Abstract
Merkel cell polyomavirus (MCPyV) was identified originally in association with a rare but aggressive skin cancer, Merkel cell carcinoma. The virus has since been found in the respiratory tract of some patients with respiratory disease. However, the role of MCPyV in the causation of respiratory disease has not been established. To determine the prevalence of MCPyV in 305 respiratory samples from immunocompetent and immunocompromised patients and evaluate their contribution to respiratory diseases, specimens were screened for MCPyV using single, multiplex, or real‐time PCR; co‐infection with other viruses was examined. Of the 305 samples tested, 10 (3.27%) were positive for MCPyV. The virus was found in two groups of patients: in 6 (2%) nasopharyngeal aspirate samples from children aged 26 days to 7 months who were immunocompetent; and in 4 (1.3%) of nasopharyngeal aspirate samples taken from patients aged 41 to 69 years who were severely immunosuppressed from leukemia or transplant therapy. Both groups had upper or lower respiratory tract infection. Co‐infections with other viruses were found in 30% of the MCPyV positive samples. The data present a pattern of infection similar to that seen with the polyomaviruses JC and BK in which the virus is acquired during childhood, probably by the respiratory route. The viruses then establish latency and become reactivated in the event of immunosuppression. J. Med. Virol. 83:2220–2224, 2011. © 2011 Wiley Periodicals, Inc.
Keywords: Merkel cell polyomavirus, respiratory infection, PCR, co‐infection, immunocompromised
INTRODUCTION
In the past few years, a number of new human polyomaviruses, KI, WU, human polyomavirus 6 (HPyV6), human polyomavirus 7 (HPyV7), trichodysplasia spinulosa virus (TSV), human polyomavirus 9 (HPyV9), and Merkel cell polyomavirus (MCPyV) have been discovered [Allander et al., 2007; Gaynor et al., 2007; Feng et al., 2008; Schowalter et al., 2010; van der Meijden et al., 2010; Scuda et al., 2011]. MCPyV was discovered by digital transcriptome subtraction from a human skin cancer, Merkel cell carcinoma [Feng et al., 2008]. The finding of MCPyV in human Merkel cell carcinoma suggests a role for this virus in the causation of this cancer.
In confirmation of this, researchers were able to detect MCPyV DNA in 77% [Germany; Kassem et al., 2008], 84.9% [Europe; Becker et al., 2009], 43% [North America; Garneski et al., 2009], and 88% [France; Foulongne et al., 2008] of Merkel cell carcinoma specimens. More recently, studies have reported detection of MCPyV in lymphoid tissues [Sharp et al., 2009] and in respiratory specimens [Bialasiewicz et al., 2009; Goh et al., 2009; Kantola et al., 2009; Babakir‐Mina et al., 2010].
The aim of this study was to determine the prevalence of MCPyV in respiratory specimens collected from immunocompetent and immunocompromised patients and evaluate the possible of contribution of the virus in respiratory disease alone, or in combination with other respiratory viruses.
METHODS
Specimen Collection and Processing
A total of 305 specimens from patients with respiratory tract disease were randomly selected from among 1,335 nasopharyngeal aspirates submitted to the Clinical Virology Laboratory, Manchester Royal Infirmary, between October 2006 and February 2007. The specimens were stored at −70°C until use. Specimens were re‐used for this study in accordance with current Royal College of Pathologists Guideline (G035) (http://www.rcpath.org/index.asp?PageID=38) on re‐use of diagnostic specimens. Specimens and associated clinical data were collected; the specimens were anonymised by renumbering and removal of all patient identifiers from the data before use in this study.
Other Virus Screening
Specimens were extracted using the QIAamp® MinElute® Virus Spin kit (Qiagen, Crawley, West Sussex, UK) following the manufacturers instructions. To allow detection of RNA viruses, viral RNA was reverse transcribed to complementary DNA (cDNA) utilizing Appplied Biosystems Taq Man® Reverse Transcription Kit (Applied Biosystems, Warrington, UK) and random hexamer priming prior to real‐time PCR amplification using in‐house assays for Influenza A, B, and C viruses; parainfluenza viruses types 1–3; PCRs for respiratory syncytial virus (RSV) types A and B, human bocavirus (hBoV), and human metapneumovirus (HMPV) [Al‐Hammadi, 2008]. For detection of the polyomaviruses KI, WU, BK, JC, SV40, and LPV in‐house assays DNA PCRs were utilized as previously described [Abedi Kiasari et al., 2008; Abedi Kiasari, 2009].
Merkel Cell Polyomavirus PCR Assay
Nucleic acid was extracted from nasopharyngeal aspirates using the QIAampDNA Blood BioRobot MDx Kit (Qiagen) according to the manufacturer's instructions and stored at −20°C until use. To avoid cross‐contamination, all pre‐PCR processing was under taken in a separate location from PCR and post‐PCR analysis. For all PCR assays, standard precautions to avoid amplicon and nucleic acid contamination were taken. All samples were tested individually adding 5 µl nucleic acid to 45 µl master mix. Each reaction mix contained 200 pmol of each primer, 200 µM of dNTPs, 1× PCR buffer, and 2.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems). PCR assays were performed on a GenAmp PCR System 9700 thermal cycler (Applied Biosystems). Samples were subjected to 1 cycle of 94°C for 10 min followed by 40 cycles of 95°C for 1 min, 56°C for 1 min, 72°C for 1 min, and a final extension of 5 min at 72°C. Negative controls were included in each experiment. Samples were initially analyzed using the LT PCR assay described by Feng et al. [2008]. Plasmid pGEMT, containing Large T antigen of MCPyV (kindly provided by Dr. Vincent Foulongne, University of Montpellier, Montpellier, France) was also used as a positive control. Amplified products were analyzed by electrophoresis on 2% agarose gel, visualized with ultraviolet (UV) light and compared for size with E‐Gel Low Range Quantitative DNA Ladder (Invitrogen, Life Technologies, Paisley, UK). A strongly positive MCPyV sample (5 × 107 molecules/µl DNA copies) was used as a positive control in subsequent experiments (VP1 PCR assay). To confirm the results, all positive specimens were subjected to a second PCR analysis using primers targeting a different genomic region of MCPyV [VP1 PCR assay; Feng et al., 2008]. Using tenfold serial dilutions of positive controls, the sensitivity of the LT PCR and VP1 PCR were determined as 1 copy/µl and 50 copies/µl, respectively. Positivity was defined as being definitely established when the results of both PCR assays agreed.
Sequencing and Phylogenetic Analysis
Sequencing was carried out using the Big Dye Terminator Cycle Sequencing kit (Applied Biosystems) and the ABI 3100 Genetic Analyzer (Applied Biosystems). Sequences were assembled, analyzed, and edited using Sequencher software version 4.6 (Gene Codes Corporation, Ann Arbor, MI). Nucleotide sequences were aligned with Clustal W (University College Dublin, Eire). Phylogenetic analysis was performed on sequences derived from the VP1 region. All phylogenetic trees were visualized using Molecular Evolutionary Genetics Analysis (Mega) version 4.0 [Tamura et al., 2007]. A bootstrap test with 1,000 replicates was used to estimate the confidence of the branching pattern of the trees.
RESULTS
Patient Characteristics
The median age of the 305 patients was 7 months (mean 7 years; range 7 days–79 years), and the male to female ratio was 1.31:1 (173:132). Specimens tested in this study were from two groups of patients; Group I consisted of 250 nasopharyngeal aspirate samples obtained from paediatric patients who were immunocompetent (age range 7 days–14 years; mean 7 months; median 4 months). Group II included 55 nasopharyngeal aspirate samples obtained from adult patients who were either solid organ or bone‐marrow transplant patients or patients being treated for leukemia (age range 15–79 years; mean 45 years; median 46 years). Both groups had upper or lower respiratory infection.
Prevalence of Merkel Cell Polyomavirus and Clinical Findings
Sixteen (5.2%) samples produced positive results by the LT PCR assay. Of these, 10 (3.27%) were confirmed by VP1 PCR assay. Positive samples were investigated further by bidirectional sequencing of their LT3 and VP1 assay amplification products. The age distribution of patients with MCPyV infection is shown in Table I. MCPyV was identified in 6 (1.96%) samples from children aged 26 days to 7 months (mean: 5 months) who were immunocompetent. MCPyV DNA was also detected in 4 (1.31%) samples taken from patients aged 41–69 years (mean: 54 years) who were immunosuppressed (Table II). The incidence of MCPyV cases showed weekly variation over the study period but there was no evidence for a higher frequency of detection of these viruses during the winter period. In three of the MCPyV positive cases (30%), a co‐infection with RSV‐A (n = 1), hMPV (n = 1), or double co‐infection with RSV‐A and hBoV (n = 1) was detected. None of the MCPyV‐positive samples were positive for influenza A, B, and C viruses, para‐influenza viruses types 1–3, RSV type B, BKV, JCV, SV40, LPV, KIV, or WUV.
Table I.
Age Distribution of MCPyV Infected Patients
| Age group (years) | No. of sample tested | MCPyV positive no. (%) |
|---|---|---|
| <1 | 188 | 6 (3.19) |
| 1–5 | 50 | 0 (0) |
| 6–14 | 12 | 0(0) |
| 15–29 | 9 | 0 (0) |
| 30–44 | 11 | 0 (0) |
| 45–60 | 20 | 3 (15) |
| >60 | 15 | 1 (6.66) |
| Total | 305 | 10 (3.27) |
Table II.
Presentation and Demographic of MCPyV Positive Patients Study Subjects
| Group | Sample tested no. | MCPyV positive no. (%) | Age range (mean) | Male/female | Respiratory disease | Co‐detected viruses |
|---|---|---|---|---|---|---|
| Immunocompetent | 250 | 6 (2.4) | 26D‐7M (5M) | 1/5 | LRTI/URTI | RSV‐A (n = 1), RSV‐A + hBoV (n = 1) |
| Immunocompromised | 55 | 4 (7.27) | 41‐69Y (54Y) | 4/0 | LRTI/URTI | hMPV (n = 1) |
Sequencing and Phylogenetic Analysis
Sequence analysis of the MCPyV PCR products from both assays revealed 99% homology with the published sequences of these viruses in GenBank (NCBI) [EU375803; EU375804; Feng et al., 2008]. The sequence products were of sufficient quality to use for phylogenetic analysis. Phylogenetic analysis revealed a high level of identity between the MCPyV isolates (GenBank accession numbers: EU375803; EU375804). Based on the VP1 gene sequences (351 bp), all of the strains found in the study were in the same cluster with over 99% DNA sequence homology (Fig. 1).
Figure 1.
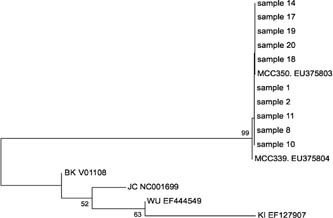
Phylogenetic analysis of nucleotide sequences of MCV VP1 partial genes from positive nasopharyngeal aspirate samples. The tree was built with the MEGA 4.0. software using the neighbor‐joining algorithm; bootstrap values were determined for 1,000 replicates; the percentage bootstrap values are shown at nodes. The horizontal scale indicates 0.5 substitutions per base pair. The analysis includes MCV references sequences from GenBank including MCC339, EU375804 and MCC350, EU375803 and representative BK, JC, KI, and WU sequences from GenBank.
DISCUSSION
MCPyV DNA was found in 3.27% of nasopharyngeal aspirate samples obtained from patients with respiratory tract disease as determined by both PCR assays (LT and VP1). We considered a sample as positive, when both PCR assays produced positive results. The prevalence reported here could therefore be an underestimate. Similar discordant LT and VP1 positive MCPyV results have been reported [Feng et al., 2008; Bialasiewicz et al., 2009; Goh et al., 2009]. This variation in detection rates could have been due to non‐specific amplification in the LT PCR or to the lower sensitivity of the VP1 PCR assay. Sample cross‐contamination is unlikely, because the samples were collected over 2 years from different hospitals. Contamination by the plasmid template is also unlikely because water controls were consistently negative and both assays were easily reproducible.
The frequency of MCPyV detection in this study was higher than that found in Australia [1.3%; Bialasiewicz et al., 2009] and Finland [2.1%; Kantola et al., 2009], but was lower than that found in Sweden [4.3%; Goh et al., 2009]. Also, the prevalence of MCPyV in this sample collection is higher than that found for KIV and WUV [2.7%, 1.08%, respectively; Abedi Kiasari et al., 2008]. Although samples tested in this study were only collected during the autumn and winter months, there was no clear peak of infection, perhaps suggesting a lack of any seasonal pattern of detection of MCPyV in association with respiratory virus infection.
Sequence data from positive specimens showed that the MCPyV found in respiratory specimens was similar to the sequence of viruses identified within Merkel cell carcinomas by Feng et al. [2008]. Despite the limited number of sequences analyzed, their minimal variation suggests the stability of the VP1 sequence of the virus genome and the global similarity of the VP1 of this virus.
The presence of MCPyV in the respiratory tract suggests that the respiratory tract may be a route of transmission just as JC and BK are suspected of being transmitted by inhalation and are occasionally detected in respiratory samples [Goudsmit et al., 1982; Sandler et al., 1997]. A relatively high co‐infection rate (30%) with other respiratory viruses was observed in this study. Several alternate respiratory pathogens including rhinovirus, adenovirus, coronavirus, and enterovirus were not tested and thus the proportion of co‐infections is possibly higher. MCPyV was however, the only virus detected in some children with evidence of respiratory tract disease, and so it is still possible that the respiratory system could be a primary target for MCPyV and that this virus contributes to respiratory tract disease in susceptible children.
In the present study, 250 specimens from immunocompetent patients and 55 specimens from immunocompromised patients were tested. Both groups had either lower or upper respiratory disease. Both immunocompetent (2.4%) and of immunocompromised (7.27%) groups were found to be MCPyV positive. The positive samples in the immunocompetent group were children age ranged 26 days–7 months. In fact, all 6 MCPyV positive patients who were not immunocompromised were <1 year of age suggesting early acquisition of MCPyV. Also, a patient aged 26 days was found to be MCPyV positive perhaps suggesting a potential vertical transmission route for MCPyV from mothers to infant. We could not however, determine whether this baby's mother had viral infection and whether the baby had viraemia. In the immunocompromised group, all four MCPyV positive patients were in an older age group (41–69 years). Higher rates of MCPyV detection in elderly immunosuppressed patients in the present study suggests an association between MCPyV and altered immune status and may mirror a similar pattern to the reactivation of BKV and JCV seen in leukemia and in transplant patients [Behzad‐Behbahani et al., 2004]. Examination of samples from control groups of patients without respiratory disease that are immunocompetent and immunosuppressed may shed further light on the association of MCPyV with disease. Further studies concerning the role of MCPyV in respiratory disease and the pathogenesis of MCPyV and its relationship with immunosuppression are clearly warranted.
Acknowledgements
We thank Dr. Vincent Foulongne from University of Montpellier, Montpellier, France for providing the Plasmid pGEMT.
REFERENCES
- Abedi Kiasari B. 2009. Molecular and serological investigation of polyomaviruses in human disease. PhD thesis, School of Medicine, University of Manchester, UK.
- Abedi Kiasari B, Vallely PJ, Corless CE, Al‐Hammadi M, Klapper PE. 2008. Age‐related pattern of KI and WU polyomavirus infection. J Clin Virol 43: 123–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Hammadi M. 2008. Molecular diagnosis and surveillance of community acquired respiratory viral infections. PhD thesis, School of Medicine, University of Manchester, UK.
- Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B. 2007. Identification of a third human polyomavirus. J Virol 81: 4130–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babakir‐Mina M, Ciccozzi M, Lo Presti A, Greco F, Perno CF, Ciotti M. 2010. Identification of Merkel cell polyomavirus in the lower respiratory tract of Italian patients. J Med Virol 82: 505–509. [DOI] [PubMed] [Google Scholar]
- Becker JC, Houben R, Ugurel S, Trefzer U, Pfohler C, Schrama D. 2009. MC polyomavirus is frequently present in Merkel cell carcinoma of European patients. Invest Dermatol 129: 248–250. [DOI] [PubMed] [Google Scholar]
- Behzad‐Behbahani A, Klapper PE, Vallely PJ, Cleator GM, Khoo SH. 2004. Detection of BK virus and JC virus DNA in urine samples from immunocompromised (HIV‐infected) and immunocompetent (HIV‐non infected) patients using polymerase chain reaction and microplate hybridisation. J Clin Virol 29: 224–229. [DOI] [PubMed] [Google Scholar]
- Bialasiewicz S, Lambert SB, Whiley DM, Nissen MD, Sloots TP. 2009. Merkel cell polyomavirus DNA in respiratory specimens from children and adults. Emerg Infect Dis 15: 492–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319: 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulongne V, Kluger N, Dereure O, Brieu N, Guillot B, Segondy M. 2008. Merkel cell polyomavirus and Merkel cell carcinoma, France. Emerg Infect Dis 14: 1491–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneski KM, Warcola AH, Feng Q, Kiviat NB, Leonard JH, Nghiem P. 2009. Merkel cell polyomavirus is more frequently present in North American than Australian Merkel cell carcinoma tumors. J Invest Dermatol 129: 246–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog 3: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh S, Lindau C, Tiveljung‐Lindell A, Allander T. 2009. Merkel cell polyomavirus in respiratory tract secretions. Emerg Infect Dis 15: 489–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudsmit J, Wertheim‐Van Dillen P, Van Strien A, Vander Noordaa J. 1982. The role of BK virus in acute respiratory tract disease and the presence of BKV DNA in tonsils. J Med Virol 10: 91–99. [DOI] [PubMed] [Google Scholar]
- Kantola K, Sadeghi M, Lahtinen A, Koskenvuo M, Aaltonen LM, Mottonen M, Rahiala J, Saarinen‐Pihkala U, Riikonen P, Jartti T, Ruuskanen O, Siderlund‐Venermo M, Hedman K. 2009. Merkel cell polyomavirus DNA in tunor‐free tonsillar tissues and upper respiratory tract samples: Implications for respiratory transmission and latency. J Clin Virol 45: 292–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassem A, Schopflin A, Diaz C, Weyers W, Stickeler E, ZurHausen A. 2008. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res 68: 5009–5013. [DOI] [PubMed] [Google Scholar]
- Sandler ES, Aquino VM, Goss‐Shohet E, Hinrichs S, Krisher K. 1997. BK papova virus pneumonia following hematopoietic stem cell transplantation. Bone Marrow Transplant 20: 163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7: 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuda N, Hofmann J, Calvignac‐Spencer S, Ruprecht K, Liman P, Kühn J, Hengel H, Ehlers B. 2011. A novel human polyomavirus closely related to the African green monkey‐derived lymphotropic polyomavirus. J Virol 85: 4586–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp CP, Norja P, Anthony I, Bell JE, Simmonds P. 2009. Reactivation and mutation of newly discovered WU, KI, and Merkel cell carcinoma polyomaviruses in immunosuppressed individuals. J Infect Dis 199: 398–404. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEG A4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog 6: e1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]


