Abstract
Human metapneumovirus is a cause of respiratory tract infections at all ages. Our objectives were to analyze the distribution of the A and B genotypes over 7 years in Dijon and to investigate a possible association between hMPV genotypes and disease severity. During 2002–2009, we genotyped the 100 isolates from children <3 years old with hMPV. Phylogenetic analysis indicated a change in the distribution of hMPV genotype over the years. Severity was then measured by detailed clinical evaluation. The hospitalization rate was greater when genotype B was involved 72.5% versus 53.3% (P = 0.054). Those infected with genotype B tended to have a higher clinical score, as measured by Vicente et al. 2006 (P = 0.07). We showed that, although clinical severity is not clearly associated with hMPV genotype in this study, pathological signs on chest X‐ray were observed more often in B subgroup (P < 0.01). J. Med. Virol. 82:1782–1789, 2010. © 2010 Wiley‐Liss, Inc.
INTRODUCTION
After being discovered in the Netherlands in 2001 [van den Hoogen et al., 2001], human metapneumovirus (hMPV) was identified in 3–10% of hospitalized young children with acute respiratory tract infection in many countries worldwide as reviewed by Kahn 2006. hMPV is a major pathogen that causes acute respiratory illness (ARI) in individuals of all ages. Higher morbidity is observed in young children, the elderly [Boivin et al., 2002; Falsey et al., 2003; van den Hoogen et al., 2003] and immunocompromised adults [Sumino et al., 2005; Williams et al., 2005; O'Gorman et al., 2006]. The clinical characteristics of hMPV infection in children range from mild upper respiratory tract to severe lower respiratory tract disease. The main clinical symptoms include rhinorrhea, cough, wheezing, and exacerbation of asthma. hMPV isolates are separated by phylogenetic analysis into two major genotypes (A and B), subdivided in four subgroups (A1, A2, B1, and B2) [Biacchesi et al., 2003; van den Hoogen et al., 2004]. Subgroup A2 has then been subdivided in two lineages A2a and A2b [Huck et al., 2006b]. Each season, hMPV circulation may vary, and hMPV genotypes circulate simultaneously in variable proportions [Williams et al., 2004; Gerna et al., 2005; Mackay et al., 2006]. In this study, we analyzed the distribution of the two hMPV genotypes and determined whether the clinical characteristics and severity of ARI episodes in children under 3 years of age correlated with a particular genotype.
MATERIALS AND METHODS
Collection and Analysis of Respiratory Specimens
The Hospital of Dijon is of medium size with a capacity of 1,700 beds, 20,000 children are admitted to the emergency department every year. During the study period, the laboratory received had 659 [95CI: 587–729] respiratory samples per year for children under 3 years old. All respiratory secretion samples (nasopharyngeal secretions, BAL, sputum) from patients presenting at Dijon University Hospital, France, for ARI from December 2002 to February 2005 sent for routine diagnosis of respiratory viruses were tested for hMPV by PCR, whereas from February 2005 to March 2009, samples were first tested by indirect immunofluorescence (IFA) using XD10 hybridoma supernatant as the primary antibody [Manoha et al., 2008]. This assay does not include pre‐screening culture methods. Respiratory syncytial virus (RSV), influenza (FLU), and parainfluenza (PI) were detected by IFA (Argène, Varilhes, France) throughout the study.
RT‐PCR for hMPV
RNA was extracted from the samples using TRIzol (Invitrogen, Cergy‐Pontoise, France). To detect hMPV, two previously described sets of primers were used. From December 2002 to December 2004, the first set of primers amplified a part of the L gene (171 bp) [van den Hoogen et al., 2001]. From 2005 to 2009, PCR was based on a second primer set which amplified a part of the F gene (750 bp) [van den Hoogen et al., 2004].
Clinical Characteristics
The analysis focused on all subjects under 3 years old who had positive results for hMPV from December 2002 to March 2009. However, those who were co‐infected with other respiratory viruses (RSV, FLU, or PI) were excluded from the analysis. Records were also excluded when medical physicians suspected respiratory bacterial infections and positive results were found. The clinical characteristics of the 100 included patients were collected mainly retrospectively through reviews of patients' medical file by a reviewer blinded to the genotyping result.
Clinical Severity Score (CSS)
The severity of the illness was assessed by two different clinical scores, CSS1 and 2. As described by Vicente et al. 2006, CSS1 ranged from 0 to 3 on the basis of the need for hospitalization, oxygen saturation <90%, and stay in an intensive care unit stay. As the clinical characteristics of hMPV infection resemble those of RSV, the second CSS was similar to that described by Martinello et al. 2002 for RSV and ranged from 0 to 6. Two points were given if the patient required mechanical ventilation during the illness, and one point was given for each of the following: hospital admission, hospitalization for >5 days, oxygen saturation <90%, and use of supplemental oxygen.
Sequencing and Phylogenetic Analysis
PCR‐amplified fragments were purified and directly sequenced in both directions using Big Dye Terminator (Applera Corporation, Foster City, CA) on an automatic DNA sequencer (Applera Corporation, Foster City, CA). Sequence comparisons were performed on a 102‐nt segment of the L gene [van den Hoogen et al., 2001] and/or a 449‐nt segment of the F gene [van den Hoogen et al., 2004]. Bionumerics software (Applied Maths, Sint‐Martens‐Latem, Belgium) was used to determine the hMPV genotypes. Multiple‐sequence alignments including some hMPV gene sequences available from GenBank were performed. Phylogenetic trees were generated by means of the UPGMA method. Bootstrap support was determined by 100 resamplings of the sequences.
Statistical Analysis
Results were analyzed using STATA version 8 (StataCorp, College Station, TX). Statistical differences in the distribution of hMPV genotypes over the years were determined by the χ 2‐test. The clinical characteristics and severity of illness associated with genotype A and B were compared using one‐way analysis' test or Kruskal–Wallis' test for continuous variables and the χ 2‐test for categorical data, as appropriate. Multivariate logistic regression analysis was conducted to estimate independence of potential confounding factors on chest radiographs, that is, signs of respiratory distress, chronic respiratory disease, gastroesophageal reflux disease (GERD), and cardiopathy.
RESULTS
Phylogenetic Analysis
On average, 179 respiratory samples [95CI: 144–213] were found to be virus positive every year. RSV was predominant, mean: n = 150 [95CI: 126–173], followed by influenza A mean: n = 26 [95CI: 3–48] and hMPV mean: n = 13.5 [95CI: 7–20]. We analyzed the 100 hMPV‐positive isolates identified in children with ARI under 3 years old presenting to the hospital during the 7 years of this study, from December 2002 to March 2009. Most cases occurred between December and April, rare cases occurred in May, June, or November (Fig. 1A). Among the 100 samples, 60% belonged to genotype A and 40% belonged to genotype B: of the 60A, seven were classified as A1 and 53 as A2; of the 40B, 12 were classified as B1 and 28 as B2 (Figs. 2 and 3). For the tree based on the L‐gene fragment (Fig. 2), which combined one third of the sequences, nucleotide identity between groups A and B was 81.4–82.4%, while it was 91.2–96.1% within group A. No B1 sequence was included in this tree, the nucleotide identity within subgroup B2 was 97.1–99.5%. For the tree based on the F‐gene fragment (Fig. 3), nucleotide identity between groups A and B was 83.2–85.1%. Within genotype A, nucleotide identity was 92.7–100%, within subgroup A1 (97.3–99.6%) and within subgroup A2 (96.4–100%); within genotype B, nucleotide identity was (95.0–95.8%), within subgroup B1 (98.9–100%), and within subgroup B2 (97.7–100%). Subgroup A was thus more divergent and the existence of two lineages within subtype A2, labelled A2a and A2b, as recently described by Huck et al. 2006b, was confirmed. As the PCR on the L gene used at the beginning of the study (2002–2004) did not allow us to distinguish lineages, only 27 A2 isolates were classified as either A2a or A2b. Five isolates (24%) belonged to A2a (97.9–99.4%), and 22 isolates (76%) belonged to A2b (98.0–99.6%). One strain isolated at the end of February 2008 (Dij‐199FEB‐08) did not fit into this dichotomy.
Figure 1.
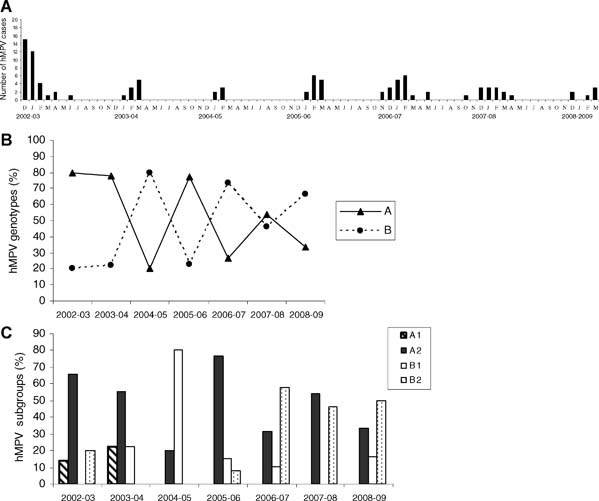
Human metapneumovirus (hMPV) in clinical specimens from children under 3 years old from December 2002 to March 2009. A: Monthly detection of hMPV. B: Distribution of hMPV genotypes over a 7‐year period (A (continuous line), B (hatched line)). C: Distribution of hMPV subgroups over a 7‐year period (A1 (hatched), A2 (black), B1 (white), B2 (with points))
Figure 2.
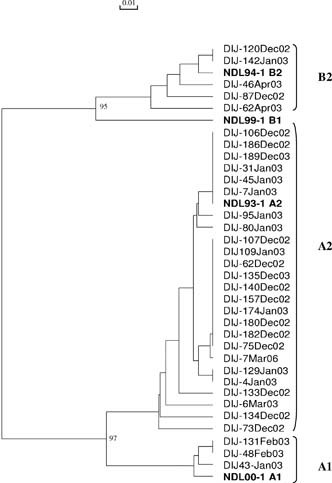
Phylogenetic analysis of nucleotide sequences from the partial L gene. Isolates are indicated by the town of origin (Dijon), order of isolate number, month and year of isolation (e.g., DIJ‐95JAN‐07). The tree generated includes four representative sequences, in bold, from isolates from the Netherlands (NL‐; GenBank accession no. AF371332, AF371333, AF371335, and AF371337).
Figure 3.
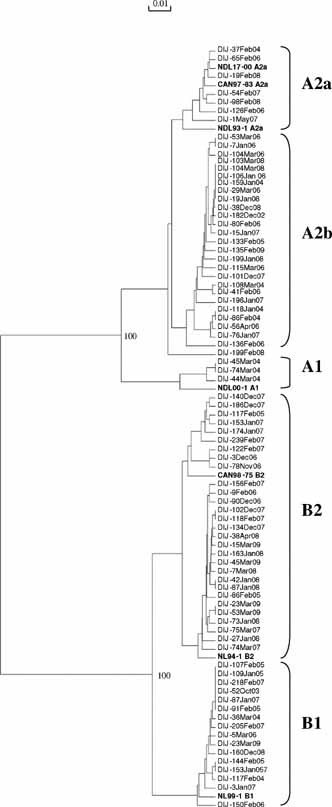
Phylogenetic analysis of nucleotide sequences from the partial F gene. Isolates are indicated by the town of origin (Dijon), order of isolate number, month and year of isolation (e.g., DIJ‐95JAN‐07). The tree generated includes five representative sequences, in bold, from isolates from the Netherlands (NL‐) and two from Canada (GenBank accession no. AY304360, AF371341, AF371337, AF371341, AY 304361, AY145289, and AY145296).
The A genotype was predominant during the first two seasons of the study, then both genotypes A and B circulated together with alternative predominance of one or the other genotype (Fig. 1B). There was not only a change in the predominant genotype but also a change within subgroups (Fig. 1C). A2 was detected throughout the study. The A1 subgroup was detected only during the first two seasons of the study, whereas the B1 subgroup emerged in 2003–2004, peaked the next winter then remained at a low level from 2005–2006 to 2008–2009. The B2 subgroup was detected mainly during the last three seasons of the study; its predominance was opposite to that of B1.
Strains of genotypes A and B circulated simultaneously throughout the studied seasons although the relative distribution of each genotype changed over the years (χ 2‐test, P = 0.001). As lower sensitivity has been reported for the primer set (L6/L7) on the reference strains NL/17//00(A2) and NL/1/99 (B1) [Maertzdorf et al., 2004], we may have missed some A2 or B1 isolates during the first two seasons. Even when we excluded this period from the analysis, the relative distribution of the genotypes (χ 2‐test, P < 0.05) and of the subgroups (χ 2‐test, P = 0.001) changed significantly in different years.
Clinical Manifestations of hMPV Infection According to Genotype
The sex and age were similar in both groups, with a median age of 5 months for children infected with hMPV genotype A and 6.5 months for children infected with hMPV genotype B (Table I). When we compared the clinical characteristics associated with each genotype, we found a similar pattern. Major clinical symptoms for both groups included cough (A: 78%; B 70%), rhinorrhea (A: 53%; B 40%), fever (A: 61%; B: 67%), and wheezing (A: 48%; B 47.5%) and the main diagnosis was lower respiratory tract disease (LRTD; A: 75; B: 80%).
Table I.
Characteristics of Patients With Human Metapneumovirus (hMPV) According to Genotype A or B
| Genotype (n = 100) | A (n = 60) | % | B (n = 40) | % | |
|---|---|---|---|---|---|
| Sex (male) | 40 | 66.7 | 22 | 55.00 | |
| Median age (months) | 5 | 6.5 | |||
| Rhinorrhea | 32 | 53.33 | 16 | 40 | |
| Cough | 47 | 78.33 | 28 | 70 | |
| Pharyngitis | 18 | 30 | 12 | 30 | |
| Otitis | 9 | 15 | 8 | 20.51 | |
| Fever | 36 | 61.02 | 26 | 66.67 | |
| Body temperature (mean) | 38.3 ± 0.9 | 38.4 ± 1 | |||
| Body temperature >38.5 | 21 | 37.5 | 18 | 47.37 | |
| Poor feeding | 23 | 38.98 | 21 | 53.85 | |
| Vomiting | 13 | 21.67 | 7 | 17.5 | |
| Diarrhea | 11 | 18.33 | 4 | 10 | |
| Wheezing | 29 | 48.33 | 19 | 47.5 | |
| Signs of Respiratory distress | 25 | 41.67 | 24 | 60 | χ 2‐test, P = 0.072 |
| Respiratory failure | 4 | 6.67 | 1 | 2.5 | |
| Oxygen saturation | 94.1 ± 4.7 | 92.6 ± 6 | |||
| Oxygen saturation <90 | 12 | 22.64 | 12 | 32.43 | |
| Oxygen treatment | 14 | 23.33 | 13 | 32.5 | |
| Duration of oxygen treatment (mean. days) | 0.76 ± 1.7 | 0.56 ± 1.1 | |||
| Bronchitis | 7 | 11.86 | 3 | 7.69 | |
| Bronchiolitis | 37 | 62.71 | 28 | 71.05 | |
| LRTD | 45 | 75 | 32 | 80 | |
| Radiological signs in chest | 6 | 14.63 | 15 | 44.12 | χ 2‐test, P = 0.005 |
| Hospitalization for ARI | 32 | 53.33 | 29 | 72.5 | χ 2‐test, P = 0.054 |
| Median duration of hospitalization (days) | 1.5 | 2.5 | |||
| Hospitalization >5days | 14 | 23.33 | 9 | 22.5 | |
| Stay in intensive care unit | 0 | 1 | |||
| Prematurity (<32 weeks) | 12 | 20 | 10 | 25 | |
| Cardiopathy | 1 | 1.67 | 5 | 12.5 | χ 2‐test, P = 0.025 |
| Asthma | 9 | 15 | 7 | 17.5 | |
| Chronic respiratory disease | 4 | 6.67 | 3 | 7.5 | |
| GERD | 2 | 3.39 | 8 | 20 | χ 2‐test, P = 0.007 |
| Smoking in family | 9 | 15 | 5 | 12.5 | |
| Asthma in family | 10 | 16.67 | 7 | 17.5 | |
| CSS1 (mean) | 0.77 | 1.07 | oneway, P = 0.068 | ||
| CSS2 (mean) | 1.2 | 1.55 |
Percentages are given relative to cases of genotype A (n = 60) or B (n = 40). For the following item, radiological signs in chest, only children who had a chest X‐ray have been taken into account. Chronic respiratory disease was measured as binary variable and included bronchopulmonary dysplasia and one case of lung agenesis; GERD, gastroesophageal reflux disease.
Asthma in the family was defined as a history of medical diagnosis of asthma in first‐degree‐relatives. Significant differences are in bold. Differences close to significance are also indicated.
The proportion of hospitalizations was higher for genotype B but without reaching statistical significance (A: 53%, B: 72.5%, χ 2‐test, P = 0.054) and the median duration of hospitalization was similar (A: 1.5 days; B: 2.5 days; Table I). Pathological findings on chest X‐ray were more frequent in genotype B (A: 15%; B 44%, P < 0.01), with most of the children showing bronchial wall thickening, alveolar condensation, perihilar haziness, or thoracic distension. There was no effect of confounding factors on chest radiographs as calculated by logistic regression analysis.
We did not find any association between genotype and any critical factors of severity such as oxygen saturation <90%, intensive care unit stay or use of supplemental oxygen. Both clinical severity scores were slightly higher for genotype B, but the differences were not statistically significant (CSS1: P = 0.07, CSS2: P = 0.24). Of note, the one and only child who did not present any risk factors (cardiopathy, chronic respiratory disease, etc.) but needed to be hospitalized in an intensive care unit had a B2 genotype. When patients with underlying diseases (cardiopathy, pulmonary disease and GERD) were removed from the analysis, the rate of hospitalization still tended to be higher for genotype B compared to genotype A (A: 48%; B 68%; P = 0.089).
When the four subgroups were compared, subgroup A1 was associated with the lowest rate and duration of hospitalization, percentage of LRTD as well as lowest values of CSS (Table II). The differences, however, did not reach statistical significance. A1 appeared to induce a milder disease than did the other subgroups. When we restricted the analysis to patients admitted to hospital (n = 61), clinical severity among genotypes was similar except for pharyngitis, which was more common in children with genotype B (P = 0.041) than in those with genotype A.
Table II.
Significant or Relevant Differences According to hMPV Subgroups
| Subgroups (n = 100) | A1 (n = 7) | % | A2 (n = 53) | % | B1 (n = 12) | % | B2 (n = 28) | % | |
|---|---|---|---|---|---|---|---|---|---|
| Sex (male) | 6 | 85.71 | 34 | 64.15 | 5 | 41.67 | 17 | 60.71 | |
| Median age (months) | 4 | 5 | 9.5 | 5 | |||||
| Rhinorrhea | 6 | 85.71 | 26 | 49.06 | 7 | 58.33 | 9 | 32.14 | χ 2‐test, P = 0.062 |
| Body temperature >38.5 | 5 | 83.33 | 16 | 32 | 8 | 66.67 | 10 | 38.46 | χ 2‐test, P = 0.024 |
| LRTD | 4 | 57.14 | 41 | 77.36 | 10 | 83.33 | 22 | 78.57 | |
| Radiological signs in chest | 0 | 0 | 6 | 17.14 | 6 | 66.67 | 9 | 36 | χ 2‐test, P = 0.008 |
| Hospitalization for ARI | 2 | 28.57 | 30 | 56.6 | 7 | 58.33 | 22 | 78.57 | χ 2‐test, P = 0.066 |
| Median duration of hospitalization (days) | 0 | 2 | 1.5 | 2.5 | |||||
| Stay in intensive care unit | 0 | 0 | 0 | 1 | |||||
| Asthma | 0 | 9 | 16.98 | 0 | 16.67 | 7 | 25.93 | ||
| GERD | 0 | 2 | 3.85 | 2 | 16.67 | 6 | 21.43 | χ 2‐test, P = 0.056 | |
| CSS1 (mean) | 0.43 | 0.81 | 1 | 1.11 | |||||
| CSS2 (mean) | 0.71 | 1.26 | 1.58 | 1.54 |
Percentages are given relative to cases of A1, A2, B1, or B2 subgroups. For the following item, radiological signs in chest, only children who had a chest X‐ray have been taken into account. Significative differences are in bold. Differences close to significance are also indicated.
DISCUSSION
hMPV are an important cause of ARI in young children, and in immunocompromised or elderly adults. To date, studies on hMPV have mainly focused on clinical characterization of hMPV infection, but only few reports have studied the influence of genotype on the severity of infection. In this report, we characterized a large cohort of hMPV isolates from children over 7 consecutive years and detailed the clinical characteristics of hMPV infection in relation to genotype.
Detection over a long period showed that most of the infections occur between December and April; these results are similar to those showing a higher frequency in late winter and spring [van den Hoogen et al., 2001; Cane et al., 2003; Esper et al., 2004; Robinson et al., 2005; Agapov et al., 2006; Rafiefard et al., 2008]. We also report on the domination of the A genotype over the B genotype (60/40). These results are consistent with those of Rafiefard et al. 2008 who also identified hMPV genotypes over a long period (5 years) in Sweden. More precisely, we showed that genotype A predominated from 2002 to 2004, then both genotypes A and B co‐circulated. In 2002–2003 a large number of specimens were positive for genotype A in our study as was the case in several countries. This result may be moderated because of the use of a primer set (originally published by Van den Hoogen 2001) located on L gene which has been shown to amplify hMPV strains of the different sublineages with variable sensitivities [Maertzdorf et al., 2004; Sarasini et al., 2006]. The assay on the F gene presented a higher sensitivity (100×) than the L assay for the four subgroups. The gain in sensitivity obtained with the second pair over the first PCR was probably sufficient to allow the detection of metapneumovirus in children presenting to the hospital with respiratory symptoms and therefore relatively high viral loads, but some B strains may have been missed during the first two seasons.
A change in the predominant genotype was observed over the years as well as a change in subgroups. Within genotypes, subgroup A2 and B2 accounted for most cases. A1 cases were detected in the early years, mainly 2001–2002 after which only a few A1 cases were reported worldwide in 2003–2004 [Ludewick et al., 2005; Mackay et al., 2006; Huck et al., 2006b; Rafiefard et al., 2008]. A1 cases were no longer reported from 2004 to 2006 [Mackay et al., 2006; Matsuzaki et al., 2008]. Concurrently, subgroup B1 emerged to become the predominant subgroup in 2004–2005 in South Africa [Ludewick et al., 2005], Australia [Mackay et al., 2006], Japan [Matsuzaki et al., 2008], and France as shown in this study. This underlines the fact that the prevalence of B1 was similar worldwide. Similar observations on changes in predominant circulating strains have led to suggestions that there is evasion of pre‐existing immunity [Ludewick et al., 2005; Agapov et al., 2006], which can be attributed to changes in immunity of the population in response to antigenic differences. Despite the stability of lineages over the years, a region of considerable variation (amino acids 260–300) on the fusion protein, analogous to an major antigenic region of the RSV fusion protein, has been identified [Yang et al., 2009]. This relative instability could be a way for the virus to escape the immune response. However, we cannot exclude the possibility that the dynamics of the virus population may vary from one genotype to another and from one year to another depending on environmental factors.
Within lineage A2, two clusters, designated A2a and A2b, have been identified [Huck et al., 2006a,b], with approximately 70% of the strains detected from 2002 to 2004 classified as A2b. Distinction between A2b and A2a strains was reported during one season in Croatia 2005/2006 [Ljubin‐Sternak et al., 2008]. In our study, RT‐PCR analysis of the F fragment gene used from 2004 to 2009 allowed us to show that A2a and A2b were prevalent during this period and co‐circulated. A majority of isolates belonged to subgroup A2b. At the end of the study, one isolate was on a separate branch from A2a and A2b. It may represent a new variant within genotype A and may signify an attempt of the virus to escape the current immune response of the population.
In the present study, we sought to determine whether the hMPV genotype was associated with disease severity. We did not find any major differences in the clinical pattern of hMPV infection in children <3 for each of the genotypes of hMPV suggesting that variations in viral genotype do not appear to be a major causal factor of differences in severity of the illness. Abnormalities on chest radiographs have been shown to occur frequently in children infected with hMPV [Esper et al., 2003; Williams et al., 2004; Wilkesmann et al., 2006]; in the present study, we showed a higher frequency for genotype B.
Three teams have screened a high number of isolates to look for correlations between genotype and disease severity in small children. None reported any significant differences between clinical data according to genotypes [Agapov et al., 2006; Sloots et al., 2006; Vicente et al., 2006]. However, a difference in the severity index as well as in the diagnosis of pneumonia was reported [Vicente et al., 2006], suggesting that hMPV genotype A may be more pathogenic than hMPV genotype B. Although we studied the same population, that is children <3 years old, and applied the same severity index as described by Vicente, we did not find a correlation between genotype and severity score. The conclusion of these studies, which included a large number of strains was that there is no clear link between viral genotype and susceptibility to more severe hMPV infection. Our study was based on metapneumovirus, and most of the main causes of coinfections, namely RSV, Flu, and PI, were excluded. We did not test for human bocavirus, adenoviruses, coronavirus, and rhinoviruses. We probably missed some co‐infections, but the clinical impact of viral co‐infection involving hMPV and these viruses is unclear. Sloots et al. 2006 found no significant difference in disease severity between hMPV infected patients with a coinfection and those without. Only hMPV‐RSV dual infections are thought to be more severe [Semple et al., 2005].
An association between hMPV infection and asthma has been reported by some authors who suggested that hMPV is a causative agent of acute wheezing in young children [Jartti et al., 2002; von Linstow et al., 2004; Manoha et al., 2007]. We did not find any difference in asthma between children infected with hMPV genotype A and children infected with genotype B. However, when we compared the four subgroups, asthma was only observed in patients with subgroup A2 and B2. Rhinoviruses are frequently associated with LRTD and asthmatic exacerbations. We found no significant difference regarding asthma according to genotype, but we cannot exclude the possibility that the presence of another virus, in particular rhinovirus, contributed to this clinical presentation. Recently, Matsuzaki et al. 2008 compared the clinical characteristics of A2, B1, and B2 hMPV infections for 85 outpatients under 15 years old over three seasons and showed that wheezing was statistically less frequent in A2 than in B1 and B2. They also reported a higher frequency of laryngitis in children with B1.
Our study included samples collected over several seasons, which comprehensively characterize the clinical presentation of hMPV infection in children who present to hospital emergency departments. However, no clear relationship between one hMPV genotype and disease severity has been established. One possibility could be that the virulence is maintained by evolution in subgroup predominance, leading to progression from rather mild infections often due to the A1 subgroup to lower RTI mainly due to A2 and B2. However, other factors such as the viral load [Bosis et al., 2008] may be involved in disease severity. Investigation of a large panel of isolates as well as predicting factors of illness severity are required to gain insights into the molecular epidemiology of hMPV and viral pathogenesis.
Acknowledgements
This work was supported by the Centre Hospitalier Universitaire of Dijon. We thank Jean‐Baptiste Bour for helpful comments, Emmanuel Florentin for statistics on virology laboratory data and Philip Bastable for editorial assistance.
Authors have no conflict of interest related to this article.
REFERENCES
- Agapov E, Sumino KC, Gaudreault‐Keener M, Storch GA, Holtzman MJ. 2006. Genetic variability of human metapneumovirus infection: Evidence of a shift in viral genotype without a change in illness. J Infect Dis 193: 396–403. [DOI] [PubMed] [Google Scholar]
- Biacchesi S, Skiadopoulos MH, Boivin G, Hanson CT, Murphy BR, Collins PL, Buchholz UJ. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315: 1–9. [DOI] [PubMed] [Google Scholar]
- Boivin G, Abed Y, Pelletier G, Ruel L, Moisan D, Cote S, Peret TC, Erdman DD, Anderson LJ. 2002. Virological features and clinical manifestations associated with human metapneumovirus: A new paramyxovirus responsible for acute respiratory‐tract infections in all age groups. J Infect Dis 186: 1330–1334. [DOI] [PubMed] [Google Scholar]
- Bosis S, Esposito S, Osterhaus AD, Tremolati E, Begliatti E, Tagliabue C, Corti F, Principi N, Niesters HG. 2008. Association between high nasopharyngeal viral load and disease severity in children with human metapneumovirus infection. J Clin Virol 42: 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cane PA, van den Hoogen BG, Chakrabarti S, Fegan CD, Osterhaus AD. 2003. Human metapneumovirus in a haematopoietic stem cell transplant recipient with fatal lower respiratory tract disease. Bone Marrow Transplant 31: 309–310. [DOI] [PubMed] [Google Scholar]
- Esper F, Boucher D, Weibel C, Martinello RA, Kahn JS. 2003. Human metapneumovirus infection in the United States: Clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics 111: 1407–1410. [DOI] [PubMed] [Google Scholar]
- Esper F, Martinello RA, Boucher D, Weibel C, Ferguson D, Landry ML, Kahn JS. 2004. A 1‐year experience with human metapneumovirus in children aged <5 years. J Infect Dis 189: 1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey AR, Erdman D, Anderson LJ, Walsh EE. 2003. Human metapneumovirus infections in young and elderly adults. J Infect Dis 187: 785–790. [DOI] [PubMed] [Google Scholar]
- Gerna G, Campanini G, Rovida F, Sarasini A, Lilleri D, Paolucci S, Marchi A, Baldanti F, Revello MG. 2005. Changing circulation rate of human metapneumovirus strains and types among hospitalized pediatric patients during three consecutive winter‐spring seasons. Brief report. Arch Virol 150: 2365–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck B, Egger M, Bertz H, Peyerl‐Hoffman G, Kern WV, Neumann‐Haefelin D, Falcone V. 2006a. Human metapneumovirus infection in a hematopoietic stem cell transplant recipient with relapsed multiple myeloma and rapidly progressing lung cancer. J Clin Microbiol 44: 2300–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck B, Scharf G, Neumann‐Haefelin D, Puppe W, Weigl J, Falcone V. 2006b. Novel human metapneumovirus sublineage. Emerg Infect Dis 12: 147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jartti T, van den Hoogen B, Garofalo RP, Osterhaus AD, Ruuskanen O. 2002. Metapneumovirus and acute wheezing in children. Lancet 360: 1393–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn JS. 2006. Epidemiology of human metapneumovirus. Clin Microbiol Rev 19: 546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljubin‐Sternak S, Santak M, Cepin‐Bogovic J, Bace A, Vojnovic G, Mlinaric‐Galinovic G, Forcic D, Drazenovic V, Falsey AR. 2008. Detection of genetic lineages of human metapneumovirus in Croatia during the winter season 2005/2006. J Med Virol 80: 1282–1287. [DOI] [PubMed] [Google Scholar]
- Ludewick HP, Abed Y, van Niekerk N, Boivin G, Klugman KP, Madhi SA. 2005. Human metapneumovirus genetic variability, South Africa. Emerg Infect Dis 11: 1074–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IM, Bialasiewicz S, Jacob KC, McQueen E, Arden KE, Nissen MD, Sloots TP. 2006. Genetic diversity of human metapneumovirus over 4 consecutive years in Australia. J Infect Dis 193: 1630– 1633. [DOI] [PubMed] [Google Scholar]
- Maertzdorf J, Wang CK, Brown JB, Quinto JD, Chu M, de Graaf M, van den Hoogen BG, Spaete R, Osterhaus AD, Fouchier RA. 2004. Real‐time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J Clin Microbiol 42: 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoha C, Espinosa S, Aho SL, Huet F, Pothier P. 2007. Epidemiological and clinical features of hMPV, RSV and RVs infections in young children. J Clin Virol 38: 221–226. [DOI] [PubMed] [Google Scholar]
- Manoha C, Bour JB, Pitoiset C, Darniot M, Aho S, Pothier P. 2008. Rapid and sensitive detection of metapneumovirus in clinical specimens by indirect fluorescence assay using a monoclonal antibody. J Med Virol 80: 154–158. [DOI] [PubMed] [Google Scholar]
- Martinello RA, Chen MD, Weibel C, Kahn JS. 2002. Correlation between respiratory syncytial virus genotype and severity of illness. J Infect Dis 186: 839–842. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Itagaki T, Abiko C, Aoki Y, Suto A, Mizuta K. 2008. Clinical impact of human metapneumovirus genotypes and genotype‐specific seroprevalence in Yamagata, Japan. J Med Virol 80: 1084–1089. [DOI] [PubMed] [Google Scholar]
- O'Gorman C, McHenry E, Coyle PV. 2006. Human metapneumovirus in adults: A short case series. Eur J Clin Microbiol Infect Dis 25: 190–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiefard F, Yun Z, Orvell C. 2008. Epidemiologic characteristics and seasonal distribution of human metapneumovirus infections in five epidemic seasons in Stockholm, Sweden, 2002–2006. J Med Virol 80: 1631–1638. [DOI] [PubMed] [Google Scholar]
- Robinson JL, Lee BE, Bastien N, Li Y. 2005. Seasonality and clinical features of human metapneumovirus infection in children in Northern Alberta. J Med Virol 76: 98–105. [DOI] [PubMed] [Google Scholar]
- Sarasini A, Percivalle E, Rovida F, Campanini G, Genini E, Torsellini M, Paolucci S, Baldanti F, Marchi A, Grazia Revello M, Gerna G. 2006. Detection and pathogenicity of human metapneumovirus respiratory infection in pediatric Italian patients during a winter–spring season. J Clin Virol 35: 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple MG, Cowell A, Dove W, Greensill J, McNamara PS, Halfhide C, Shears P, Smyth RL, Hart CA. 2005. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J Infect Dis 191: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloots TP, Mackay IM, Bialasiewicz S, Jacob KC, McQueen E, Harnett GB, Siebert DJ, Masters BI, Young PR, Nissen MD. 2006. Human metapneumovirus, Australia, 2001–2004. Emerg Infect Dis 12: 1263–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumino KC, Agapov E, Pierce RA, Trulock EP, Pfeifer JD, Ritter JH, Gaudreault‐Keener M, Storch GA, Holtzman MJ. 2005. Detection of severe human metapneumovirus infection by real‐time polymerase chain reaction and histopathological assessment. J Infect Dis 192: 1052–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, van Doornum GJ, Fockens JC, Cornelissen JJ, Beyer WE, de Groot R, Osterhaus AD, Fouchier RA. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J Infect Dis 188: 1571–1577. [DOI] [PubMed] [Google Scholar]
- van den Hoogen BG, Herfst S, Sprong L, Cane PA, Forleo‐Neto E, de Swart RL, Osterhaus AD, Fouchier RA. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis 10: 658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente D, Montes M, Cilla G, Perez‐Yarza EG, Perez‐Trallero E. 2006. Differences in clinical severity between genotype A and genotype B human metapneumovirus infection in children. Clin Infect Dis 42: e111–e113. [DOI] [PubMed] [Google Scholar]
- von Linstow ML, Larsen HH, Eugen‐Olsen J, Koch A, Nordmann Winther T, Meyer AM, Westh H, Lundgren B, Melbye M, Hogh B. 2004. Human metapneumovirus and respiratory syncytial virus in hospitalized Danish children with acute respiratory tract infection. Scand J Infect Dis 36: 578–584. [DOI] [PubMed] [Google Scholar]
- Wilkesmann A, Schildgen O, Eis‐Hubinger AM, Geikowski T, Glatzel T, Lentze MJ, Bode U, Simon A. 2006. Human metapneumovirus infections cause similar symptoms and clinical severity as respiratory syncytial virus infections. Eur J Pediatr 165: 467–475. [DOI] [PubMed] [Google Scholar]
- Williams JV, Harris PA, Tollefson SJ, Halburnt‐Rush LL, Pingsterhaus JM, Edwards KM, Wright PF, Crowe JE, Jr. 2004. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med 350: 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JV, Martino R, Rabella N, Otegui M, Parody R, Heck JM, Crowe JE, Jr. 2005. A prospective study comparing human metapneumovirus with other respiratory viruses in adults with hematologic malignancies and respiratory tract infections. J Infect Dis 192: 1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CF, Wang CK, Tollefson SJ, Piyaratna R, Lintao LD, Chu M, Liem A, Mark M, Spaete RR, Crowe JE, Jr. , Williams JV. 2009. Genetic diversity and evolution of human metapneumovirus fusion protein over twenty years. Virol J 6: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]


