable only in this tissue or when the agent was also detectable in other sites, such as palatine tonsils and NPS. K, HAdV viral loads in NPS from patients with this virus detectable only in this site or when the agent was also detectable in other tissues, including adenoids and palatine tonsils. L, Association of HAdV viral loads in palatine tonsils with the presence of viral coinfection or with the detection of the mRNA of the HAdV hexon gene. M, Association of HAdV viral loads in adenoids with the presence of viral coinfection or with the detection of the mRNA of the HAdV hexon gene. N, Association of HAdV viral loads in NPS with the presence of viral coinfection or with the detection of the mRNA of the HAdV hexon gene. The red line in all graphs represents the median of the viral load in the analyzed condition. ADs, adenoids; HAdV, human adenovirus; NPS, nasopharyngeal secretions; PTs, palatine tonsils; qPCR, quantitative real‐time PCR
In general, the HAdV loads in the adenoids, PTs, and NPS were not significantly different among the patients with and without any of the several clinical features analyzed in the present study (Figure 1C‐H). Of note, the HAdV viral loads were significantly higher in patients with sleep apnea (1.4 × 107 ± 6.7 × 107 copies/g) than in those without the condition (2.4 × 106 ± 7.3 × 106 copies/g; P = 0.03), although lower in patients with OME (1.6 × 106 ± 5.6 × 106 copies/g) than in those without the disease (1.07 × 107 ± 5.6 × 107 copies/g; P = 0.006). The HAdV load was also significantly lower in PTs from patients with OME (1.6 × 105 ± 3.1 × 105 copies/g) than those without the condition (1.1 × 106 ± 2.4 × 106 copies/g; P = 0.02), suggesting that higher HAdV load may be a possible protective factor against the development of OME. However, since the number of patients in this group is very small and there is overlap between these two populations, including patients with recurrent tonsillitis and adenotonsillar hypertrophy in both groups, any kind of conclusion about this finding is very risky.
HAdV loads between patients with and without simultaneous detection of the virus in other sampling sites were also compared, as well as the influence of codetection of other respiratory viruses on HAdV loads. The analysis suggested an apparent trend for patients with HAdV in multiple infection sites with higher HAdV loads than individuals with HAdV detection in only one infection site, although the differences were not significant by one‐way analysis of variance applying Bonferroni as post‐test (Figure 1I ‐K). Therefore, the HAdV loads in adenotonsillar tissues were not significantly associated with the detection of the virus in NPS or with the simultaneous detection of other respiratory viruses (Figure 1L‐N and Figure 2).
Figure 2.
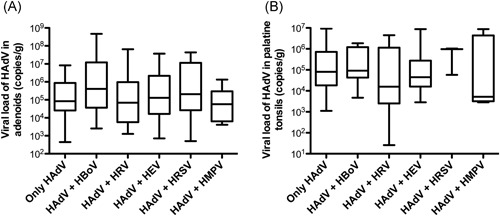
HAdV viral loads in adenoids (A) and palatine tonsils (B) from patients with HAdV as a single agent or in dual infection with other respiratory viruses: human bocavirus (HBoV), human rhinovirus (HRV), human enterovirus (HEV), human respiratory syncytial virus (HRSV), and human metapneumovirus (HMPV). Boxes extend from the 25th to 75th percentiles, middle whiskers mark median values, and upper and lower whiskers mark the highest and the lowest values, respectively. HAdV, human adenovirus
3.3. Productive infections by HAdV
The high frequency (27%) of patients with significant viral loads in the adenotonsillar tissues (>106 copies/g tissue), along with the high rate of HAdV detection in NPS (45.3%), is indicative that some of the enrolled patients had a productive infection. Thus, to verify the presence of active viral gene expression, suggestive of viral replication in the adenoids and tonsils, we attempted to detect the mRNA of the hexon gene in the tissues by real‐time RT‐PCR. Hexon gene mRNA was found in 12 (14.1%) of the HAdV‐positive adenoids, 4 (8.2%) of the HAdV‐positive PTs, and 2 (4.6%) of the HAdV‐positive NPS. Importantly, the presence of HAdV mRNA was correlated with high viral load, mainly in adenoids (Figure 1L‐N), indicating that the latter appears to be the primary site of HAdV replication in patients with tonsillar hypertrophy.
The presence of HAdV hexon gene mRNA in the adenoid was not associated with age, sex, or any specific clinical feature analyzed in the present study, including the presence of sleep apnea, OME, recurrent tonsillitis or the intensity of airway obstruction (Table 3). Remarkably, mRNA for the HAdV hexon gene was detected in one of the 3 (33.3%) HAdV‐positive adenoid biopsies obtained from control patients without adenotonsillar diseases (Table 4), indicating that the replicative activity of HAdV in tonsils does not necessarily lead to the development of the chronic adenotonsillar disease.
Table 3.
Clinical and demographic data of patients with replicant and persistent HAdV detected in adenoids
| Disease patients group | Control patients group | |||
|---|---|---|---|---|
| mRNA Hexon + | mRNA Hexon − | mRNA Hexon + | mRNA Hexon − | |
| Patients | 12 (14.1%) | 73 (85.9%) | 1 (33.3%) | 2 (66.6%) |
| Males | 6 (50.0%) | 34 (46.6%) | 0 (0.0%) | 1 (50.0%) |
| Age (median of years) | 4.0 | 6.0 | 2.0 | 3.0 |
| Viral coinfection | 62 (72.9%) | 45 (50.6%) | 1 (100%) | 1 (50.0%) |
| Viral load (median copies/g)* | 1.08 × 107 | 4.31 × 104 | 1.1 × 108 | 1.3 × 106 |
| High viral load (>106)* | 10 (83.3%) | 13 (17.8%) | 1 (100%) | 1 (50.0%) |
| Detection in several sites | 9 (75.0%) | 50 (68.5%) | 1 (100%) | 1 (50%) |
| Spreading of HAdV in NPS | 7 (58.3%) | 34 (46.6%) | ||
| 0%‐50% nasal obstruction | 1 (8.3%) | 7 (9.5%) | – | – |
| 50%‐75% nasal obstruction | 3 (25.0%) | 33 (45.2%) | – | – |
| 75%‐100% nasal obstruction | 8 (66.6%) | 33 (45.2%) | – | – |
| Sleep apnea | 9 (75.0%) | 41 (56.2%) | – | – |
| Otitis media with effusion | 1 (8.3%) | 11 (15.1%) | – | – |
| Allergy | 2 (16.6%) | 25 (34.2%) | – | – |
Abbreviations: HAdV, human adenovirus; NPS, nasopharyngeal secretion.
P < 0.05.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Table 4.
Clinical and demographic data of patients with replicant and persistent HAdV detected in palatine tonsils
| Disease patients group | Control patients group | |||
|---|---|---|---|---|
| Replicant HAdV | Persistent HAdV | Replicant HAdV | Persistent HAdV | |
| Patients | 4 (8.2%) | 45 (91.8%) | 0 (0.0%) | 2 (100.0%) |
| Males | 2 (50.0%) | 21 (42.3%) | – | 1 (50.0%) |
| Age (median of years) | 4.0 | 5.0 | – | 3.0 |
| Viral coinfection | 3 (75.0%) | 30 (66.6%) | – | 0 (0.0%) |
| Viral load (median copies/g) | 9.56 × 105 | 4.50 × 104 | – | 1.8 × 104 |
| High viral load (>106)* | 2 (50.0%) | 5 (11.1%) | – | 0 (0.0%) |
| Detection in several sites | 4 (100.0%) | 37 (82.2%) | – | 2 (100%) |
| Spreading of HAdV in NPS | 3 (75.0%) | 20 (44.4%) | ||
| Recurrent tonsillitis | 3 (75.0%) | 26 (57.7%) | – | – |
| Tonsillar hypertrophy | 4 (100.0%) | 37 (82.2%) | – | – |
| Sleep apnea | 4 (100.0%) | 25 (55.5%) | – | – |
| Otitis media with effusion | 0 (0.0%) | 8 (17.7%) | – | – |
| Allergy | 1 (25.0%) | 13 (28.8%) | – | – |
Abbreviations: HAdV, human adenovirus; NPS, nasopharyngeal secretion.
P < 0.05.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.4. HAdV typing by DNA sequencing
To demonstrate which HAdV species can replicate or establish persistence in the adenoids and PTs of the enrolled patients, the amplification and sequencing of a hypervariable region of the hexon gene were attempted. Of the 95 HAdV‐positive patients by real‐time PCR, 20 (21%) were positive for HAdV by conventional nested‐PCR, with the visualization of DNA products in agarose gels ranging from 688 to 821 bp in size (Figure 3A), always using DNA obtained from adenoid tissue. The palatine and nasopharyngeal secretion samples were not positive by this nested‐PCR. Based on the phylogenetic analysis (Figure 3B), 15 isolates (75%) were classified as human mastadenovirus C (13 related with HAdV type 1, and the others clustered with HAdV type 5 or HAdV type 6). Also, four isolates (20%) grouped with HAdV type 3 (human mastadenovirus B), and one was classified as human mastadenovirus E, clustering with HAdV type 4. Interestingly, viral replication was only detected in human mastadenovirus C‐infected adenoids, indicating that this species is able to replicate efficiently in tonsillar tissues. In contrast, the tissues infected with HAdV B and E did not show any sign of viral replication, indicating that adenotonsillar tissue can sustain nonproductive infections of HAdV B and E.
Figure 3.
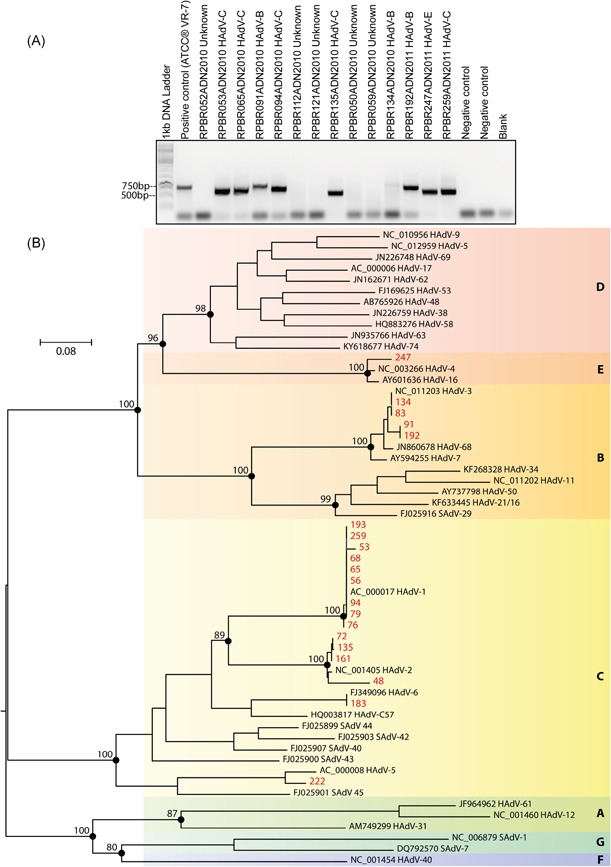
HAdV typing by conventional nested‐PCR and DNA sequencing. A, Representative agarose gel electrophoresis denoting the detection of genomic hexon gene sequences after nested‐PCR. B, Maximum likelihood phylogeny of strains of adenoviruses identified in this study within representative members of the Adenoviridae family. The tree was inferred using nucleotide alignments of the partial hexon gene based on TIM2+F+I+G4 of the DNA substitution model. Phylogeny is midpoint rooted. Scale bar indicates evolutionary distance in numbers of substitutions per nucleotide sites. Bootstrap values of 1000 replicates are shown in primary nodes. The “isolated” adenovirus sequences obtained herein are shown in red. Viral species are indicated by color and letter on the right. HAdV, human adenovirus; SAdV, simian adenovirus
3.5. Immunodetection of HAdV in adenotonsillar tissues
To localize the sites of HAdV replication in vivo, histological sections of HAdV PCR‐positive and negative tissues were tested by immunohistochemistry using anti‐HAdV antibodies (Figure 4). In the presence of the positive control, which consisted of HAdV‐infected Hep‐2 cells, viral structural proteins were detected in the epithelial layer of adenoids from four patients with HAdV detectable by PCR. Also, HAdV was simultaneously detected in the subepithelial layer and lymphoid parenchyma of a PT from one patient. All positive immunohistochemistry patients were also positive for viral mRNA detection by real‐time RT‐PCR. Thus, it can be concluded that HAdV C can replicate in epithelial and lymphoid cells from adenoids and PTs.
Figure 4.
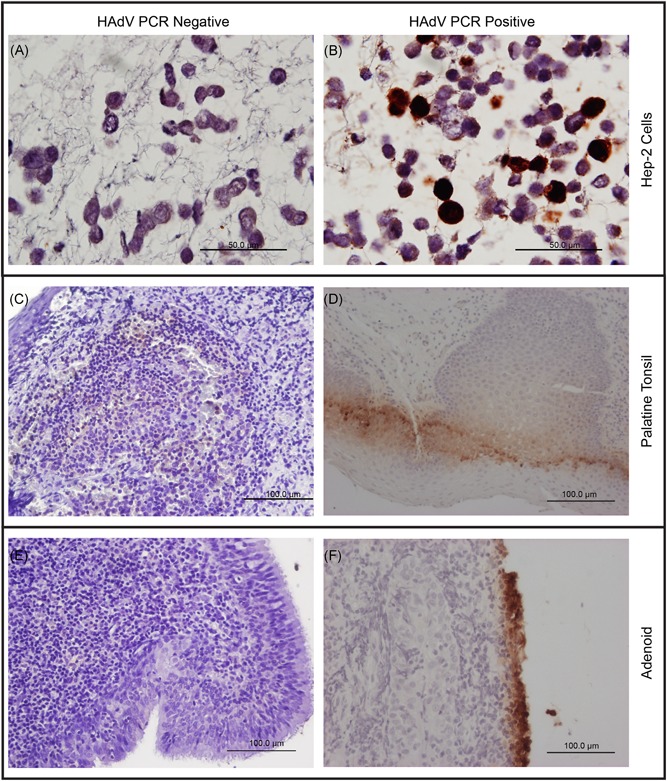
Immunohistochemistry for HAdV of noninfected and infected Hep‐2 cells, palatine tonsils and adenoids from patients with adenotonsillar chronic diseases. A, Noninfected Hep‐2 cells as negative controls counterstained with Hematoxylin and Eosin. B, HAdV‐infected Hep‐2 cells as positive controls counterstained with Hematoxylin and Eosin. C, Representative palatine tonsil from a patient without HAdV detectable by qPCR. D, Representative palatine tonsil from an HAdV‐positive patient. E, Representative adenoid from a patient without HAdV detectable by qPCR. F, Representative adenoid from a patient with HAdV detectable by qPCR, illustrating the presence of viral antigens in superficial cells. The positive signal is visible as brown color. The adenoids and palatine tonsils shown here were obtained from the same patient. HAdV, human adenovirus; qPCR, real‐time PCR
4. DISCUSSION
HAdV is among the leading causative agents of ARI in humans.4 In addition to causing ARI, a previous study by our group showed that HAdV is one of the most frequent respiratory viruses detected in children with chronic adenotonsillar disease in the absence of ARI symptoms.23 The near 50% detection rate of the HAdV genome reported herein confirms previous findings and agrees with adenoids being preferred sites of HAdV infection when compared with PTs.16, 18
HAdV has been detected in PB from patients with HAdV‐related tonsillitis in the presence of interleukin‐6 production by endothelial cells, fibroblasts or activated T lymphocytes, an essential mechanism for the persistence of fever.6 In the present study of patients without symptoms of ARI or acute tonsillitis, HAdV was undetectable in PB, suggesting that asymptomatic viremia is not frequent in asymptomatic HAdV carriers.
As previously published by our group in a small cohort of patients,23 HAdV was detected more frequently in tonsil tissues where codetection of other respiratory viruses was present. However, we demonstrated herein that such codetection is not linked to higher HAdV loads (>106 copies/g), nor with the detection of mRNA of the HAdV hexon gene. These findings indicate that the presence of HAdV in tonsils, with or without evidence of structural viral protein production, is not associated with a simultaneous increase in permissiveness of adenotonsillar tissues to other respiratory viruses. Furthermore, this observation infers that HAdV replication is not activated, nor reduced, by the presence of coinfection with other respiratory viruses.
Although adenovirus DNA is frequently found in tonsils, adenoids, and intestinal tissues (varying from 30%‐80% of cases), infectious viruses are rarely detected in these tissues, as measured by in situ hybridization or coculture with permissive cells.16, 29 Corroborating these findings, we were able to detect HAdV‐specific mRNA (signaling productive infection) in tonsillar tissue from 12 (14.1%) patients, suggesting that the majority of HAdV PCR‐positive patients with the chronic adenotonsillar disease have a persistent nonproductive infection.
Among the HAdV‐associated respiratory diseases, viruses of the species HAdV B (HAdV‐3, ‐7, ‐11, ‐14, ‐16, ‐21, ‐34, ‐35, ‐50, ‐55, and ‐66), HAdV C (HAdV‐1, ‐2, ‐5, and ‐6), and HAdV E (HAdV‐4) are frequently described as capable of replicating in the respiratory tract.30 As expected, we found HAdV‐1, ‐5, and ‐6 (species HAdV C), HAdV‐3 (HAdV B) and HAdV‐4 (HAdV E) in adenoids obtained from the studied patients. Interestingly, viral mRNA was detected only in adenoids from patients infected with HAdV‐1, indicating that HAdV C was able to replicate in the chronically inflamed adenoids analyzed.
Recent studies have pointed out the possibility of adenotonsillar tissue to act as a site for DNA respiratory virus production, helping viral spreading between healthy individuals, since HAdV or HBoV are frequently undetectable in asymptomatic individuals in adenotonsillectomy follow‐ups.31 In fact, some published data have demonstrated that lymphoid cells from adenoids, PTs, and intestinal lamina propria are the main sites of HAdV latency in humans, while epithelial cells from these tissues are essential for virus production and shedding in NPS or stools.16, 30 In addition, corroborating with these findings, HAdV antigen was detected in the epithelial layer of adenoids from 4 patients by immunohistochemistry in this study, suggesting that epithelial cells from tonsillar tissue comprise a site of viral proliferation preceding viral dissemination. We also detected HAdV antigen in the lymphoid parenchyma of one patient, indicating that other cells, aside from epithelial cells, can sustain HAdV replication in tonsillar tissue.
Persistent infection by HAdV has been associated with chronic airway obstructive diseases in children, such as asthma.32, 33 In those studies, HAdV antigen or genome was found in bronchoalveolar lavage from more than 75% of children with asthma, respectively, by immunohistochemistry32 or PCR.33 In the present study, the detection of HAdV was not significantly correlated with chronic adenotonsillar disease, respiratory symptoms or OME, nor with any other detectable disease phenotype.
Some clinical studies have associated the detection of respiratory viruses with OME. Viral infections caused by respiratory syncytial virus, influenza virus (types A and B), and adenovirus have been shown to increase the risk of OME, which can be in part attributed to these viral infections facilitating colonization of the nasopharynx by Streptococcus pneumoniae, Haemophilus influenzae, and M. catarrhalis,34, 35 and the adhesion of S. pneumoniae to epithelial cells of the respiratory tract.36 In contrast, the development of sleep apnea is partially associated with upper airway obstruction due to enlargement of the PTs and adenoids, seen significantly more often in obese patients with asymptomatic viral infections, such as those caused by adenoviruses.37, 38 Proinflammatory cytokines released by visceral adipocytes seem to contribute to tonsillar inflammation and the development of sleep apnea.39, 40, 41 Although no substantial association of HAdV and the severity of adenotonsillar enlargement was found in this study, a significant correlation was observed regarding HAdV quantities and the presence of sleep apnea or OME.
The present study has shown that patients without OME had significantly higher viral loads than individuals with the condition. Some viruses, such as human cytomegalovirus, are known to target dendritic cells, subverting and compromising the host's adaptive immunity by interfering with the cellular transport of major histocompatibility complex molecules.42, 43 Dendritic cells infected with HAdV strongly stimulate T cell proliferation,44 which may result in increased cellular response to other infectious agents, protecting the host from the development of OME. In addition, persistent adenoviral infection, with small HAdV loads, could function as a chronic stimulus for the development of OME.
In contrast, patients with sleep apnea exhibited significantly higher HAdV loads than individuals lacking the condition. Cellular and humoral responses are critical for the control of HAdV infection. The recruitment of macrophages and natural killer cells leads to the release of a range of proinflammatory cytokines, stimulating both CD4+ and CD8+ T cells, and, consequently, B cell proliferation with humoral antibody response.45 Thus, in keeping with this idea, it is reasonable to consider that high levels of HAdV may induce the production of proinflammatory and vasoactive cytokines, which increase chances of developing apnea.
In conclusion, the present study demonstrated that a high proportion of patients with the chronic adenotonsillar disease had persistent HAdV infection in the adenoids and tonsils. However, the presence of productive HAdV infection was not associated with the severity of nasal obstruction, recurrent tonsillitis or viral coinfections. The presence of higher HAdV loads in patients with apnea, in parallel with a protective effect against secretory otitis media, indicates that additional studies are required to provide a definitive role for HAdV during chronic adenotonsillar diseases.
ACKNOWLEDGMENTS
The authors thank Maria Cecılia Onofre and Helder G. de Souza for their secretarial assistance and Geraldo Cassio dos Reis for the statistical analysis. This study was supported by the São Paulo Research Foundation – FAPESP (Grant No. 2009/51818‐8 and 2016/00194‐8).
Proenca‐Modena JL, de Souza Cardoso R, Criado MF, et al. Human adenovirus replication and persistence in hypertrophic adenoids and palatine tonsils in children. J Med Virol. 2019;91:1250‐1262. 10.1002/jmv.25441
Contributor Information
José Luiz Proenca‐Modena, Email: jlmodena@unicamp.br.
Eurico Arruda, Email: eaneto@fmrp.usp.br.
References
REFERENCES
- 1. Gonçalves MA, de Vries AA. Adenovirus: from foe to friend. Rev Med Virol. 2006;16:167‐186. [DOI] [PubMed] [Google Scholar]
- 2. Rowe WP, Hubner RJ, Gilmore LK, Parrott RH, Ward TG. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc Soc Exp Biol Med. 1953;84:570‐573. [DOI] [PubMed] [Google Scholar]
- 3. Hilleman MR, Werner JH. Recovery of new agent from patients with acute respiratory illness. Proc Soc Exp Biol Med. 1954;85:183‐188. [DOI] [PubMed] [Google Scholar]
- 4. Lynch J, Fishbein M, Echavarria M. Adenovirus. Semin Respir Crit Care Med. 2011;32:494‐511. [DOI] [PubMed] [Google Scholar]
- 5. Rebelo‐de‐Andrade H, Pereira C, Gíria M, et al. Outbreak of acute respiratory infection among infants in Lisbon, Portugal, caused by human adenovirus serotype 3 and a new 7/3 recombinant strain. J Clin Microbiol. 2010;48:1391‐1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kohdera U, Kino M, Ito M. Detection of adenovirus DNA in throat swabs and blood by SYBR green real‐time PCR assay in patients with adenovirus‐associated tonsillitis. Jpn J Infect Dis. 2006;59:394‐396. [PubMed] [Google Scholar]
- 7. Adhikary AK, Banik U. Human adenovirus type 8: the major agent of epidemic keratoconjunctivitis (EKC). J Clin Virol. 2014;61:477‐486. [DOI] [PubMed] [Google Scholar]
- 8. Antoine JC, Pozetto B, Lucht F, Michel D, Gaudin OG, Rousset H. Acute adenovirus encephalitis diagnosed by prolonged intrathecal antibody production. Lancet (London, England). 1987;1:1382. [DOI] [PubMed] [Google Scholar]
- 9. Treacy A, Carr MJ, Dunford L, et al. First report of sudden death due to myocarditis caused by adenovirus serotype 3. J Clin Microbiol. 2010;48:642‐645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Terasako K, Oshima K, Wada H, et al. Fulminant hepatic failure caused by adenovirus infection mimicking peliosis hepatitis on abdominal computed tomography images after allogeneic hematopoietic stem cell transplantation. Intern Med. 2012;51:405‐411. [DOI] [PubMed] [Google Scholar]
- 11. Evans AS. Latent adenovirus infections of the human respiratory tract. Am J Hyg. 1958;67:256‐266. [DOI] [PubMed] [Google Scholar]
- 12. Neumann R, Genersch E, Eggers HJ. Detection of adenovirus nucleic acid sequences in human tonsils in the absence of infectious virus. Virus Res. 1987;7:93‐97. [DOI] [PubMed] [Google Scholar]
- 13. Fox JP, Brandt CD, Wassermann FE, et al. The virus watch program: a continuing surveillance of viral infections in metropolitan New York families. VI. Observations of adenovirus infections: virus excretion patterns, antibody response, efficiency of surveillance, patterns of infections, and relation to illness. Am J Epidemiol. 1969;89:25‐50. [DOI] [PubMed] [Google Scholar]
- 14. Fox JP, Hall CE, Cooney MK. The Seattle Virus Watch. VII. Observations of adenovirus infections. Am J Epidemiol. 1977;105:362‐386. [DOI] [PubMed] [Google Scholar]
- 15. Adrian T, Schäfer G, Cooney M, Fox J, Wigand R. Persistent enteral infections with adenovirus types 1 and 2 in infants: no evidence of reinfection. Epidemiol Infect. 1988;3:503‐509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garnett CT, Erdman D, Xu W, Gooding LR. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J Virol. 2002;76:10608‐10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van der Veen J, Lambriex M. Relationship of adenovirus to lymphocytes in naturally infected human tonsils and adenoids. Infect Immun. 1973;7:604‐609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garnett CT, Talekar G, Mahr JA, et al. Latent species C adenoviruses in human tonsil tissues. J Virol. 2009;83:2417‐2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flomenberg P, Piaskowski V, Harb J, Segura A, Casper JT. Spontaneous, persistent infection of a B‐cell lymphoma with adenovirus. J Med Virol. 1996;48:267‐272. [DOI] [PubMed] [Google Scholar]
- 20. Andiman WA, Miller G. Persistent infection with adenovirus types 5 and 6 in lymphoid cells from humans and woolly monkeys. J Infect Dis. 1982;145:83‐88. [DOI] [PubMed] [Google Scholar]
- 21. McNees AL, Mahr JA, Ornelles D, Gooding LR. Postinternalization inhibition of adenovirus gene expression and infectious virus production in human T‐cell lines. J Virol. 2004;78:6955‐6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paradise JL, Bluestone CD, Bachman RZ, et al. Efficacy of tonsillectomy for recurrent throat infection in severely affected children. Results of parallel randomized and nonrandomized clinical trials. N Engl J Med. 1984;310:674‐683. [DOI] [PubMed] [Google Scholar]
- 23. Proenca‐Modena JL, Pereira Valera FC, Jacob MG, et al. High rates of detection of respiratory viruses in tonsillar tissues from children with chronic adenotonsillar disease. PLoS One. 2012;7:e42136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Proença‐Modena JL, Gagliardi TB, Paula FE, et al. Detection of human bocavirus mRNA in respiratory secretions correlates with high viral load and concurrent diarrhea. PLoS One. 2011;6:e21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu X, Erdman D. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch Virol. 2006;151:1587‐1602. [DOI] [PubMed] [Google Scholar]
- 26. Katoh K, Standley D, M. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 2014;1079:131‐146. [DOI] [PubMed] [Google Scholar]
- 27. Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ‐TREE: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Mol Biol Evol. 2015;32:268‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587‐589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kosulin K, Geiger E, Vécsei A, et al. Persistence and reactivation of human adenoviruses in the gastrointestinal tract. Clin Microbiol Infect. 2016;22:381.e1‐381.e8. [DOI] [PubMed] [Google Scholar]
- 30. Dou Y, Li Y, Ma C, et al. Rapid diagnosis of human adenovirus B, C and E in the respiratory tract using multiplex quantitative polymerase chain reaction. Mol. Med. Report. 2018;18:2889‐2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martins RB, Rocha LP, Prates MM, et al. Respiratory DNA viruses are undetectable in nasopharyngeal secretions from adenotonsillectomized children. PLoS One. 2017;12:e0174188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Macek V, Sorli J, Kopriva S, Marin J. Persistent adenoviral infection and chronic airway obstruction in children. Am J Respir Crit Care Med. 1994;150:7‐10. [DOI] [PubMed] [Google Scholar]
- 33. Marin J, Jeler‐Kacar D, Levstek V, Macek V. Persistence of viruses in upper respiratory tract of children with asthma. J Infect. 2000;41:69‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henderson FW, Collier A, M, Sanyal MA, et al. A longitudinal study of respiratory viruses and bacteria in the etiology of acute otitis media with effusion. N Engl J Med. 1982;306:1377‐1383. [DOI] [PubMed] [Google Scholar]
- 35. Buzatto GP, Tamashiro E, Proenca‐Modena JL, et al. The pathogens profile in children with otitis media with effusion and adenoid hypertrophy. PLoS One. 2017;12:e0171049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Håkansson A, Kidd A, Wadell G, Sabharwal H, Svanborg C. Adenovirus infection enhances in vitro adherence of Streptococcus pneumoniae. Infect Immun. 1994;62:2707‐2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Faden H, Callanan V, Pizzuto M, et al. The ubiquity of asymptomatic respiratory viral infections in the tonsils and adenoids of children and their impact on airway obstruction. Int J Pediatr Otorhinolaryngol. 2016;90:128‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yeshuroon‐Koffler K, Shemer‐Avni Y, Keren‐Naus A, Goldbart AD. Detection of common respiratory viruses in tonsillar tissue of children with obstructive sleep apnea. Pediatr Pulmonol. 2015;50:187‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gaines J, Vgontzas AN, Fernandez‐Mendoza J, et al. Inflammation mediates the association between visceral adiposity and obstructive sleep apnea in adolescents. Am J Physiol Endocrinol Metab. 2016;311:E851‐E858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang Y‐S, Guilleminault C, Hwang FM, et al. Inflammatory cytokines in pediatric obstructive sleep apnea. Medicine (Baltimore). 2016;95:e4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mutlu M, Vuralkan E, Yardim Akaydin S, Akin I, Miser E. Effects of adenoid/tonsillectomy on inflammatory response in snoring children with witnessed apnoea. Clin Otolaryngol. 2014;39:266‐271. [DOI] [PubMed] [Google Scholar]
- 42. Tomazin R, Boname J, Hegde NR, et al. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat Med. 1999;5:1039‐1043. [DOI] [PubMed] [Google Scholar]
- 43. Beck K, Meyer‐König U, Weidmann M, Nern C, Hufert FT. Human cytomegalovirus impairs dendritic cell function: a novel mechanism of human cytomegalovirus immune escape. Eur J Immunol. 2003;33:1528‐1538. [DOI] [PubMed] [Google Scholar]
- 44. Keßler T, Hamprecht K, Feuchtinger T, Jahn G. Dendritic cells are susceptible to infection with wild‐type adenovirus, inducing a differentiation arrest in precursor cells and inducing a strong T‐cell stimulation. J Gen Virol. 2010;91:1150‐1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zuckerman AJ, Banatvala JE, Pattison JR, Griffiths PD, Schoub BD. Principles and Practice of Clinical Virology West Sussex, England: John Wiley & Sons Ltd; 2004. [Google Scholar]


