Abstract
Nonhuman primates (NHPs) play an indispensable role in biomedical research because of their similarities in genetics, physiological, and neurological function to humans. Proteomics profiling of monkey heart could reveal significant cardiac biomarkers and help us to gain a better understanding of the pathogenesis of heart disease. However, the proteomic study of monkey heart is relatively lacking. Here, we performed the proteomics profiling of the normal monkey heart by measuring three major anatomical regions (vessels, valves, and chambers) based on iTRAQ‐coupled LC‐MS/MS analysis. Over 3,200 proteins were identified and quantified from three heart tissue samples. Furthermore, multiple bioinformatics analyses such as gene ontology analysis, protein–protein interaction analysis, and gene‐diseases association were used to investigate biological network of those proteins from each area. More than 60 genes in three heart regions are implicated with heart diseases such as hypertrophic cardiomyopathy, heart failure, and myocardial infarction. These genes associated with heart disease are mainly enriched in citrate cycle, amino acid degradation, and glycolysis pathway. At the anatomical level, the revelation of molecular characteristics of the healthy monkey heart would be an important starting point to investigate heart disease. As a unique resource, this study can serve as a reference map for future in‐depth research on cardiac disease‐related NHP model and novel biomarkers of cardiac injury.
Keywords: bioinformatics analysis, gene‐disease associations, monkey heart, proteomics
Cardiovascular disease is one of the leading causes of mortality worldwide, but very little is known about the constituent proteins of the nonhuman primates (NHPs). Here, we determine the spatial‐resolved proteomic map of the healthy monkey heart by mass spectrometry‐based proteomics. This study will be beneficial to future research into cardiac disease‐related NHP model and novel biomarkers of cardiac injury.

1. INTRODUCTION
The heart, which composed of vessels, valves, and chambers, pumps adequate blood to organs and tissues, then provides oxygen and nutrients to the whole body, as well as assists in the removal of metabolic wastes. Periodic dilatation and contraction of the heart is caused by a complicated interaction between biological electrocardiac signals and mechanical forces (Kwon et al., 2016). The heart vessels consist largely of endothelium, vascular smooth muscle and adventitia. The four valves open or close precisely on differential blood pressure, which is mainly made up of endothelial cells (ECs) and interstitial cells (ICs; Liu, Joag, & Gotlieb, 2007; Taylor, Batten, Brand, Thomas, & Yacoub, 2003). The atria and ventricles are divided by valves to prevent blood reflux, and the principal cell type of the heart chambers is cardiomyocytes (CMs). Besides, cardiac fibroblasts (CFs) produce the extracellular matrix (ECM) scaffold of the heart and are estimated to constitute more than half of total heart cells (Bergmann et al., 2015).
Cardiovascular disease is one of the leading causes of mortality. The inflammatory reaction in the vessel walls is working to promote atherosclerosis (Bryan et al., 2014; Gimbrone & Garcia‐Cardena, 2016; McAlpine & Swirski, 2016). Valve disorder, especially mitral valve disease, could contribute to cardiac dysfunction, and ultimately lead to heart failure (Levine et al., 2015). Atrial and ventricular hypertrophy can also cause pathological conditions, including hypertension and valvular insufficiency (Angeli, Reboldi, & Verdecchia, 2014; Bekeredjian & Grayburn, 2005; Cuspidi, Negri, Tadic, Sala, & Parati, 2014).
So far, molecular mechanisms in several cardiac diseases are relatively unknown. Proteomic technology, which aimed to reveal intrinsic relationships between genotype and phenotype, provides useful tools for the diagnosis and treatment of heart‐related diseases. A draft of the human proteome assembled from human tissues, cell lines, and body fluids helps us to enhance the understanding of the human body at the molecular level (Kim et al., 2014; Wilhelm et al., 2014). Recently, the protein composition of the dissimilar regions and cell types of the healthy human heart has been revealed, and offer a valuable reference for future research aimed to uncover biomarkers indicative of cardiac compartments deterioration (Doll et al., 2017). Transcriptomic (Ounzain et al., 2015), proteomic (Lau et al., 2016; Lu, Sinha, Sharma, Kislinger, & Gramolini, 2014; Xiang et al., 2013), and phosphoproteomic (Lundby & Olsen, 2013; Peng et al., 2014) studies have also been applied to the analysis of mammalian hearts, and provide attractive candidate biomarkers for the clinical study.
Nonhuman primates (NHPs) play an important role in biomedical research because of their similarities in genetics, physiological, and neurological function to humans. The differential proteins of serum and heart tissue in NHP models associated with cardiac disorder provide significant insight to continue the cardiac biomarker discovery and understand the cellular mechanism of drug‐induced heart injury (Liu et al., 2013; Song et al., 2014). At the anatomical level, the revelation of molecular characteristics of the monkey heart, particularly the healthy state monkey heart, would be an important starting point to investigate heart disease. But, the study about protein composition in dissimilar regions of the monkey heart is relatively less.
Here, we conducted a systematic quantitative proteomics analysis on monkey heart proteins from three compartments, including vessels, valves, and chambers. The high‐sensitivity sample preparation, peptide fractionation, and an advanced iTRAQ‐coupled LC‐MS workflow were used to quantify a total of more than 3,200 proteins. Our results establish proteomic and functional differences between normal monkey heart compartments and pinpoint potential cardiac diseases‐related proteins in different regions.
2. MATERIALS AND METHODS
2.1. Animal information and tissue collection
Three non‐naive rhesus macaques (male) with ages ranging from 4 to 6 years were assigned to the study. The selected animal was killed under deep anesthesia with pentobarbital. Then the heart with proximal aortic artery and pulmonary artery connected was excised. The study was approved by the Ethics Committee on Animal Experimentation of the Hunan Normal University (Protocol 259/2012) and the experiments were carried out according to the ethical guidelines for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006). The heart anatomy of rhesus monkey was previously reported to be similar to that of human (Sleeper, Gaughan, Gleason, & Burkett, 2008). The harvest of vessels, valves, and myocardial tissues was done by an experienced cardiologist under the assistance of a veterinary. At first, discreetly isolated coronary vessels ran at the surface of heart from myocardium and then harvested them. After that, opened the left atrium (LA) and the left ventricular (LV) wall was dissected starting at the anterolateral commissure to harvest LA and LV myocardial tissues, and anterior, and posterior mitral leaflets. The similar process was performed for the right heart to collect the right atrium and ventricular myocardial tissues. The chordae tendineae consisted in atrioventricular valves (mitral and tricuspid) was not to be taken as the valve samples, as its distinctive organizational structure and cell types differed from semilunar aortic and pulmonary valves. Finally, the rest of valves were obtained after opening the aorta and pulmonary artery. Besides, the heart was identified with normal size and structure and without heart disease, that is, no signs of hypertrophy, atrophy or scar of myocardium, thickening, shortening, or stiffening of valves and atherosclerosis of great vessels, by using visual inspection of the gross appearance of explanted tissue by the cardiologist during the tissue harvest process. All tissues were snap frozen in liquid nitrogen and stored at −80°C until analysis.
2.2. Sample preparation
Tissue samples were washed in phosphate‐buffered saline (PBS) for three times and ground in liquid nitrogen. The powdered tissues were resuspended dissolved in lysis buffer (7 M urea, 2 M thiourea, 65 mM dithiothreitol, and 0.1 mM phenylmethylsulfonyl fluoride) and ultra‐sonicated for 3 min followed by dissolving for 1 hr at 4°C. After centrifugation at 14,000 × g for 1 hr at 4°C, the supernatant was collected and the protein concentration was determined by the 2D Quantification Kit (Amersham Biosciences, Freiburg, Baden‐Württemberg, Germany).
Trypsin digestion and iTRAQ labeling were performed according to the manufacturer's protocol (Sicex, Framingham, MA). In brief, 100 μg proteins of each sample were reduced, alkylated and precipitated using acetone and resuspended using 50 mM triethylammonium bicarbonate (TEAB), pH 8.5. Then the protein samples were digested by trypsin (Promega, Madison, WI) overnight at 37°C. The samples were subsequently labeled with iTRAQ reagents as following; Vessel (MO1) with 115 tags; Vessel (MO2) with 116 tags; Vessel (MO3) with 116 tags; Mix (MO1–3) with 114 tags. And, the same tag method was used in the other two tissues (valve and chamber). A total of six sets of iTRAQ analysis (per regional sample was repeated two times) were constructed in each monkey. Each set of iTRAQ labeled peptides was mixed, dried, and desalted for further analysis.
2.3. 2D‐LC‐MS/MS analysis and database searching
The labeled peptides were diluted with the loading buffer (5 mM ammonium formate containing 2% acetonitrile, pH 10) and fractionated by using high pH reversed phase liquid chromatography on a UPLC system (Waters, Milford, MA). A C‐18 column (2.1 × 250 mm X Bridge BEH300) was used for the separation. The gradient elution was performed by 0–25% B (5 mM ammonium formate containing 98% acetonitrile, pH 10, 5–35 min) and 25–45% B (35–48 min) on high pH RPLC column (Waters, Xbridge C18 3.5 µm, 150 × 2.1 mm, Milford, MA) with a flow rate of 300 µl/min. A total of 20 fractions were collected which were then combined to 10 fractions.
Each fraction was dried, dissolved in buffer A (0.1% FA, H2O) and analyzed on a LC‐MS/MS platform which was equipped with an Orbitrap Fusion™ mass spectrometry (Thermo Fisher Scientific, San Jose, CA) and a nano Easy‐Spray (Thermo Fisher Scientific, San Jose, CA). Peptides were separated with a linear gradient from 5% B (98% ACN with 0.1% formic acid) to 40% B in 90 min within a total analysis of 120 min. Orbitrap Fusion™ mass spectrometer was operated in the data‐dependent mode to switch automatically between MS and MS/MS acquisition. Survey full‐scan MS spectra (m/z 300–1200) were acquired with a mass resolution of 70 K, followed by 10 sequential high energy collisional dissociation (HCD) MS/MS scans (100–2000) with a resolution of 17.5 K. The dynamic exclusion time was set as 30 s. For MS/MS, precursor ions were activated using 27% normalized collision energy.
All the raw data were submitted to Proteome Discoverer software (version 1.3, Thermo scientific, Sunnyvale, CA) using the MASCOT search engine for protein identification and quantitation. The data was search against rhesus Uniprot database with 161,948 entries. Parameters searching were set as following: Quantitation: iTRAQ‐8plex; enzyme: Trypsin; variable modification: Oxidation (Met); fixed modification: Methyl methanethiosulfonate (Cys). MS1 tolerance: 20 ppm; MS/MS tolerance: 20 mmu; with one missed cleavage on trypsin was allowed. The false discovery rate (FDR) of 1% was used to control protein level identification based on the target‐decoy strategy. The peptides which were unique to the given proteins and with a confidence higher than 95% were used for the calculation of the protein ratios.
2.4. Functional annotation and enrichment analysis
Based on cytoscape platform, the plugin ClueGO in conjunction with CluePedia created a dynamic and visualized network to analyze biological functions and signaling pathways of monkey cardiac proteins. All the significant pathways or terms with a statistical threshold (p < 0.05) were functionally grouped and highlighted with different colors (Bindea et al., 2009; Bindea, Galon, & Mlecnik, 2013). The gene lists were imported into the Cytoscape plugins to match the Macaca mulatta species [9,544 genes] database and enriched in specific terms with diverse functional analyses (including the biological process [BP], molecular function [MF], and Kyoto Encyclopedia of Genes and Genomes [KEGG] analysis). The topological information was presented in charts and tables. Annotations of cellular component (CC) were based on the analysis using the DAVID online software (https://david.ncifcrf.gov/; Huang, Sherman, & Lempicki, 2009a, 2009b).
2.5. Signaling pathway enrichment analysis
The Reactome database can be obtained from http://www.reactome.org/, which comprises a set of core modules called the reaction (Fabregat et al., 2018). Entities (nucleic acids, proteins, complexes, etc.) participating in reactions form a network of biological interactions and are grouped into several pathways like classical intermediary metabolism, signaling, transcriptional regulation, apoptosis, and disease (Matthews et al., 2009). To illuminate the correlation between monkey heart genes and human pathways, we imported the healthy monkey cardiac gene list into Reactome online tools for pathway analysis.
2.6. Protein–protein interaction analysis and network reconstruction
The STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) online engine, which aimed to assemble, evaluate, and disseminate protein–protein association information, was used to estimate the interactions between proteins in each area (Matthews et al., 2009). To narrow down the lists and enhance the readability of the result, we merely selected the core proteins which were contained in principal biological processes. Be worth what carry is, the STRING data lists were imported into Cytoscape for network reconstruction and visualization, and were emphasized by the level of the combined score. Furthermore, the interaction information between proteins in dissimilar heart regions was presented in Figure 5 and Supporting Information S3–4.
Figure 5.
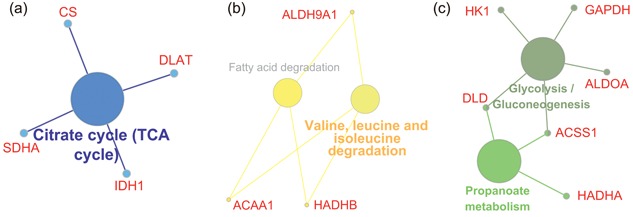
Regional signaling pathway analysis of the genes associated with heart diseases. (a) The genes related to heart diseases in cardiac vessels such as CS, SDHA, IDH1, and DLAT worked together to regulate TCA cycle pathway; (b) The disease‐related genes in valves, like ACAA1, ALDH9A1, and HADHB, performed their functions in amino acid and fatty acid degradation pathway; (c) The genes in chambers were mainly involved in multiple pathways including glycolysis and propanoate metabolism pathway. TCA: citrate cycle [Color figure can be viewed at wileyonlinelibrary.com]
2.7. Gene‐disease association analysis
To better understand the association between cardiac genes and diseases, the core gene lists from protein–protein association analysis in each heart region above were put through the DisGeNET database (http://www.disgenet.org/web/DisGeNET/menu/home) for gene‐disease association analysis. The associations are all supported by experimental evidence or previous publications. The associations from the DisGeNET are all supported by expert curated repositories, GWAS catalogs, animal models, and the scientific literature (Pinero et al., 2017). Then, we mapped our lists to the knowledge base (more than 500 thousand gene‐disease associations [GDAs]), so that all the GDAs could be extracted; one gene may be involved in several diseases, and one disease may include numerous genes. Finally, the cardiovascular disorder‐related genes in each heart region were further enriched by signaling pathways involved.
2.8. Statistical analysis
To ensure the reliability of the reported data acquired from LC‐MS/MS, we performed the following steps for data quality control. First, 1% FDR was performed to control protein identification and quantification before mass spectra data retrieval. Second, unused ≥ 1.3 was set to ensure the confidence of their proteins higher than 95% after database searching. Third, we in turn removed the results identified by reverse database, the proteins with extremely high or low ratios, and the proteins with abnormal quantification between biological repetition and technical repetition. Statistical significance of DAVID bioinformatics enrichment analysis was assumed when p‐values were < 0.05. Meanwhile, multiple statistical values were estimated for the purpose of enrichment evaluation in each term, such as count and percentage of enriched genes, p‐value, fold enrichment score, Bonferroni value, and false discovery rate (FDR).
3. RESULTS
3.1. Establishing a proteomic draft of the normal monkey heart
Three anatomically compartments from the normal monkey heart were separated for MS analysis: The vessels, valves, and chambers. To obtain accurate proteome information, all the sample preparation was performed by reducing sample loss and increasing quantification accuracy after tissue homogenization in liquid nitrogen. The workflow of this study was presented in Figure 1a, including experimental design (upper panel), protein sample collection, and bioinformatics analysis (lower panel). A total of 3,226 proteins were identified and quantified from all three heart tissue samples. And, the number of identified proteins in each region ranged from 1,600 to 2,100 (Figure 1b). Over 700 identified proteins were coexpressed in three heart compartments. Besides, a larger number of proteins (486–806) were found to be unique proteins of three regions (Figure 1c). The quantitative information of the data set was included in the Supporting Information Supplementary File 1.
Figure 1.
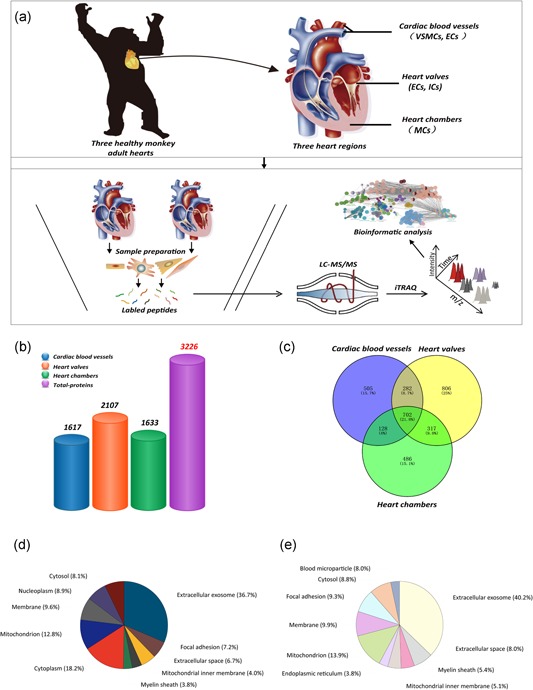
The schematic and protein distributions of the monkey heart proteome. (a) Schematic depiction of cardiac proteomic analysis of the normal monkey heart tissues, including experimental design (upper panel), protein sample collection, and bioinformatics analysis (lower panel); (b) Bar plot comparing the total number of cardiac proteins in dissimilar heart regions; (c) Venn diagram showing the count of respective and common proteins in each cardiac region; (d) The subcellular localization of total heart proteins in normal monkey; (e) The top 10 terms illustrating the subcellular distribution of cardiac proteins in vessels [Color figure can be viewed at wileyonlinelibrary.com]
3.2. Gene ontology (GO) analysis and signaling pathways enrichment of monkey heart proteins
To gain a better understanding of proteins from normal monkey heart, we analyzed their biological characteristics and signaling pathways enrichment (Supporting Information Supplementary File 2). For an overall assessment of subcellular distribution of total cardiac proteins, the top 10 terms with subcellular localization annotations were used by the counts of enriched genes, which represent the majority of protein distribution in monkey heart. Among all heart proteins, 36.7% were extracellular proteins, 18.2% were related to the cytoplasm, 12.8% located in mitochondrion, 9.6% functioned in the membrane, 8.9% were distributed in the nucleoplasm, and 7.1% were found in the cytosol (Figure 1d). There are a large number of proteins from all enriched terms than the number of identified proteins in heart region, suggesting that part of proteins was identified in two or more organelles. Specific details about the term, count, gene name, and statistical values of cellular component in total heart proteins are contained in Supporting Information Supplementary File 2.
As displayed in Figure 2a, all the monkey heart genes were annotated into 37 categories and over 231 functional terms of diverse biological processes (each dot represents one functional term). Meanwhile, for each term, a series of statistical values such as the count of enriched genes, p‐value and Bonferroni value were taken for the purpose of enrichment evaluation. More than half of all biological terms were focused on metabolism‐related processes, including ATP metabolic process, organonitrogen compound metabolic process, and small molecule metabolic process. Myocardial contraction‐related processes, such as heart contraction, myofibril assembly, and fibril organization, are also thought as an essential part in all biological terms. Besides, other processes like transcription processes also played a vital role in regulating heart function.
Figure 2.
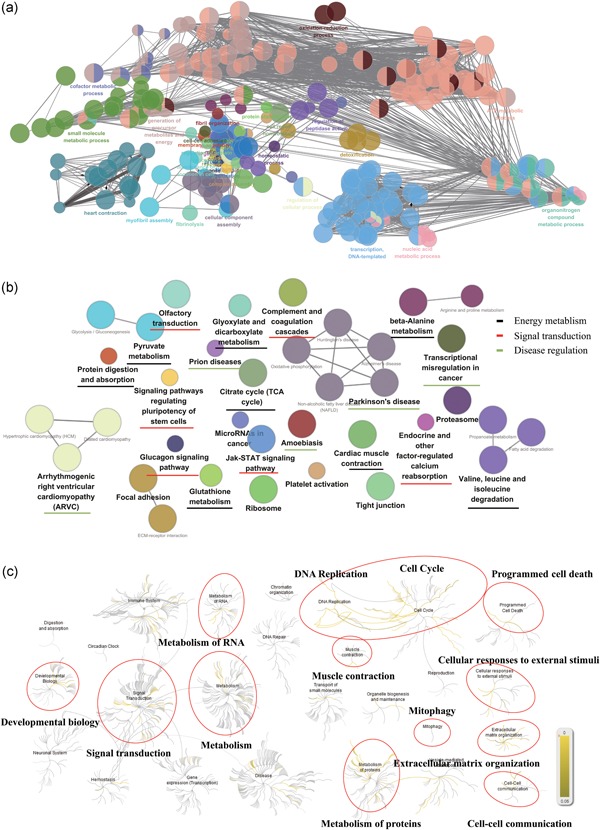
The bioinformatics analysis of the healthy monkey heart genes. (a) Enrichment network of functional terms from monkey cardiac proteome (p < 0.05). Each dot represents one functional term, and the same color dots stands for terms which have similar biological functions; (b) The pathway annotation network in total heart proteins was classified into 25 signaling pathways (p < 0.05), which mainly contain three major pathway categories including energy metabolism, signal transduction, and disease regulation; (c) Main groups and hierarchical relationship of Reactome pathways. Based on the similarity of their physiological functions, Reactome pathways were grouped into several categories [Color figure can be viewed at wileyonlinelibrary.com]
To investigate the signaling pathways involved in the corresponding proteins of monkey heart, all identified proteins were submitted to Cytoscape platform for KEGG pathway analysis and functional annotation clustering (FAC). The pathway annotation network in total heart proteins was classified into 25 signaling pathways. Three major types of pathways involving energy metabolism, signal transduction, and disease regulation were considered as the important parts (Figure 2b and Supporting Information S2). Monkey cardiac proteins were partially enriched into multiple metabolic pathways such as carbon metabolism, fatty acid degradation, and amino acid degradation. Twenty‐two cardiac proteins were clustered into the citrate cycle (TCA cycle) pathway, which account for 65.71% of the total proteins in this signaling pathway. The genetic information associated with each pathway was provided in Supporting Information Table S1.
To further elucidate the correlation between monkey heart proteins and human pathways, Reactome pathways analysis was used on the basis of the orthologous correspondences informed from the ENSEMBL Compara database. According to physiological similarity and hierarchy, human biological pathways were classified into different groups including immune system, signal transduction, metabolism, disease, and cell cycle and hemostasis (Figure 2c). As shown in Figure 2c, 1,330 matched genes were enriched in 1,535 specific pathways, which were accompanied by several statistical values including Entities p value and Entities FDR. Based on the degree of Entities FDR, these genes were mainly involved in several pathway categories such as extracellular matrix organization, muscle contraction, and programmed cell death (Table 1). More narrowly, most of these genes are enriched in the citric acid cycle, respiratory electron transport, Complex I biogenesis, and other pathways (Supporting Information Table S2).
Table 1.
Relevant main groups of Reactome pathways involved in the normal monkey heart genes
| Pathway name | Entities found | Entities total | Entities FDR* |
|---|---|---|---|
| Extracellular matrix organization | 92 | 329 | 1.18E−3 |
| Muscle contraction | 64 | 216 | 2.73E−3 |
| Programmed cell death | 56 | 185 | 3.48E−3 |
| Organelle biogenesis and maintenance | 84 | 334 | 2.48E−2 |
| Cellular responses to external stimuli | 132 | 598 | 1.19E−1 |
| Mitophagy | 10 | 29 | 2.64E−1 |
| Cell‐cell communication | 33 | 133 | 3.17E−1 |
| DNA replication | 30 | 122 | 3.59E−1 |
| Cell cycle | 119 | 680 | 7.38E−1 |
| Developmental biology | 215 | 1,150 | 7.38E−1 |
| Metabolism of proteins | 422 | 2,325 | 7.38E−1 |
| VesiclE‐1mediated transport | 143 | 827 | 7.65E−1 |
| Metabolism of RNA | 134 | 782 | 7.91E−1 |
| Reproduction | 19 | 123 | 7.98E−1 |
| Hemostasis | 129 | 801 | 9.39E−1 |
| Circadian clock | 9 | 99 | 9.93E−1 |
| Chromatin organization | 21 | 257 | 10.00E−1 |
| Digestion and absorption | 1 | 71 | 10.00E−1 |
| Disease | 206 | 1,503 | 10.00E−1 |
| DNA repair | 34 | 348 | 10.00E−1 |
| Gene expression (Transcription) | 136 | 1,686 | 10.00E−1 |
| Immune system | 387 | 2,437 | 10.00E−1 |
| Metabolism | 554 | 3,458 | 10.00E−1 |
| Neuronal system | 37 | 435 | 10.00E−1 |
| Signal transduction | 289 | 3,024 | 10.00E−1 |
| Transport of small molecules | 132 | 963 | 10.00E−1 |
Note: FDR, false discovery rate.
The results are sorted by the value of Entities FDR in each pathway group, which are thought to reflect the degree of the correlation between the input gene list and this pathway.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.3. Biological characteristics of spatial‐specific proteins
Gene Ontology (GO) analysis of unique proteins in each region was used to elucidate their characteristic functions of heart tissue (Supporting Information Supplementary File 3). The contribution of main organelles to the amount of cellular protein in each region is similar to the cellular component in total heart proteins. Similar to the cellular component in total heart proteins, more than half of all spatial‐specific cardiac proteins were expressed in extracellular exosome, mitochondrion, plasma membrane, cytosol, and endoplasmic reticulum (Figure 1e and Supporting Information S1a,b).
Proteomics analysis of heart vessels mainly detected the genes among aorta and pulmonary artery that does not distinguish between the concave and convex areas. In heart vessels, the region‐specific proteins were enriched in multiple molecular functions categories such as oxidoreductases activity, RNA binding, cytoskeletal protein binding, and cell adhesion molecule binding, and were distributed on 12 specific terms (Figure 3a–c). Further, the BP enrichment network of heart vessel‐specific proteins was displayed in Figure 3d. These proteins were implicated in numerous biological functions including aerobic respiration, cellular component assembly, generation of precursor metabolite and energy, and organic acid metabolic process (Supporting Information Table S3). For example, up to 41 vessel‐expressed genes are involved in regulation of oxidation–reduction process in vivo, including UQCRC2, EIF6, ALDOC, and ADSSL1. In addition, ALDOC, EIF6, and UQCRC2 have the potential to play a significant role in multiple metabolic processes involving nucleoside monophosphate, ribonucleoside triphosphate, and ATP metabolic process.
Figure 3.

GO analysis of unique genes from dissimilar heart regions (p < 0.05). (a) The count of vessel‐specific genes in monkey heart was highlighted in red; (b) The enrichment of molecular functions in vessel‐specific genes; (c) Histogram plot displaying the terms and number of vessel‐specific genes in monkey heart; (d) The enriched biological processes network of heart vessel‐specific genes; (e–h) The number (e), molecular function categories (f) and terms (g), and biological processes (h) of heart valves‐specific proteins were showed, respectively; (i–l) Similar GO visualized network specific for monkey heart chambers‐related genes were also displayed including the number (i), molecular function categories (j) and terms (k), and biological processes (l). GO: Gene Ontology [Color figure can be viewed at wileyonlinelibrary.com]
As shown in Figure 3e–g, the unique proteins in cardiac valves were focused on multiple MF categories involving oxidoreductases activity, RNA binding, cytoskeletal protein binding, and cell adhesion molecule binding. Our BP analysis shows that most valves‐specific proteins contributed to metabolic processes (Supporting Information Table S4). Cellular component assembly, protein activation cascade and organelle organization processes were also prominent in heart valves (Figure 3h). At the genetic level, over 100 genes specifically expressed in heart valves are associated with regulation of organelle organization, 75 genes with proteolysis process, and 132 genes with metabolic processes.
In atria and ventricles, the molecular functions of specific proteins mainly were involved in activate various enzymes, and bind related molecules and complexes (Figure 3i–k). Furthermore, BP analysis of the chambers‐specific proteins highlighted the characteristic processes about multiple generating energy and metabolism, response to oxidative stress, and protein complex subunit organization (Figure 3l and Supporting Information Table S5). Among these, up to 20 metabolism‐related biological processes and 83 chambers‐specific genes are enriched in the Group 7.
3.4. Biological network reconstruction and visualization of region‐specific proteins
The forming protein–protein interactions network is rather intricate because of the counts of unique proteins in each region varied from 400 to 600. To narrow down the list and enhance the readability of the result, the specific proteins of each region were categorized by diverse biological processes in which these proteins participate. The target gene lists, derived from diverse biological processes in each heart region, were imported into STRING and Cytoscape for protein–protein association network analysis, reconstruction, and visualization. As showed in Figure 4a–e and Supporting Information S3–4, over 20 specific protein interaction networks with diverse biological processes were generated. There is a closely relationship between proteins in the same biological process. For example, the relativity of vessel‐specific proteins enriched in aerobic respiration was showed in Figure 4a. Among these genes, the genes with a large size, like Dlat, Sdha, and Uqcrc2, occupy an important position in aerobic respiration. But, other genes with a bold line, including Ndufab1, Ndufv1, Ndufa10, mt‐Nd5, and Uqcrc2, have close relation with each other. Similar to Figure 4a, the vessel‐expressed proteins, such as Itgb1, Fn1, and Kdr, remains a pivotal agent in cellular component assembly with a weaker interaction, whereas the generation of precursor metabolite and energy‐regulated protein Idh1, Idh3a, and Aco2 are important factors in this process with an intricate association each other.
Figure 4.
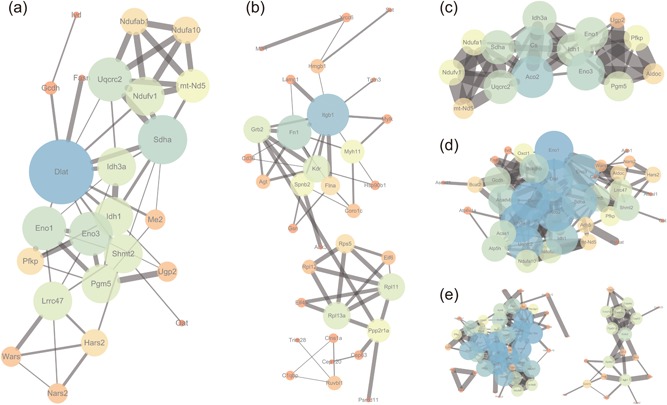
Protein–protein interactions network simplification and reconstruction of vessel‐specific proteins based on different biological processes. The relativity between proteins was expressed by the size of the dot and the thickness of the line based on the combined score. (a–e) Vessel‐specific proteins interaction networks were generated by enriched biological processes including aerobic respiration (a), cellular component assembly (b), generation of precursor metabolite and energy (c), organic acid metabolic process (d), and oxidation–reduction process (e) [Color figure can be viewed at wileyonlinelibrary.com]
3.5. Monkey cardiac genes involved in heart disorders
To gain deeper insights into the association between cardiac genes and diseases, we intentionally investigated the crucial gene from protein–protein interaction networks above which are most likely to reflect the biological functions in each heart region. A total of 5,522 GDAs were obtained by matching the core gene lists to the entire GDA data set (Supporting Information Supplementary File 4). Among these associations, many diseases such as cardiovascular diseases, nutritional and metabolic diseases and endocrine system diseases were highly associated with the physiology and pathology of different heart regions.
From pathophysiological perspective, it would be desirable to further elucidate the relevance between monkey heart genes and cardiac‐related illnesses. As displayed as Table 2, more than 66 genes related to heart diseases (17, 26, and 23 genes in vessels, valves and chambers, respectively) were described in three dissimilar heart regions, which are involved in several cardiac‐related illnesses including hypertrophic cardiomyopathy, heart failure, and myocardial infarction.
Table 2.
Correlation analysis between region‐specific genes and cardiac‐related diseases
| Gene* | Gene full name | Cardiac‐related diseases ** | Region | |
|---|---|---|---|---|
| ACADVL * | Acyl‐CoA Dehydrogenase, Very Long Chain | Hypertrophic cardiomyopathy; cardiac arrest; cardiomyopathies; sudden cardiac death; | Vessel | |
| ALDH1B1 | Aldehyde Dehydrogenase 1 Family Member B1 | Coronary heart disease; coronary artery disease | Vessel | |
| BCKDHB | Branched Chain Keto Acid Dehydrogenase E1 Subunit Beta | Multiple myeloma | Vessel | |
| CS | Citrate Synthase | Cerebrovascular accident; angiomatosis, bacillary; cataract and cardiomyopathy; heart failure; cardiomyopathies | Vessel | |
| DLAT | Dihydrolipoamide S‐acetyltransferase | Myocardial Ischemia | Vessel | |
| ENO1 | Enolase 1 | Left ventricular hypertrophy; bacterial endocarditis; myocardial infarction; Behcet syndrome; acute coronary syndrome *** | Vessel | |
| FN1 | Fibronectin 1 | Myocardial infarction; left ventricular hypertrophy; increase in blood pressure; aortic valve insufficiency; hypertensive disease | Vessel | |
| IDH1 | Isocitrate Dehydrogenase (NADP(+)) 1, Cytosolic | Venous thrombosis; vascular neoplasms; hemangioma; hemangiomatosis; visceral angiomatosis | Vessel | |
| ITGB1 | Integrin Subunit Beta 1 | Multiple myeloma; myocardial infarction; cardiomyopathy, dilated; heart failure; cardiomyopathies | Vessel | |
| NDUFAB1 | NADH: Ubiquinone Oxidoreductase Subunit AB1 | Hypertensive disease; coronary artery disease | Vessel | |
| NDUFS7 | NADH:Ubiquinone Oxidoreductase Core Subunit S7 | Cardiomyopathies | Vessel | |
| NDUFV1 | NADH:Ubiquinone Oxidoreductase Core Subunit V1 | Hypertrophic cardiomyopathy | Vessel | |
| PPP2R1A | Protein Phosphatase 2 Scaffold Subunit Alpha | Heart failure; congestive heart failure | Vessel | |
| RPL11 | Ribosomal Protein L11 | Conduction disorder of the heart; electrocardiogram abnormal; electrocardiogram change; EKG abnormalities; cardiac conduction abnormalities | Vessel | |
| SDHA | Succinate Dehydrogenase Complex Flavoprotein Subunit A | Hypertrophic cardiomyopathy; left ventricular noncompaction; gastrointestinal hemorrhage; cardiomyopathy, dilated, 1GG; cardiomyopathy, dilated | Vessel | |
| TUBA1A | Tubulin Alpha 1A | Myocardial ischemia | Vessel | |
| TXNRD2 | Thioredoxin Reductase 2 | Cardiomyopathy, dilated; myocardial infarction; cardiomyopathy, familial idiopathic; diabetic retinopathy; cerebrovascular accident | Vessel | |
| ACAA1 | Acetyl‐CoA Acyltransferase 1 | Carotid atherosclerosis | Valves | |
| ACAD9 | Acyl‐CoA Dehydrogenase Family Member 9 | Cardiomyopathy, dilated; hypertrophic cardiomyopathy; heart failure; congestive heart failure; cerebrovascular accident | Valves | |
| ALDH9A1 | Aldehyde Dehydrogenase 9 Family Member A1 | Hypertensive disease | Valves | |
| C1S | Complement C1s | Hereditary angioedema types I and II | Valves | |
| CAPZB | Capping Actin Protein of Muscle Z‐line Beta Subunit | Acute coronary syndrome | Valves | |
| CCT7 | Chaperonin Containing TCP1 Subunit 7 | Myocardial infarction | Valves | |
| COPS5 | COP9 Signalosome Subunit 5 | Arteriosclerosis; atherosclerosis | Valves | |
| DENR | Density Regulated RE‐1initiation and Release Factor | Heart failure; congestive heart failure; brain infarction; idiopathic pulmonary arterial hypertension | Valves | |
| EIF2S2 | Eukaryotic Translation Initiation Factor 2 Subunit Beta | Multiple myeloma | Valves | |
| EIF3E | Eukaryotic Translation Initiation Factor 3 Subunit E | Brain ischemia; intermittent claudication | Valves | |
| FLNA | Filamin A | Heart valve disease; mitral valve prolapse syndrome; aortic valve insufficiency; heart failure; congestive heart failure | Valves | |
| TPM3 | Tropomyosin 3 | Conduction disorder of the heart; electrocardiogram abnormal; electrocardiogram change; EKG abnormalities; cardiac conduction abnormalities | Valves | |
| FXN | Frataxin | Cardiomyopathies; hypertrophic cardiomyopathy; heart failure; congestive heart failure; echocardiogram abnormal | Valves | |
| HADHB | Hydroxyacyl‐CoA Dehydrogenasebeta Subunit | Cardiomyopathy, dilated; heart diseases; heart failure; congestive heart failure; cardiomyopathy, hypertrophic, familial | Valves | |
| HSP90B1 | Heat Shock Protein 90 Beta Family Member 1 | Multiple myeloma; cerebrovascular disorders; atrial fibrillation; chronic atrial fibrillation | Valves | |
| KIF5B | Kinesin Family Member 5B | Henoch‐Schoenlein purpura | Valves | |
| LONP1 | Lon Peptidase 1, Mitochondrial | Endocardial cushion defects; atrial septal defects; ventricular septal defects; atrioventricular canal defect; heart failure | Valves | |
| MYH11 | Myosin Heavy Chain 11 | Heart failure; congestive heart failure | Valves | |
| NDUFS3 | NADH:Ubiquinone Oxidoreductase Core Subunit S3 | Cardiomyopathy, familial Idiopathic hypertrophic, 3 (disorder); cardiomyopathy, dilated, 1y; hypertrophic cardiomyopathy; cardiomyopathy, dilated; cardiomyopathy, hypertrophic, familial | Valves | |
| PFKL | Phosphofructokinase, Liver Type | Cardiomyopathy, dilated; hypertrophic cardiomyopathy | Valves | |
| PSMC5 | Proteasome 26S Subunit, ATPase 5 | Patent ductus arteriosus; hypertensive disease; aortic valve insufficiency; left‐sided heart failure; coronary artery disease | Valves | |
| RPL11 | Ribosomal Protein L11 | Hypertrophic cardiomyopathy | Valves | |
| SPTBN1 | Spectrin Beta, Non‐erythrocytic 1 | Ventricular septal defects; tetralogy of fallot | Valves | |
| TPM1 | Tropomyosin 1 (alpha) | Arteriosclerosis; atherosclerosis | Valves | |
| TPM3 | Tropomyosin 3 | Conduction disorder of the heart; electrocardiogram abnormal; electrocardiogram change; EKG abnormalities; cardiac conduction abnormalities | Valves | |
| ACADVL | Acyl‐CoA Dehydrogenase, Very Long Chain | Hypertrophic cardiomyopathy; cardiac arrest; cardiomyopathies; sudden cardiac death | Cavities | |
| ACSS1 | Acyl‐CoA Synthetase Short‐chain Family Member 1 | Cerebrovascular accident; subarachnoid hemorrhage; intracranial hemorrhages; cerebral hemorrhage | Cavities | |
| AK1 | Adenylate Kinase 1 | Myocardial ischemia; myocardial infarction; infarction, middle cerebral artery | Cavities | |
| ALDOA | Aldolase, FructosE‐1bisphosphate A | Myocardial ischemia | Cavities | |
| CMPK1 | Cytidine/Uridine Monophosphate Kinase 1 | Kartagener syndrome | Cavities | |
| DLD | Dihydrolipoamide Dehydrogenase | Hypertrophic cardiomyopathy; hypertensive disease; atrial fibrillation; atrial premature complexes | Cavities | |
| EIF4E | Eukaryotic Translation Initiation Factor 4E | Multiple myeloma; myocardial ischemia; dementia, vascular | Cavities | |
| EIF4H | Eukaryotic Translation Initiation Factor 4H | Williams syndrome; hypertensive disease; coronary artery disease | Cavities | |
| GAPDH | GlyceraldehydE‐13‐phosphate Dehydrogenase | Bacterial endocarditis; ischemic cardiomyopathy; multiple myeloma; cardiomyopathy, familial idiopathic; acute coronary syndrome | Cavities | |
| HADHA | Hydroxyacyl‐CoA Dehydrogenase, Alpha Subunit | Congestive heart failure; myocardial ischemia; cardiomyopathy, hypertrophic, familial; cardiomyopathy, dilated; cardiomyopathies | Cavities | |
| HK1 | Hexokinase 1 | Myocardial ischemia; pulmonary hypertension | Cavities | |
| HRG | Histidine Rich Glycoprotein | Thrombosis; myocardial infarction | Cavities | |
| IDH1 | Isocitrate Dehydrogenase (NADP(+)) 1, Cytosolic | Venous thrombosis; vascular neoplasms; hemangioma; hemangiomatosis; visceral angiomatosis | Cavities | |
| PDK2 | Pyruvate Dehydrogenase Kinase 2 | Cardiomyopathies | Cavities | |
| PSMB1 | Proteasome Subunit Beta 1 | Acute coronary syndrome | Cavities | |
| RPL28 | Ribosomal Protein L28 | Infarction, middle cerebral artery | Cavities | |
| RPS2 | Ribosomal Protein S2 | Turner syndrome; Gonadal Dysgenesis, 45,X | Cavities | |
| RPS9 | Ribosomal Protein S9 | Takayasu arteritis | Cavities | |
| TSFM | Ts Translation Elongation Factor, Mitochondrial | Cardiomyopathies; patent ductus arteriosus; foramen ovale, patent; cardiomyopathy, concentric hypertrophic | Cavities | |
| TXNRD2 | Thioredoxin Reductase 2 | Cardiomyopathy, dilated; myocardial infarction; cardiomyopathy, familial idiopathic; diabetic retinopathy; cerebrovascular accident | Cavities | |
| VAMP3 | Vesicle Associated Membrane Protein 3 | Coronary artery disease | Cavities |
The region‐specific genes of monkey heart enriched in protein–protein interactions above were imported to a gene‐disease association analysis.
These genes associated with heart diseases were listed and involved in one or more cardiac‐related illnesses.
We only displayed the five major heart‐related disorders for one gene when exceeding this limit.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
To illuminate potential differences of the heart diseases‐related genes in different heart regions, the gene lists mentioned above were enriched into diverse signaling pathways. The enrichment genes such as CS, SDHA, IDH1, and DLAT in cardiac vessels worked together to regulate the citrate cycle (TCA cycle) pathway (Figure 5a). And, the genes in heart valves, like ACAA1, ALDH9A1, and HADHB, were mainly functional in amino acid and fatty acid degradation pathway (Figure 5b). However, the genes in atria and ventricles were mainly involved in glycolysis and propanoate metabolism pathway (Figure 5c).
4. DISCUSSION
As the important part of studying the heart function, the region‐resolved proteomics profiling of monkey heart is comparatively lacking. In this study, we established the first spatial‐resolved proteomic map of the healthy monkey heart. A total of 3,226 proteins were cataloged in monkey heart, and conveyed multiple information for these genes, including biological characteristics, protein–protein interaction network and genotype–phenotype relationships. Among these genes, more than 60 regional‐specific genes are implicated with heart diseases such as hypertrophic cardiomyopathy, heart failure, and myocardial infarction. These genes are mainly enriched in citrate cycle, amino acid degradation, and glycolysis pathway.
Although the orthologous genes among mammals are not fully equal to the genes in human, the homology models in mammals (i.e., mouse, rat, and NHP) are widely used to deduce the function of human genes (Alessio et al., 2017; Gharib & Robinson‐Rechavi, 2011). The NHP models have long been idealized as a valid pathological model of the human disease to study the pathogenesis of various diseases such as severe acute respiratory syndrome (SARS), AIDS disease, and transthyretin (TTR) amyloidosis. The common marmoset with SARS‐associated coronavirus infection is suited for understanding SARS pathogenesis, and will allow for efficient testing of vaccines and various treatment modalities (Greenough et al., 2005). Simian immunodeficiency virus (SIV)‐infected and chimeric SIVHIV (SHIV)‐infected macaques have become a well‐established primate model to study AIDS and pulmonary arterial disease pathogenesis (George et al., 2013; Letvin et al., 1985; Voelkel, Cool, & Flores, 2008). Aged vervet monkeys have been seen as an ideal animal model of TTR amyloidosis, and have the TTR Ile122 allele, which is well known as a frequent mutation causing the leptomeningeal phenotype of human TTR amyloidosis (Ueda et al., 2012).
Besides, multiple nonhuman primate models of heart disease afford an opportunity to understand the pathology of human heart disease. Monkey model of myocardial infarction is generated by ischemia followed by reperfusion to study the extent of remuscularization of the infarcted heart by transplanting human cardiomyocyte (Chong et al., 2014). Acute myocardial infarction is also induced in monkey by left anterior descending artery ligation (Yang et al., 2011; Yoshioka et al., 2005). Adhesion molecule P‐selectin of coronary endothelium is overexpressed in a monkey heart ischemia reperfusion model, which allow stable adherence with leukocytes that provide possibility for detecting myocardial ischemia by molecular imaging of P‐selectin with targeted ultrasound contrast agents (Kaufmann et al., 2010; Kaufmann, Lewis, Xie, Mirza‐Mohd, & Lindner, 2007; Thomas et al., 2010). Sensitive proarrhythmia model establishing by a chronic atrioventricular block in cynomolgus monkey possesses essentially the same pathophysiological adaptations with human (Sugiyama, 2008). CCR5 inhibition observably impacts myocardial viral load and prevents cardiomyocyte functional impairment in SIV‐infected macaque model of diastolic dysfunction (Kelly et al., 2014). CCR5 blockade also attenuates postsurgical stress responses and favorably modulates pathogenic alloimmunity in nonhuman primate cardiac allograft model (Schroder et al., 2007). Monkey models of heart disease also serve as a suitable large‐animal model to examine diabetic cardiomyopathy (Gong et al., 2013; Qian et al., 2015), postmenopausal atherosclerosis (Melendez et al., 2015), and heart failure (Qiu et al., 2008). Interestingly, the major pathological process of heart disease is different in humans and chimpanzees, although heart disease is common in the two species (Varki et al., 2009). The commonest cause of heart disease in human arises predominantly from coronary artery atherosclerosis, whereas sudden death of cardiac etiology in chimpanzees is generally related to myocardial fibrosis (Varki et al., 2009).
Dissecting monkey heart proteins may provide a fresh perspective into understanding heart‐related biological functions. In our current study, there is a greater proportion of monkey heart proteins from extracellular exosome (36.7%), plasma membrane (9.6%), and nucleoplasm (8.9%) compared with human heart proteome (Doll et al., 2017). But, the count of monkey heart proteins and the detected monkey heart proteins in myofibril, mitochondrion, and endoplasmic reticulum less than in healthy human heart (Doll et al., 2017). In addition, the biological processes of identified proteins in normal monkey heart were classified into metabolism and myocardial contraction‐related processes. The metabolism processes detected in monkey heart consists mainly of multiple metabolic processes, which regulate the chemical reactions and pathways involving ATP, any low molecular weight molecule, and organonitrogen compound. Heart contraction processes are primarily comprised of regulation of heart contraction, heart rate, and blood circulation. Moreover, fibril organization is mainly implicated with the assembly, arrangement, and disassembly of several fibrils. These major biological processes play an equally important role in the regulation of heart function.
Mitochondrial dysfunction would lower the rate of oxidative phosphorylation and cause energy deficiency, which have been implicated with many diseases, including heart disease and neurological disorder (Fillmore, Mori, & Lopaschuk, 2014; Ryan, Hoek, Fon, & Wade‐Martins, 2015; Subramaniam & Chesselet, 2013). Previous studies showed that several proteins, like ATP5B, CYC1, and COX5A, involved in the oxidative phosphorylation pathway are significantly downregulated in myocardial infarct‐like monkey heart (Song et al., 2014). In addition to these proteins, more than 100 proteins, which are associated with oxidative phosphorylation and neurological disorders pathway, were identified in normal monkey heart tissue. Similarly, Reactome pathways analysis showed that most of these identified proteins are enriched in energy metabolic pathway including TCA cycle, respiratory electron transport and mitochondrial protein import pathway.
The disturbance in the energy metabolism is inextricably linked with cardiac disorders (Lopaschuk, Ussher, Folmes, Jaswal, & Stanley, 2010). In the GDA study, various genes from heart regions were implicated with several heart illnesses including hypertrophic cardiomyopathy, heart failure, and myocardial infarction. Among these, vessel‐expressed genes like CS, SDHA, IDH1, and DLAT are enriched in TCA cycle pathway. Citrate synthase (CS), as the rate‐limiting enzyme of TCA cycle, reflects mitochondrial oxidative capacity in cardiac tissue (de Castro Bras et al., 2014; Jaenisch, Bertagnolli, Borghi‐Silva, Arena, & Lago, 2017). ACAA, ALDH9A1, and HADHB expressed in heart valves can perform these functions in metabolism pathways including amino acid and fatty acid degradation. Besides, the genes in chambers were mostly involved in metabolism pathways including glycolysis and propanoate metabolism pathway.
In our study, we identified some proteins implicated in the signaling pathway of heart disorders such as hypertrophic cardiomyopathy (HCM), arrhythmogenic right ventricular cardiomyopathy (ARVC), and dilated cardiomyopathy (DCM) pathway. Among these proteins, ACTC1, MYBPC, MYL, TNC, TNI, and TPM belonged to sarcomeric or intrasarcomeric protein which enhances Ca2+ sensitivity of ATPase activity and force production (Gangadharan et al., 2017; Gomez et al., 2014; Sequeira et al., 2013), PRKCG, and RYR consisted in mitochondria and sarcoplasmic reticulum, respectively, which increases cytosolic free Ca2+ level, take mutual effort in left ventricle hypertrophy and can potentially lead to heart failure (Rouet‐Benzineb et al., 1996; Uenoyama et al., 2010). In terms of ARVC mechanisms, DSG and DSP, which induce loss of electrical coupling between cardiac myocytes and lead to myocyte cell death (Azaouagh, Churzidse, Konorza, & Erbel, 2011; Pilichou et al., 2006), and RYR2, which disrupts normal Ca2+ cycling (Kannankeril et al., 2006; Tang, Tian, Wang, Fill, & Chen, 2012; Yano, Yamamoto, Kobayashi, & Matsuzaki, 2009), indirectly participate in symptomatic right ventricular arrhythmias mediation. In addition, protein kinase A (PKA) and sarcoplasmic protein RYR and PLN destroy intracellular Ca2+ homeostasis resulting in further left ventricle chamber dilation and systolic dysfunction (Gramolini et al., 2008; Sun et al., 2010; Yaniv et al., 2011).
In summary, we have applied an iTRAQ‐coupled LC‐MS/MS method to identify cardiac region‐specific mapping from healthy monkeys. Results from multiple bioinformatics analysis enable us to better understand the relationship between biological function, molecule mechanism, and disease development in cardiac proteins, which may help other researchers to develop comprehensive cognition on physiology and etiology studies regarding cardiac proteins. Furthermore, our study provided a reference map for future in‐depth research on cardiac disease‐related NHP model and novel biomarkers of cardiac injury.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China [grant numbers 81573320, 81470508]. Ming‐Qiang Rong, Ren Lai, and Qin Zhang contributed to the conceptual framework of the study and experimental design. Hao‐Liang Hu, Yu Kang, Yong Zeng, Min Zhang, and Qiong Liao participated in data collection and data analysis. Hao‐Liang Hu and Ming‐Qiang Rong drafted the manuscript. All investigators contributed to critical revision of the article.
Hu H-L, Kang Y, Zeng Y, et al. Region‐resolved proteomics profiling of monkey heart. J Cell Physiol. 2018;234:13720–13734. 10.1002/jcp.28052
Contributor Information
Ming‐Qiang Rong, Email: rongmq@hunnu.edu.cn.
Qin Zhang, Email: qzhang2000cn@163.com.
Ren Lai, Email: rlai@mail.kiz.ac.cn.
References
REFERENCES
- Alessio, N. , Capasso, S. , Ferone, A. , Di Bernardo, G. , Cipollaro, M. , Casale, F. , … Galderisi, U. (2017). Misidentified human gene functions with mouse models: The case of the retinoblastoma gene family in senescence. Neoplasia, 19(10), 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeli, F. , Reboldi, G. , & Verdecchia, P. (2014). Hypertension, inflammation and atrial fibrillation. Journal of Hypertension, 32(3), 480–483. [DOI] [PubMed] [Google Scholar]
- Azaouagh, A. , Churzidse, S. , Konorza, T. , & Erbel, R. (2011). Arrhythmogenic right ventricular cardiomyopathy/dysplasia: A review and update. Clinical Research in Cardiology: Official Journal of the German Cardiac Society, 100(5), 383–394. [DOI] [PubMed] [Google Scholar]
- Bekeredjian, R. , & Grayburn, P. A. (2005). Valvular heart disease: Aortic regurgitation. Circulation, 112(1), 125–134. [DOI] [PubMed] [Google Scholar]
- Bergmann, O. , Zdunek, S. , Felker, A. , Salehpour, M. , Alkass, K. , Bernard, S. , … Frisén, J. (2015). Dynamics of cell generation and turnover in the human heart. Cell, 161(7), 1566–1575. [DOI] [PubMed] [Google Scholar]
- Bindea, G. , Galon, J. , & Mlecnik, B. (2013). CluePedia cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics, 29(5), 661–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea, G. , Mlecnik, B. , Hackl, H. , Charoentong, P. , Tosolini, M. , Kirilovsky, A. , … Galon, J. (2009). ClueGO: A cytoscape plug‐in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics, 25(8), 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, M. T. , Duckles, H. , Feng, S. , Hsiao, S. T. , Kim, H. R. , Serbanovic‐Canic, J. , & Evans, P. C. (2014). Mechanoresponsive networks controlling vascular inflammation. Arteriosclerosis, Thrombosis, and Vascular Biology, 34(10), 2199–2205. [DOI] [PubMed] [Google Scholar]
- de Castro Brás, L. E. , Cates, C. A. , DeLeon‐Pennell, K. Y. , Ma, Y. , Iyer, R. P. , Halade, G. V. , … Lindsey, M. L. (2014). Citrate synthase is a novel in vivo matrix metalloproteinase‐9 substrate that regulates mitochondrial function in the postmyocardial infarction left ventricle. Antioxidants & Redox Signaling, 21(14), 1974–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, J. J. H. , Yang, X. , Don, C. W. , Minami, E. , Liu, Y. W. , Weyers, J. J. , … Murry, C. E. (2014). Human embryonic‐stem‐cell‐derived cardiomyocytes regenerate non‐human primate hearts. Nature, 510(7504), 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuspidi, C. , Negri, F. , Tadic, M. V. , Sala, C. , & Parati, G. (2014). Left atrial enlargement and right ventricular hypertrophy in essential hypertension. Blood Pressure, 23(2), 89–95. [DOI] [PubMed] [Google Scholar]
- Doll, S. , Dreßen, M. , Geyer, P. E. , Itzhak, D. N. , Braun, C. , Doppler, S. A. , … Mann, M. (2017). Region and cell‐type resolved quantitative proteomic map of the human heart. Nature Communications, 8(1), 1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat, A. , Jupe, S. , Matthews, L. , Sidiropoulos, K. , Gillespie, M. , Garapati, P. , … D'eustachio, P. (2018). The Reactome pathway knowledgebase. Nucleic Acids Research, 46(D1), D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillmore, N. , Mori, J. , & Lopaschuk, G. D. (2014). Mitochondrial fatty acid oxidation alterations in heart failure, ischaemic heart disease and diabetic cardiomyopathy. British Journal of Pharmacology, 171(8), 2080–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangadharan, B. , Sunitha, M. S. , Mukherjee, S. , Chowdhury, R. R. , Haque, F. , Sekar, N. , … Mercer, J. A. (2017). Molecular mechanisms and structural features of cardiomyopathy‐causing troponin T mutants in the tropomyosin overlap region. Proceedings of the National Academy of Sciences of the United States of America, 114(42), 11115–11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, M. P. , Champion, H. C. , Simon, M. , Guyach, S. , Tarantelli, R. , Kling, H. M. , … Norris, K. A. (2013). Physiologic changes in a nonhuman primate model of HIV‐associated pulmonary arterial hypertension. American Journal of Respiratory Cell and Molecular Biology, 48(3), 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharib, W. H. , & Robinson‐Rechavi, M. (2011). When orthologs diverge between human and mouse. Briefings in Bioinformatics, 12(5), 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimbrone, M. A., Jr. , & García‐Cardeña, G. (2016). Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circulation Research, 118(4), 620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez, J. , Reguero, J. R. , Moris, C. , Martin, M. , Alvarez, V. , Alonso, B. , … Coto, E. (2014). Mutation analysis of the main hypertrophic cardiomyopathy genes using multiplex amplification and semiconductor next‐generation sequencing. Circulation Journal: Official Journal of the Japanese Circulation Society, 78(12), 2963–2971. [DOI] [PubMed] [Google Scholar]
- Gong, L. , Zeng, W. , Yang, Z. , Chen, Z. , Cheng, A. , Shen, Y. , … Yang, Y. (2013). Comparison of the clinical manifestations of type 2 diabetes mellitus between rhesus monkey (Macaca mulatta lasiotis) and human being. Pancreas (New York, NY), 42(3), 537–542. [DOI] [PubMed] [Google Scholar]
- Gramolini, A. O. , Kislinger, T. , Alikhani‐Koopaei, R. , Fong, V. , Thompson, N. J. , Isserlin, R. , … Emili, A. (2008). Comparative proteomics profiling of a phospholamban mutant mouse model of dilated cardiomyopathy reveals progressive intracellular stress responses. Molecular & Cellular Proteomics: MCP, 7(3), 519–533. [DOI] [PubMed] [Google Scholar]
- Greenough, T. C. , Carville, A. , Coderre, J. , Somasundaran, M. , Sullivan, J. L. , Luzuriaga, K. , & Mansfield, K. (2005). Pneumonitis and multi‐organ system disease in common marmosets (Callithrix jacchus) infected with the severe acute respiratory syndrome‐associated coronavirus. The American Journal of Pathology, 167(2), 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D. W. , Sherman, B. T. , & Lempicki, R. A. (2009a). Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research, 37(1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D. W. , Sherman, B. T. , & Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4(1), 44–57. [DOI] [PubMed] [Google Scholar]
- Jaenisch, R. B. , Bertagnolli, M. , Borghi‐Silva, A. , Arena, R. , & Lago, P. D. (2017). Respiratory muscle training improves diaphragm citrate synthase activity and hemodynamic function in rats with heart failure. Brazilian Journal of Cardiovascular Surgery, 32(2), 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannankeril, P. J. , Mitchell, B. M. , Goonasekera, S. A. , Chelu, M. G. , Zhang, W. , Sood, S. , … Hamilton, S. L. (2006). Mice with the R176Q cardiac ryanodine receptor mutation exhibit catecholamine‐induced ventricular tachycardia and cardiomyopathy. Proceedings of the National Academy of Sciences of the United States of America, 103(32), 12179–12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, B. A. , Carr, C. L. , Belcik, J. T. , Xie, A. , Yue, Q. , Chadderdon, S. , … Lindner, J. R. (2010). Molecular imaging of the initial inflammatory response in atherosclerosis: Implications for early detection of disease. Arteriosclerosis, Thrombosis, and Vascular Biology, 30(1), 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, B. A. , Lewis, C. , Xie, A. , Mirza‐Mohd, A. , & Lindner, J. R. (2007). Detection of recent myocardial ischaemia by molecular imaging of P‐selectin with targeted contrast echocardiography. European Heart Journal, 28(16), 2011–2017. [DOI] [PubMed] [Google Scholar]
- Kelly, K. M. , Tocchetti, C. G. , Lyashkov, A. , Tarwater, P. M. , Bedja, D. , Graham, D. R. , … Mankowski, J. L. (2014). CCR5 inhibition prevents cardiac dysfunction in the SIV/macaque model of HIV. Journal of the American Heart Association, 3(2), e000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. S. , Pinto, S. M. , Getnet, D. , Nirujogi, R. S. , Manda, S. S. , Chaerkady, R. , … Pandey, A. (2014). A draft map of the human proteome. Nature, 509(7502), 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, H. B. , Wang, S. , Helker, C. S. M. , Rasouli, S. J. , Maischein, H. M. , Offermanns, S. , … Stainier, D. Y. R. (2016). In vivo modulation of endothelial polarization by Apelin receptor signalling. Nature Communications, 7, 11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, E. , Cao, Q. , Ng, D. C. M. , Bleakley, B. J. , Dincer, T. U. , Bot, B. M. , … Ping, P. (2016). A large dataset of protein dynamics in the mammalian heart proteome. Scientific Data, 3, 160015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin, N. , Daniel, M. , Sehgal, P. , Desrosiers, R. , Hunt, R. , Waldron, L. , … King, N. (1985). Induction of AIDS‐like disease in macaque monkeys with T‐cell tropic retrovirus STLV‐III. Science, 230(4721), 71–73. [DOI] [PubMed] [Google Scholar]
- Levine, R. A. , Hagége, A. A. , Judge, D. P. , Padala, M. , Dal‐Bianco, J. P. , Aikawa, E. , … Yacoub, M. H. , Leducq Mitral Transatlantic N. (2015). Mitral valve disease‐‐morphology and mechanisms. Nature Reviews Cardiology, 12(12), 689–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, A. C. , Joag, V. R. , & Gotlieb, A. I. (2007). The emerging role of valve interstitial cell phenotypes in regulating heart valve pathobiology. The American Journal of Pathology, 171(5), 1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Parman, T. , Schneider, B. , Song, B. , Galande, A. K. , Anderson, D. , & Mirsalis, J. (2013). Serum biomarkers reveal long‐term cardiac injury in isoproterenol‐treated African green monkeys. Journal of Proteome Research, 12(4), 1830–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopaschuk, G. D. , Ussher, J. R. , Folmes, C. D. L. , Jaswal, J. S. , & Stanley, W. C. (2010). Myocardial fatty acid metabolism in health and disease. Physiological Reviews, 90(1), 207–258. [DOI] [PubMed] [Google Scholar]
- Lu, Z. Q. , Sinha, A. , Sharma, P. , Kislinger, T. , & Gramolini, A. O. (2014). Proteomic analysis of human fetal atria and ventricle. Journal of Proteome Research, 13(12), 5869–5878. [DOI] [PubMed] [Google Scholar]
- Lundby, A. , & Olsen, J. V. (2013). Phosphoproteomics taken to heart. Cell Cycle, 12(17), 2707–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, L. , Gopinath, G. , Gillespie, M. , Caudy, M. , Croft, D. , de Bono, B. , … D'Eustachio, P. (2009). Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Research, 37(Database issue), D619–D622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlpine, C. S. , & Swirski, F. K. (2016). Circadian influence on metabolism and inflammation in atherosclerosis. Circulation Research, 119(1), 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meléndez, G. C. , Register, T. C. , Appt, S. E. , Clarkson, T. B. , Franke, A. A. , & Kaplan, J. R. (2015). Beneficial effects of soy supplementation on postmenopausal atherosclerosis are dependent on pretreatment stage of plaque progression. Menopause, 22(3), 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ounzain, S. , Micheletti, R. , Beckmann, T. , Schroen, B. , Alexanian, M. , Pezzuto, I. , … Pedrazzini, T. (2015). Genome‐wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart‐specific long non‐coding RNAs. European Heart Journal, 36(6), 353–368a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Y. , Gregorich, Z. R. , Valeja, S. G. , Zhang, H. , Cai, W. , Chen, Y. C. , … Ge, Y. (2014). Top‐down proteomics reveals concerted reductions in myofilament and Z‐disc protein phosphorylation after acute myocardial infarction. Molecular & Cellular Proteomics: MCP, 13(10), 2752–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilichou, K. , Nava, A. , Basso, C. , Beffagna, G. , Bauce, B. , Lorenzon, A. , … Rampazzo, A. (2006). Mutations in desmoglein‐2 gene are associated with arrhythmogenic right ventricular cardiomyopathy. Circulation, 113(9), 1171–1179. [DOI] [PubMed] [Google Scholar]
- Piñero, J. , Bravo, À. , Queralt‐Rosinach, N. , Gutiérrez‐Sacristán, A. , Deu‐Pons, J. , Centeno, E. , … Furlong, L. I. (2017). DisGeNET: A comprehensive platform integrating information on human disease‐associated genes and variants. Nucleic Acids Research, 45(D1), D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, C. , Gong, L. , Yang, Z. , Chen, W. , Chen, Y. , Xu, Z. , … Zeng, W. (2015). Diastolic dysfunction in spontaneous type 2 diabetes rhesus monkeys: A study using echocardiography and magnetic resonance imaging. BMC Cardiovascular Disorders, 15, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, H. , Dai, H. , Jain, K. , Shah, R. , Hong, C. , Pain, J. , … Depre, C. (2008). Characterization of a novel cardiac isoform of the cell cycle‐related kinase that is regulated during heart failure. The Journal of Biological Chemistry, 283(32), 22157–22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet‐Benzineb, P. , Mohammadi, K. , Pérennec, J. , Poyard, M. , El houda bouanani, N. , & Crozatier, B. (1996). Protein kinase C isoform expression in normal and failing rabbit hearts. Circulation Research, 79(2), 153–161. [DOI] [PubMed] [Google Scholar]
- Ryan, B. J. , Hoek, S. , Fon, E. A. , & Wade‐Martins, R. (2015). Mitochondrial dysfunction and mitophagy in Parkinson's: From familial to sporadic disease. Trends in Biochemical Sciences, 40(4), 200–210. [DOI] [PubMed] [Google Scholar]
- Schroder, C. , Pierson, R. N., 3rd , Nguyen, B. N. H. , Kawka, D. W. , Peterson, L. B. , Wu, G. , … DeMartino, J. A. (2007). CCR5 blockade modulates inflammation and alloimmunity in primates. Journal of Immunology, 179(4), 2289–2299. [DOI] [PubMed] [Google Scholar]
- Sequeira, V. , Wijnker, P. J. M. , Nijenkamp, L. L. A. M. , Kuster, D. W. D. , Najafi, A. , Witjas‐Paalberends, E. R. , … van der Velden, J. (2013). Perturbed length‐dependent activation in human hypertrophic cardiomyopathy with missense sarcomeric gene mutations. Circulation Research, 112(11), 1491–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeper, M. M. , Gaughan, J. M. , Gleason, C. R. , & Burkett, D. E. (2008). Echocardiographic reference ranges for sedated healthy cynomolgus monkeys (Macaca fascicularis). Journal of the American Association for Laboratory Animal Science: JAALAS, 47(1), 22–25. [PMC free article] [PubMed] [Google Scholar]
- Song, B. , Liu, Y. , Parman, T. , Liu, S. , Miller, J. K. , Liu, X. , … Mirsalis, J. (2014). Quantitative proteomics for cardiac biomarker discovery using isoproterenol‐treated nonhuman primates. Journal of Proteome Research, 13(12), 5909–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, S. R. , & Chesselet, M. F. (2013). Mitochondrial dysfunction and oxidative stress in Parkinson's disease. Progress in Neurobiology, 106‐107, 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama, A. (2008). Sensitive and reliable proarrhythmia in vivo animal models for predicting drug‐induced torsades de pointes in patients with remodelled hearts. British Journal of Pharmacology, 154(7), 1528–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. , Ai, J. , Wang, N. , Zhang, R. , Li, J. , Zhang, T. , … Yang, B. (2010). Cerebral ischemia elicits aberration in myocardium contractile function and intracellular calcium handling. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology, 26(3), 421–430. [DOI] [PubMed] [Google Scholar]
- Tang, Y. , Tian, X. , Wang, R. , Fill, M. , & Chen, S. R. W. (2012). Abnormal termination of Ca2 + release is a common defect of RyR2 mutations associated with cardiomyopathies. Circulation Research, 110(7), 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, P. M. , Batten, P. , Brand, N. J. , Thomas, P. S. , & Yacoub, M. H. (2003). The cardiac valve interstitial cell. The International Journal of Biochemistry & Cell Biology, 35(2), 113–118. [DOI] [PubMed] [Google Scholar]
- Thomas, R. , Cheng, Y. , Yan, J. , Bettinger, T. , Broillet, A. , Rioufol, G. , & Nunn, A. D. (2010). Upregulation of coronary endothelial P‐selectin in a monkey heart ischemia reperfusion model. Journal of Molecular Histology, 41(4‐5), 277–287. [DOI] [PubMed] [Google Scholar]
- Ueda, M. , Ageyama, N. , Nakamura, S. , Nakamura, M. , Chambers, J. K. , Misumi, Y. , … Ando, Y. (2012). Aged vervet monkeys developing transthyretin amyloidosis with the human disease‐causing Ile122 allele: A valid pathological model of the human disease. Laboratory Investigation; A Journal of Technical Methods and Pathology, 92(3), 474–484. [DOI] [PubMed] [Google Scholar]
- Uenoyama, M. , Ogata, S. , Nakanishi, K. , Kanazawa, F. , Hiroi, S. , Tominaga, S. , … Suzuki, S. (2010). Protein kinase C mRNA and protein expressions in hypobaric hypoxia‐induced cardiac hypertrophy in rats. Acta Physiologica, 198(4), 431–440. [DOI] [PubMed] [Google Scholar]
- Varki, N. , Anderson, D. , Herndon, J. G. , Pham, T. , Gregg, C. J. , Cheriyan, M. , … Varki, A. (2009). Heart disease is common in humans and chimpanzees, but is caused by different pathological processes. Evolutionary Applications, 2(1), 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel, N. F. , Cool, C. D. , & Flores, S. (2008). From viral infection to pulmonary arterial hypertension: A role for viral proteins? AIDS, 22(Suppl 3), S49–S53. [DOI] [PubMed] [Google Scholar]
- Wilhelm, M. , Schlegl, J. , Hahne, H. , Gholami, A. M. , Lieberenz, M. , Savitski, M. M. , … Kuster, B. (2014). Mass‐spectrometry‐based draft of the human proteome. Nature, 509(7502), 582–587. [DOI] [PubMed] [Google Scholar]
- Xiang, F. , Guo, X. , Chen, W. , Wang, J. , Zhou, T. , Huang, F. , … Chen, X. (2013). Proteomics analysis of human pericardial fluid. Proteomics, 13(17), 2692–2695. [DOI] [PubMed] [Google Scholar]
- Yang, P. , Han, P. , Hou, J. , Zhang, L. , Song, H. , Xie, Y. , … Kang, Y. J. (2011). Electrocardiographic characterization of rhesus monkey model of ischemic myocardial infarction induced by left anterior descending artery ligation. Cardiovascular Toxicology, 11(4), 365–372. [DOI] [PubMed] [Google Scholar]
- Yaniv, Y. , Juhaszova, M. , Lyashkov, A. E. , Spurgeon, H. A. , Sollott, S. J. , & Lakatta, E. G. (2011). Ca2 + ‐regulated‐cAMP/PKA signaling in cardiac pacemaker cells links ATP supply to demand. Journal of Molecular and Cellular Cardiology, 51(5), 740–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M. , Yamamoto, T. , Kobayashi, S. , & Matsuzaki, M. (2009). Role of ryanodine receptor as a Ca(2)( + ) regulatory center in normal and failing hearts. Journal of Cardiology, 53(1), 1–7. [DOI] [PubMed] [Google Scholar]
- Yoshioka, T. , Ageyama, N. , Shibata, H. , Yasu, T. , Misawa, Y. , Takeuchi, K. , … Hanazono, Y. (2005). Repair of infarcted myocardium mediated by transplanted bone marrow‐derived CD34 + stem cells in a nonhuman primate model. Stem Cells, 23(3), 355–364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information
Supporting information


