Abstract
Respiratory syncytial virus (RSV) infection is the leading cause of acute respiratory tract disease in children less than 5 years old. The aim of this study was to further elucidate the molecular properties and clinical characteristics of RSV infection. The study sample included 238 patients <5 years old who were hospitalized with clinical symptoms of upper or lower respiratory tract infection (URTI or LRTI) in the Pediatric Department at the First People's Hospital of Chenzhou, South China in 2014. We subjected nasopharyngeal aspirate (NPA) or nasal swab (NS) samples from the patients to indirect fluorescence assay screens. RSV G genes were amplified by reverse transcription‐PCR (RT‐PCR) and sequenced. Of the 238 patients screened, 64 (26.8%) were confirmed to have RSV infections. Of those 64 confirmed RSV infection cases, 39 (60.9%) had subtype BA9, 13 (20.3%) had the recently identified subtype ON1, 11 (17.2%) had subtype NA1, and 1 (1.6%) had subtype GB2. The predominant presentation was LRTI with coughing, sputum production, fever, and wheezing. RSV subtype NA1 and BA9 infections were found mostly in infants, whereas the age distribution of subtype ON1 infections was more uniform across the age bands. Phylogenetic analysis indicated that, compared with the prototype strain A2, all ON1 and most NA1 isolates had lost one potential N‐glycosylation site at amino acid 251 and 249 due to T251K and N249Y substitution, respectively. These findings suggest that NA1, BA9, and ON1 are the dominant RSV subtypes causing respiratory tract infections in young children presenting to the hospital in South China. J. Med. Virol. 89:213–221, 2017. © 2016 Wiley Periodicals, Inc.
Keywords: respiratory syncytial virus (RSV, lower respiratory tract infections (LRTIs), subtype ON1, subtype NA1, subtype BA9, attachment glycoprotein (G) gene
Abbreviations
- RSV
respiratory syncytial virus
- RT‐PCR
reverse transcription‐PCR
- URTI
upper respiratory tract infection
- LRTI
lower respiratory tract infection
- RSV‐A
respiratory syncytial virus A
- RSV‐B
respiratory syncytial virus B
- NPAs
nasopharyngeal aspirates
- NSs
nasal swabs
- IFA
indirect immunofluorescence assay
INTRODUCTION
Respiratory syncytial virus (RSV) is a common pathogen that induces acute lower respiratory tract infections (LRTIs) in the elderly and in children, especially infants [Nair et al., 2010]. A meta‐analysis estimated that RSV accounts for 3.4 million hospitalizations and 66,000–199,000 deaths due to acute LRTIs among children less than 5 years old worldwide [Anderson, 2013]. Due to the lack of available vaccines, RSVs circulate every season and induce mild to serious disease in children and elderly adults.
RSV is an enveloped, single‐stranded, negative‐sense RNA virus that belongs to the genus Pneumovirus of the Paramyxoviridae family [Collins and Melero, 2011]. Based on the genetic characteristics of the attachment protein (G protein) and monoclonal antibody reactions, RSVs are subdivided into two genotypes: genotype A (RSV‐A) and genotype B (RSV‐B) [Mufson et al., 1985]. To date, 14 RSV‐A subtypes (GA1–7, SAA1–2, NA1–4, and ON1 [Peret et al., 2000, 1998; Venter et al., 2001; Shobugawa et al., 2009; Eshaghi et al., 2012; Pretorius et al., 2013; Ren et al., 2014]) and 22 RSV‐B subtypes (GB1–5, SAB1–4, URU1–2, BA1–10, and THB [Peret et al., 2000, 1998; Venter et al., 2002; Dapat et al., 2010; Trento et al., 2010; Auksornkitti et al., 2014; Ren et al., 2015]) have been described. The newly identified ON1 subtype of RSV‐A, which was first reported in Canada in 2010 [Eshaghi et al., 2012], contains a unique 72‐nucleotide duplication in the second hyper‐variable region of the G gene and may be spreading rapidly, perhaps replacing NA1, another prevalent RSV‐A subtype, in countries such as China, South Korea, Germany, and Italy [Kim et al., 2014; Pierangeli et al., 2014; Tabatabai et al., 2014; Cui et al., 2015]. A similar duplication phenomenon in the variable region of the G protein was also observed in the BA subtype of RSV‐B in 1999 in Buenos Aries, Argentina [Trento et al., 2003]; this strain spread rapidly to additional countries, resulting in the current prevalence of RSV‐B infections [Dapat et al., 2010; Trento et al., 2010].
The molecular epidemiological characteristics of RSV have been described previously for southwestern [Zhang et al., 2010b; Qin et al., 2013], northwestern [Zhang et al., 2010a; Cui et al., 2015] and eastern China [Liu et al., 2014]. However, there is limited information regarding the molecular epidemiology of RSV in South China. South China has a humid climate and a high frequency of acute respiratory infections in young children. In the present study, we screened hospitalized children under 5 years old with LRTIs or upper respiratory tract infections (URTIs) in Chenzhou, China, and analyzed the molecular epidemiological characteristics of the RSV strains identified.
MATERIALS AND METHODS
Patients and Clinical Samples
The study was performed in the Department of Pediatric, The First People's Hospital of Chenzhou, University of South China in 2014, and the study protocol was approved by the Hospital Ethics Committee. A total of 238 patients who met the following criteria were enrolled: (i) age <5 years old; (ii) hospitalized with one or more clinical symptoms of URTI or LRTI (i.e., cough, fever, wheezing, expectoration, anhelation, etc). Demographic and clinical characteristic data were collected. Written informed consent for participation in this study was obtained from the patients’ parents.
Prior to admission, a nasopharyngeal aspirate (NPA) or nasal swab (NS) was obtained from each patient to screen for RSV and other respiratory viruses with an indirect immunofluorescence assay (IFA) kit (D3 Ultra DFA Respiratory Virus Screen and ID Kit, Diagnostic Hybrids, Inc., OH); RSV‐negative patients were excluded from subsequent data analysis. All RSV positive NPAs or NSs were stored at −80°C for further molecular analysis by RSV reverse transcription‐PCR (RT‐PCR) and phylogenetic analysis.
RNA Extraction and RT‐PCR
RNA was extracted from NPA/NS samples which were identified as RSV infections by IFA using a TIANamp Virus DNA/RNA Kit (TIANGEN, Beijing, China). cDNA was synthesized by using random hexamer primers and the TIANScriptII RT Kit (TIANGEN). RSV G gene amplification was performed by nested PCR. First‐round amplification (sense primer, 5′‐CAA GCA AAT TYT GGC C‐3′; anti‐sense primer, 5′‐CCT YTG CTA ACT GCA C‐3′) was conducted under the following thermocycling conditions: 94°C for 3 min, followed by thirty 45‐sec cycles at 94°C, 48°C for 30 sec, and 72°C for 1 min, followed by a final extension at 72°C for 5 min. Second‐round amplification (sense primer, 5′‐GGG CAA ATG CAA ACA TGT CC‐3′; anti‐sense primer 5′‐TCC ATT GTT ATT TGC CCC AG‐3′) was conducted with the same thermocycling conditions that were applied in the first round, except that the annealing temperature was increased to 54°C. All PCR amplifications were performed with the TIANGEN 2× Taq Plus MasterMix Kit (TIANGEN).
Co‐Infection Detection
Detection of other pathogens was performed in parallel using specific laboratory tests, including immunofluorescence to detect influenza A and B, parainfluenza viruses 1–3, and adenoviruses; enzyme‐linked immunosorbent assays to detect human coronavirus and Mycoplasma pneumonia; and PCR to detect human bocavirus and human metapneumovirus. Sputum and blood cultures for bacterial detection were also performed. All sputum samples were obtained on all children once before treatment by aspiration, unless the patient was suffering from severe progressive disease or ineffective treatment. And blood for bacterial culture was obtained from patients who had a fever.
Nucleotide Sequence Analysis
All positive PCR products were sequenced by the Genscript (Nanjing) company. The sequences were determined and analyzed using the Basic Local Alignment Search Tool (BLAST) program available on the NCBI homepage (http://blast.ncbi.nlm.nih.gov/Blast.cgi). A maximum likelihood tree was constructed in MEGA‐5.1 software. The deduced amino acid sequences were analyzed with MEGA‐5.1 and BioEdit software. Potential N‐glycosylation (Asn‐Xaa‐Ser/Thr) sites were predicted using the NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/). Alignments of the second hyper‐variable region of the G protein were compared to those available from GenBank.
RESULTS
Characterization of RSV Infections in Hospitalized Children
IFA revealed that 64 of the 238 hospitalized children with respiratory tract infections who were screened (27%) had RSV infections. Of those 64 patients, 48 (75%) were male, 16 (25%) were female. Most, 63/64 (98.4%), were diagnosed with LRTIs at the time of admission. Cough (n = 64, 100%), sputum production (n = 61, 95.3%), wheezing (n = 49, 76.5%), and fever (n = 30, 46.9%) were the most common symptoms observed (Table I). Previous allergies, including eczema, urticarial dermatitis, and asthma, were reported in 25 (39%) patients.
Table I.
Demographic and Clinical Characteristics of Patients Infected With Respiratory Syncytial Virus
| RSV‐A | RSV‐B | |||
|---|---|---|---|---|
| Characteristic | ON1 (N = 13) | NA1 (N = 11) | BA9 (N = 39) | GB2* (N = 1) |
| Demographics | ||||
| Age, median (range), months | 14 (2–36) | 10 (1–72) | 9 (1–52) | 8 |
| Age band, no. (%) | ||||
| 0–6 months | 3 (23.1) | 9 (81.8) | 22 (55.0) | 0 |
| 6–12 months | 6 (46.2) | 1 (9.1) | 7 (18.0) | 1 (100) |
| 12–24 months | 2 (15.4) | 0 | 8 (20.0) | 0 |
| >24 months | 2 (15.4) | 1 (9.1) | 2 (5.0) | 0 |
| Male, no. (%) | 12 (92.3) | 8 (72.7) | 28 (70) | 1 (100) |
| Symptoms | ||||
| Fever, any, no. (%) | 8 (61.5) | 4 (36.4) | 18 (45) | 1 (100) |
| Maximal temperature | ||||
| Subgroup, no. (%) | ||||
| 37.3–38.0°C | 0 | 1 (25) | 3 (16.7) | 0 |
| 38.1–39.0°C | 4 (50) | 3 (75) | 9 (50) | 1 (100) |
| >39.0°C | 4 (50) | 0 | 6 (33.3) | 0 |
| Cough, no. (%) | 13 (100) | 11 (100) | 40 (100) | 1 (100) |
| Wheezing, no. (%) | 12 (92.3) | 8 (72.7) | 29 (72.5) | 1 (100) |
| Sputum production, no. (%) | 12 (92.3) | 11 (100) | 38 (95) | 1 (100) |
| Shortness of breath, no. (%) | 3 (23.1) | 2 (18.2) | 8 (20) | 0 |
| Allergic history, no. (%) | 3 (23.1) | 3 (27.3) | 19 (47.5) | 0 |
| Clinical diagnosis, no. (%) | ||||
| URTI | 0 | 1 (9.1) | 0 | 0 |
| LRTI | 13 (100) | 10 (90.9) | 40 (100) | 1 (100) |
| Mean (±SD) disease course disease, days* | 14 ± 6 | 11 ± 2 | 16 ± 12 | 12 |
RSV, respiratory syncytial virus; URTI, upper respiratory tract infection; LRTI, lower respiratory tract infection; SD, standard deviation.
Days from symptom onset to recovery was calculated as course of disease.
Sequencing of the RSV G gene in the patient samples indicated that 24/64 (37.5%) and 40/64 (62.5%) patients had RSV genotypes A and B, respectively, demonstrating simultaneous circulation of RSV‐A and ‐B in the study area. Furthermore, in the 24 RSV‐A‐infected patients, subtypes ON1 (n = 13, 54%) and NA1 (n = 11, 46%) were predominant; and 39 of the 40 RSV‐B‐infected patients were infected with RSV subtype BA9. Thus, subtypes ON1, NA1, and BA9 are the dominant circulating subtypes that cause respiratory infections in children in this geographical area.
ON1 is a newly identified RSV‐A subtype [Eshaghi et al., 2012], and our data indicate that ON1 infections exhibit some differences in the age of onset compared to NA1. NA1 mostly infected children less than 6 months old (81.8%), whereas ON1 did not show this trend.
Co‐Infection
In total, co‐infecting viruses were detected in 9 (15.7%) of the 64 RSV‐infected patient samples, with human coronavirus being the most common (Table II). Bacteria were isolated from 14 (21.9%) of the patient samples, with Escherichia coli accounting for six of those 14. Mycoplasma was detected in 3 (4.5%) of the patient samples. Bacteria were mainly isolated from sputum (N = 9), including E. coli (N = 4), Klebsiella pneumoni (N = 2), Straphylococcus aureus, Streptococcus pneumonia, and Serratia marcescens (each N = 1). In the remaining five cases, bacteria were detected in blood cultures (Table II).
Table II.
Co‐Infection in RSV‐Infected Patients
| Co‐infection type | No. cases (%) |
|---|---|
| Viral | |
| Parainfluenza 1 virus | 1 (1.6) |
| Human coronavirus | 7 (10.9) |
| Human bocavirus | 1 (1.6) |
| Human metapneumovirus | 1 (1.6) |
| Bacterial | |
| Escherichia coli a | 6 (9.4) |
| Klebsiella pneumonia b | 2 (3.1) |
| Straphylococcus aureus c | 2 (3.1) |
| Streptococcus pneumonia d | 1 (1.6) |
| Staphylococcus epidermidis e | 1 (1.6) |
| Staphylococcus capitis f | 1 (1.6) |
| Serratia marcescens g | 1 (1.6) |
| Mycoplasma | 3 (4.7) |
RSV, respiratory syncytial virus.
Two results were based on blood culture, and four based on sputum culture.
b,d,gThe results were based on sputum culture.
One result based on blood culture, one based on sputum culture.
e,fThe result were based on blood culture.
Phylogenetic Analyses
Sequences of the G gene from 24 RSV‐A and 40 RSV‐B isolates were successfully obtained and aligned with representative GenBank sequences of previously published genotypes. The RSV‐A strains were clustered into two subtypes: ON1 with a 72 nucleotide G gene duplication and NA1 (Fig. 1A). The RSV‐B strains were clustered into two subtypes: BA9 with a duplication of 60 nucleotides and GB2 (Fig. 1B).
Figure 1.
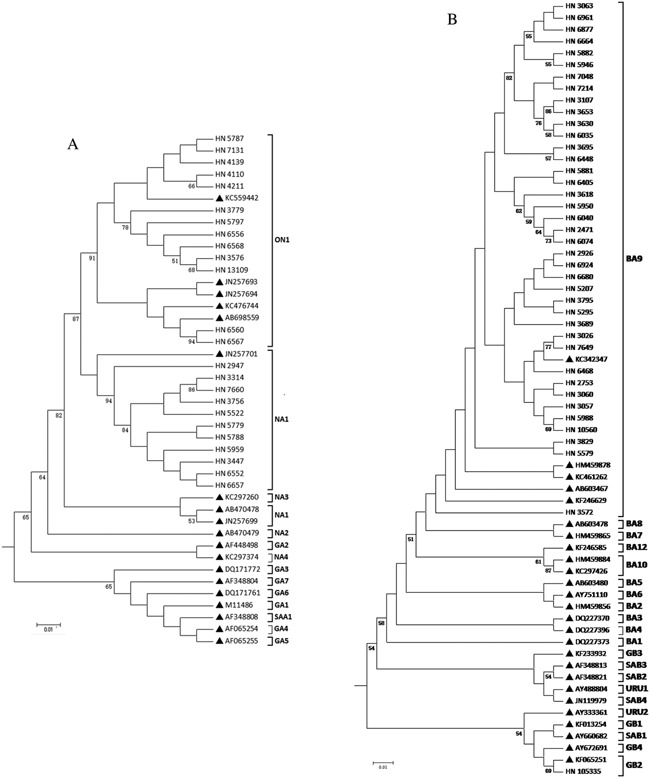
Phylogenetic tree of RSV‐A and RSV‐B isolates and reference sequences of identified genotypes. Phylogenetic trees for RSV‐A and RSV‐B isolates were constructed with maximum‐likelihood methods using MEGA 5.1 software. Hunan/China RSV strains are indicated by “HN” followed by their strain identification number. Reference strains representing known genotypes were retrieved from GenBank and indicated by solid triangles followed by their accession number. The genotype assignment is shown on the right by brackets. Tree topology was supported with 1,000 bootstrap replicates. Bootstrap values greater than 50 are shown at branch nodes.
Predominant Genotype in China
To evaluate the circulating of RSV in China, we analyzed the predominant genotype of human RSV in children from northeastern (Beijing), eastern (Suzhou and Shanghai), southwestern (Chongqing), and northwestern (Lanzhou) China in the most recent 8 years (Table III). These regions have experienced infections with similar subtypes of RSV (ABBAABA) in the last 8 years [Zhang et al., 2010a,2013, 2010b; Cui et al., 2015, 2013; Qin et al., 2013; Liu et al., 2014; Ren et al., 2014]. However, we lacked the complete datasets from Lanzhou and Chenzhou. Additionally, the NA1 and GA2 subtypes have been the predominant RSV‐A subtypes present, and the BA9 subtype has been the predominant RSV‐B subtype present. There are distinct subtypes of RSV‐A present in Beijing (NA1), Shanghai (NA1), and Chongqing (GA2), and Lanzhou (GA2) from 2007 to 2011. The predominant RSV genotype in the present study sample was RSV‐B, whereas that identified in a study in Beijing in the same time period was RSV‐A (Table III).
Table III.
The Predominant Genotype of Human RSV in Children From Four Cities in China in the Most Recent 8 Years
| Epidemic season | Beijing a (Northeast China) | Chongqing b (Southwest China) | Suzhou and Shanghai c (East China) | Lanzhou d (Northwest China) | Chenzhou (South China, this study) |
|---|---|---|---|---|---|
| 2007/2008 | RSV‐A/NA1 | RSV‐A/GA2 | RSV‐A* | RSV‐A/GA2 | ND |
| 2008/2009 | RSV‐B/BA9 | RSV‐B/BA* | RSV‐B* | RSV‐B/BA* | ND |
| 2009/2010 | RSV‐B/BA9 | RSV‐B/BA* | RSV‐B/BA | ND | ND |
| 2010/2011 | RSV‐A/NA1 | RSV‐A/GA2 | RSV‐A/NA1 | ND | ND |
| 2011/2012 | RSV‐A/NA1 | RSV‐A/NA1 | RSV‐A/NA1 | ND | ND |
| 2012/2013 | RSV‐B/BA9 | RSV‐B/BA* | RSV‐B/BA* | ND | ND |
| 2013/2014 | RSV‐A/ON1 | ND | ND | ND | RSV‐B/BA9 |
Deduced Amino Acid Sequence Analyses
We aligned and compared 19 Hunan RSV genotype A (ON1 and NA1) and 21 genotype B, with the prototype strain A2 (GenBank accession number M11486) (Fig. 2) and CH10B (GB1) strain (GenBank accession number AF065250) (Fig. 3), respectively. The deduced amino acid sequences of subgroup NA1 and ON1 observed in this study had two predicted lengths (297 and 321 aa) due to the duplication of amino acids 261–283 (i.e., QEETLHSTTSEGYLSPSQVYTTS). And the predicted lengths were 298 and 320 for the CH10B and BA9 subtypes due to the duplication of amino acids 241–260 (i.e., ERDTSTPQSTVLDTTTSKHT) and the loss of amino acids 167–168 (i.e., PK; data are not shown), respectively.
Figure 2.
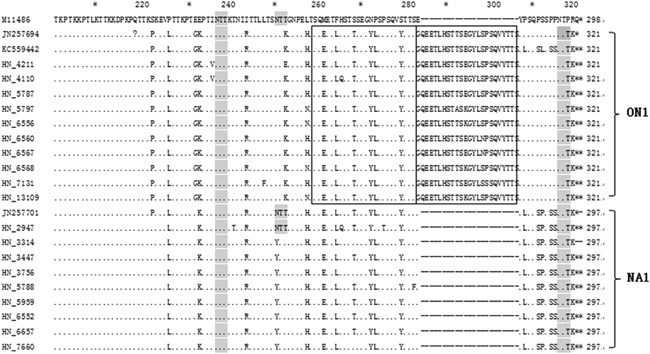
Alignment of deduced amino acid sequences of RSV‐A strains. Amino acid sequence alignment of the second variable region of the Hunan RSV‐A strains with the A2 prototype strain (GenBank accession number M11486), ON1 strains (GenBank accession numbers JN257694 and KC559442), and the NA1 strain (GenBank accession number JN257701). Identical residues are indicated by dots, stop codons are indicated by asterisks, and N‐linked glycosylation sites (NXT, where X is not a proline) are indicated by light gray. For the ON1 strain, the duplicated regions are framed by a rectangle. Genotype names are indicated to the right of the brackets.
Figure 3.
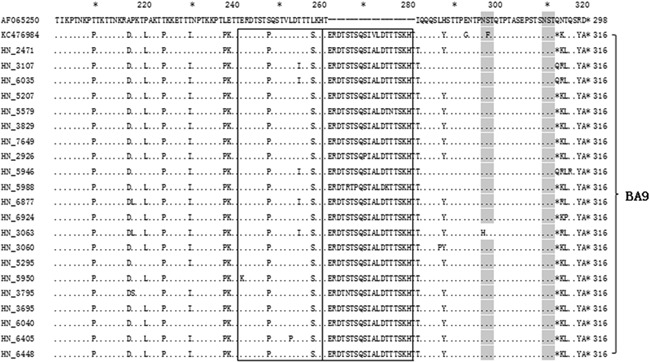
Alignment of deduced amino acid sequences of RSV‐B strains. Amino acid sequence alignment of the second variable region of the Hunan RSV‐B strains with the CH10B (GB1) (GenBank accession number AF065250) and BA9 strains (GenBank accession number KC476984). Identical residues are indicated by dots, stop codons are indicated by asterisks, and N‐linked glycosylation sites (NXT, where X is not a proline) are indicated by light gray. The duplicated regions are framed by a rectangle. Genotype names are indicated to the right of the brackets.
Compared to the A2 prototype, ON1 subtype‐specific substitutions (S222P, E232G, T253K, and P292L) were observed in all of the ON1 subtypes. There were six unique amino acid substitutions for NA1: N251Y, P310L, P313S, S314P, P316S, and P317S. In the A2 prototype strain, there are three potential N‐glycosylation sites (NXT, where X is not a proline) in the second variable region of the G protein. However, all of the ON1 isolates and most of the NA1 isolates had lost the N‐glycosylation site at amino acid 251and 249 due to T251K and N249Y substitutions, respectively (Fig. 2). Compared to the GB1, All BA9 strains had T207P, A215D, P219L, T223P, T229I, L237P, E238K, S247P, L257S, V270A, and L287Y substitutions (Fig. 3).
Genbank Nucleotide Sequence Accession Numbers
Sequences of G‐protein genes were submitted to Genbank (accession numbers KT781344–KT781406).
DISCUSSION
RSV is the most common pathogen that causes LRTIs in infants and young children [Eshaghi et al., 2012], but few studies of the molecular epidemiology of RSV have been conducted in China, especial South China. In this study, we analyzed the genetic diversity and patterns of co‐circulating genotypes of both RSV‐A and RSV‐B subtypes in Chenzhou, South China, in 2014. Of the 64 RSV positive cases, 24 were RSV‐A and 40 were RSV‐B, respectively. Phylogenetic analyses confirmed that the majority of the RSV‐B strains clustered with BA9 and that the RSV‐A strains clustered with strains of the new genotype, ON1 and its ancestor, NA1. In line with other studies in China and elsewhere, this study reports BA (BA9) as the predominant genotype [Zhang et al., 2010b; Rebuffo‐Scheer et al., 2011; Yamaguchi et al., 2011; Ren et al., 2015], suggesting that this subgroup has undergone adaptation and rapid spreading. Interestingly, the predominant subtype in Chenzhou from 2013 to 2014 was different from that in Beijing, which may be due to the differing climates between the two regions.
The novel ON1 genotype, which has a 72‐nucleotide duplication in the C‐terminal third of the G‐protein gene, was first detected during the winter of 2010–2011 in Ontario, Canada [Eshaghi et al., 2012]. A similar duplication phenomenon in the variable region of the G‐protein was also observed in the 1999BA genotype of RSV‐B [Trento et al., 2010]. Over the last 15 years, the BA subtype became the globally dominant RSV‐B strain [Dapat et al., 2010; Trento et al., 2010; Zhang et al., 2010b; Rebuffo‐Scheer et al., 2011; Yamaguchi et al., 2011; Ren et al., 2015]. Now, ON1 has thus far been detected in many countries, including Canada [Eshaghi et al., 2012], Italy [Pierangeli et al., 2014], Germany [Tabatabai et al., 2014], China [Cui et al., 2013; Liu et al., 2014; Ren et al., 2014], Japan [Hirano et al., 2014], Malaysia [Khor et al., 2013], India [Choudhary et al., 2013], South Korea [Lee et al., 2012], Thailand [Auksornkitti et al., 2014], Kenya [Agoti et al., 2014], and South Africa [Valley‐Omar et al., 2013]. These studies suggest that the ON1 subtype was transmitted globally and rapidly. Supporting this notion, many countries have reported that ON1 has become the predominant subtype [Lee et al., 2012; Agoti et al., 2014; Pierangeli et al., 2014; Tabatabai et al., 2014]. Thus, whether the ON1 subtype will follow the pattern of the BA subtype and become the predominant strain of RSV‐A worldwide is a question of great interest.
In China, the ON1 strain was first reported in Beijing (BJ/35320) in 2012 [Zhang et al., 2010a]. Later, circulation of the ON1 genotype was reported in Chongqing [Ren et al., 2014], and Shanghai [Liu et al., 2014]. According to the circulating pattern of the RSV subtypes in Chongqing, the NA1 genotype has been the predominant RSV‐A strain for the past 8 years, and the ON1 genotype will be the predominant RSV‐A subtype in 2014–2015 (Table III) [Ren et al., 2014]. Our results, which are in line with this circulation pattern, indicate that the ON1 genotype was the predominant RSV‐A subtype in 2014, suggesting that a rapid spread of this emerging RSV strain had occurred. Thus, the ON1 genotype may become the predominant strain of RSV in China, but further study will be needed to confirm this possibility.
Phylogenetic and deduced amino acid sequence analyses of the C‐terminal hyper‐variable region of the G protein among RSV strains revealed that ongoing changes characterized the different clusters within the RSV‐A subtype. Similar to previous studies conducted in China, the ON1 genotype was characterized by E232G, T253K, and P292L substitutions [Ren et al., 2014]. Additionally, an S222P substitution was also observed in all ON1 genotypes. Interestingly, all of the ON1 strains and almost all of the NA1 strains had lost the N‐glycosylation site at amino acid 251 due to T251K and N249Y substitutions, respectively [Auksornkitti et al., 2014]. However, it is not clear how these changes favored evasion of the host immune response.
In accordance with a prior study [Hall et al., 2009], most of the RSV‐infected children in this study were diagnosed with bronchiolitis or pneumonia after presenting with the symptoms of cough, persistent wheezing, fever, and sputum production. Presentation of clinical symptoms did not differ between patients with RSV‐A versus RSV‐B infections. Our age‐group analysis revealed that infants under 1 year of age accounted for 75.6% of the detected RSV infections and infants less than 6 months old accounted for ∼53%. Interestingly, the age distribution of patients with subtype ON1 infections was more uniform across age groups, whereas the NA1 and BA9 subtypes were found mostly in infants less than 6 months old. Thus, we hypothesized that the ON1 genotype is a novel RSV genotype, and given that most children were infected with ON1 for the first time, we reasoned that the organism must have no specific protective immunity. Conversely, NA1 and BA9 mainly infected infants less than 6 months of age likely because infection provides protective immunity thereafter, although limited. Thus, the vast majority of RSV reinfections are with genetically distinct strains [Agoti et al., 2012]. Unfortunately, the sample size of this study was not large enough to confirm this reasoning.
In summary, the molecular characterization of RSV in this geographical area in 2014 indicated that genotypes BA9, NA1, and ON1 co‐circulated and that the predominant genotype was BA9. In line with other studies in China and elsewhere, it can be hypothesized that subgroup ON1 will become the predominant strain of RSV‐A worldwide similar to the emergence of the BA genotype. These findings will improve the understanding of the molecular characterization of RSV in South China. However, further surveillance of circulating RSV genotypes, genetic diversity, and the corresponding clinical data is needed to better understand the antigenic mechanisms of RSV infections.
ACKNOWLEDGMENTS
We thank all family members for their enrollment in this study. We appreciate the Department of pediatrics of The First People's Hospital of Chenzhou for sample collection.
Conflicts of interest: All authors have no conflicts of interest.
REFERENCES
- Agoti CN, Mwihuri AG, Sande CJ, Onyango CO, Medley GF, Cane PA, Nokes DJ. 2012. Genetic relatedness of infecting and reinfecting respiratory syncytial virus strains identified in a birth cohort from rural Kenya. J Infect Dis 206:1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agoti CN, Otieno JR, Gitahi CW, Cane PA, Nokes DJ. 2014. Rapid spread and diversification of respiratory syncytial virus genotype ON1, Kenya. Emerg Infect Dis 20:950–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LJ. 2013. Respiratory syncytial virus vaccine development. Semin Immunol 25:160–171. [DOI] [PubMed] [Google Scholar]
- Auksornkitti V, Kamprasert N, Thongkomplew S, Suwannakarn K, Theamboonlers A, Samransamruajkij R, Poovorawan Y. 2014. Molecular characterization of human respiratory syncytial virus, 2010‐2011: Identification of genotype ON1 and a new subgroup B genotype in Thailand. Arch Virol 159:499–507. [DOI] [PubMed] [Google Scholar]
- Choudhary ML, Anand SP, Wadhwa BS, Chadha MS. 2013. Genetic variability of human respiratory syncytial virus in Pune, Western India. Infect Genet Evol 20:369–377. [DOI] [PubMed] [Google Scholar]
- Collins PL, Melero JA. 2011. Progress in understanding and controlling respiratory syncytial virus: Still crazy after all these years. Virus Res 162:80–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Zhu R, Deng J, Zhao L, Sun Y, Wang F, Qian Y. 2015. Rapid replacement of prevailing genotype of human respiratory syncytial virus by genotype ON1 in Beijing, 2012–2014. Infect Genet Evol 33:163–168. [DOI] [PubMed] [Google Scholar]
- Cui G, Zhu R, Qian Y, Deng J, Zhao L, Sun Y, Wang F. 2013. Genetic variation in attachment glycoprotein genes of human respiratory syncytial virus subgroups a and B in children in recent five consecutive years. PLoS ONE 8:e75020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapat IC, Shobugawa Y, Sano Y, Saito R, Sasaki A, Suzuki Y, Kumaki A, Zaraket H, Dapat C, Oguma T, Yamaguchi M, Suzuki H. 2010. New genotypes within respiratory syncytial virus group B genotype BA in Niigata, Japan. J Clin Microbiol 48:3423–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshaghi A, Duvvuri VR, Lai R, Nadarajah JT, Li A, Patel SN, Low DE, Gubbay JB. 2012. Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: A novel genotype with a 72 nucleotide G gene duplication. PLoS ONE 7:e32807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, Auinger P, Griffin MR, Poehling KA, Erdman D, Grijalva CG, Zhu Y, Szilagyi P. 2009. The burden of respiratory syncytial virus infection in young children. N Engl J Med 360:588–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano E, Kobayashi M, Tsukagoshi H, Yoshida LM, Kuroda M, Noda M, Ishioka T, Kozawa K, Ishii H, Yoshida A, Oishi K, Ryo A, Kimura H. 2014. Molecular evolution of human respiratory syncytial virus attachment glycoprotein (G) gene of new genotype ON1 and ancestor NA1. Infect Genet Evol 28:183–191. [DOI] [PubMed] [Google Scholar]
- Khor CS, Sam IC, Hooi PS, Chan YF. 2013. Displacement of predominant respiratory syncytial virus genotypes in Malaysia between 1989 and 2011. Infect Genet Evol 14:357–360. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Kim DW, Lee WJ, Yun MR, Lee HY, Lee HS, Jung HD, Kim K. 2014. Rapid replacement of human respiratory syncytial virus A with the ON1 genotype having 72 nucleotide duplication in G gene. Infect Genet Evol 26:103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, Kim YJ, Kim DW, Lee HS, Lee HY, Kim K. 2012. Complete genome sequence of human respiratory syncytial virus genotype A with a 72‐nucleotide duplication in the attachment protein G gene. J Virol 86:13810–13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Mu Y, Dong W, Yao F, Wang L, Yan H, Lan K, Zhang C. 2014. Genetic variation of human respiratory syncytial virus among children with fever and respiratory symptoms in Shanghai, China, from 2009 to 2012. Infect Genet Evol 27:131–136. [DOI] [PubMed] [Google Scholar]
- Mufson MA, Orvell C, Rafnar B, Norrby E. 1985. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol 66:2111–2124. [DOI] [PubMed] [Google Scholar]
- Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: A systematic review and meta‐analysis. Lancet 375:1545–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peret TC, Hall CB, Hammond GW, Piedra PA, Storch GA, Sullender WM, Tsou C, Anderson LJ. 2000. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J Infect Dis 181:1891–1896. [DOI] [PubMed] [Google Scholar]
- Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. 1998. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol 79:2221–2229. [DOI] [PubMed] [Google Scholar]
- Pierangeli A, Trotta D, Scagnolari C, Ferreri ML, Nicolai A, Midulla F, Marinelli K, Antonelli G, Bagnarelli P. 2014. Rapid spread of the novel respiratory syncytial virus A ON1 genotype, central Italy, 2011 to 2013. Euro Surveill 19:20843. [DOI] [PubMed] [Google Scholar]
- Pretorius MA, van Niekerk S, Tempia S, Moyes J, Cohen C, Madhi SA, Venter M, Group SS. 2013. Replacement and positive evolution of subtype A and B respiratory syncytial virus G‐protein genotypes from 1997–2012 in South Africa. J Infect Dis 208:S227–S237. [DOI] [PubMed] [Google Scholar]
- Qin X, Zhang C, Zhao Y, Zhao X. 2013. Genetic variability of subgroup A and B respiratory syncytial virus strains circulating in southwestern China from 2009 to 2011. Arch Virol 158:1487–1495. [DOI] [PubMed] [Google Scholar]
- Rebuffo‐Scheer C, Bose M, He J, Khaja S, Ulatowski M, Beck ET, Fan J, Kumar S, Nelson MI, Henrickson KJ. 2011. Whole genome sequencing and evolutionary analysis of human respiratory syncytial virus A and B from Milwaukee, WI 1998‐2010. PLoS ONE 6:e25468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Xia Q, Xiao Q, Zhou L, Zang N, Long X, Xie X, Deng Y, Wang L, Fu Z, Tian D, Zhao Y, Zhao X, Li T, Huang A, Liu E. 2014. The genetic variability of glycoproteins among respiratory syncytial virus subtype A in China between 2009 and 2013. Infect Genet Evol 27:339–347. [DOI] [PubMed] [Google Scholar]
- Ren L, Xiao Q, Zhou L, Xia Q, Liu E. 2015. Molecular characterization of human respiratory syncytial virus subtype B: A novel genotype of subtype B circulating in China. J Med Virol 87:1–9. [DOI] [PubMed] [Google Scholar]
- Shobugawa Y, Saito R, Sano Y, Zaraket H, Suzuki Y, Kumaki A, Dapat I, Oguma T, Yamaguchi M, Suzuki H. 2009. Emerging genotypes of human respiratory syncytial virus subgroup A among patients in Japan. J Clin Microbiol 47:2475–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabatabai J, Prifert C, Pfeil J, Grulich‐Henn J, Schnitzler P. 2014. Novel respiratory syncytial virus (RSV) genotype ON1 predominates in Germany during winter season 2012–13. PLoS ONE 9:e109191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trento A, Casas I, Calderon A, Garcia‐Garcia ML, Calvo C, Perez‐Brena P, Melero JA. 2010. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60‐nucleotide duplication in the G protein gene. J Virol 84:7500–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trento A, Galiano M, Videla C, Carballal G, Garcia‐Barreno B, Melero JA, Palomo C. 2003. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J Gen Virol 84:3115–3120. [DOI] [PubMed] [Google Scholar]
- Valley‐Omar Z, Muloiwa R, Hu NC, Eley B, Hsiao NY. 2013. Novel respiratory syncytial virus subtype ON1 among children, Cape Town, South Africa, 2012. Emerg Infect Dis 19:668–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter M, Collinson M, Schoub BD. 2002. Molecular epidemiological analysis of community circulating respiratory syncytial virus in rural South Africa: Comparison of viruses and genotypes responsible for different disease manifestations. J Med Virol 68:452–461. [DOI] [PubMed] [Google Scholar]
- Venter M, Madhi SA, Tiemessen CT, Schoub BD. 2001. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: Identification of new subgroup A and B genotypes. J Gen Virol 82:2117–2124. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Sano Y, Dapat IC, Saito R, Suzuki Y, Kumaki A, Shobugawa Y, Dapat C, Uchiyama M, Suzuki H. 2011. High frequency of repeated infections due to emerging genotypes of human respiratory syncytial viruses among children during eight successive epidemic seasons in Japan. J Clin Microbiol 49:1034–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RF, Jin Y, Xie ZP, Liu N, Yan KL, Gao HC, Song JR, Yuan XH, Xiao NG, Guo MW, Zhou QH, Hou YD, Duan Z. 2010a. Human respiratory syncytial virus in children with acute respiratory tract infections in China. J Clin Microbiol 48:4193–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XL, Shao XJ, Wang J, Guo WL. 2013. Temporal characteristics of respiratory syncytial virus infection in children and its correlation with climatic factors at a public pediatric hospital in Suzhou. J Clin Virol 58:666–670. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Du LN, Chen X, Zhao Y, Liu EM, Yang XQ, Zhao XD. 2010b. Genetic variability of respiratory syncytial viruses (RSV) prevalent in Southwestern China from 2006 to 2009: Emergence of subgroup B and A RSV as dominant strains. J Clin Microbiol 48:1201–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]


