Abstract
Coronaviruses (CoVs) are by far the largest group of known positive‐sense RNA viruses having an extensive range of natural hosts. In the past few decades, newly evolved Coronaviruses have posed a global threat to public health. The immune response is essential to control and eliminate CoV infections, however, maladjusted immune responses may result in immunopathology and impaired pulmonary gas exchange. Gaining a deeper understanding of the interaction between Coronaviruses and the innate immune systems of the hosts may shed light on the development and persistence of inflammation in the lungs and hopefully can reduce the risk of lung inflammation caused by CoVs. In this review, we provide an update on CoV infections and relevant diseases, particularly the host defense against CoV‐induced inflammation of lung tissue, as well as the role of the innate immune system in the pathogenesis and clinical treatment.
Keywords: chemokine, coronavirus, cytokines, inflammation, interferon
Highlights
This highlights the importance of immune responses under coronavirus infection and improve the understanding of the features of CoV‐induced inflammatory response.
1. INTRODUCTION
During the end of 2019 and the beginning of 2020, multiple human cases of novel coronavirus infection were reported in relation to the Huanan Seafood Wholesale Market (South China Seafood City Food Market) in Wuhan, China. At 9 O'clock, 7 January 2020, the virus was identified as a novel coronavirus and officially named by the WHO as 2019‐nCoV, the new coronavirus in 2019. 1 On 22 January 2020, a total of 314 confirmed case have been reported, and 6 patients were reported to have died. 2 On 13, 16, and 21 January, respectively, Thailand, Japan, and Korea confirmed the detection of a human infection with 2019‐nCoV from China. 2 In recent years, novel coronaviruses emerge periodically in different areas around the world. Severe acute respiratory syndrome coronavirus (SARS‐CoV) occurred in 2002, which reportedly infected 8422 people and caused 916 deaths worldwide during the epidemic. Middle East respiratory syndrome coronavirus (MERS‐CoV) was first identified in 2012, bringing a total of 1401 MERS‐CoV infections, and 543 (~39%) of which died. 3 , 5 All the infection cases and recent epidemics show that coronaviruses impose a continuous threat to human beings and the economy as they emerge unexpectedly, spread easily, and lead to catastrophic consequences.
Coronaviruses are enveloped, nonsegmented, positive‐sense single‐stranded RNA virus genomes in the size ranging from 26 to 32 kilobases, the largest known viral RNA genome. The virion has a nucleocapsid composed of genomic RNA and phosphorylated nucleocapsid (N) protein, which is buried inside phospholipid bilayers and covered by two different types of spike proteins: the spike glycoprotein trimmer (S) that can be found in all CoVs, and the hemagglutinin‐esterase (HE) that exists in some CoVs. The membrane (M) protein (a type III transmembrane glycoprotein) and the envelope (E) protein are located among the S proteins in the virus envelope. CoVs were given their name based on the characteristic crown‐like appearance. The structure of CoV virion is shown in Figure 1.
Figure 1.
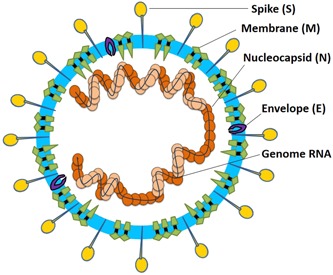
Coronavirus particle. Coronaviruses are enveloped, nonsegmented, positive‐sense single‐stranded RNA virus genomes in the size ranging from 26 to 32 kilobases. The virion has a nucleocapsid composed of genomic RNA and phosphorylated nucleocapsid (N) protein, which is buried inside phospholipid bilayers and covered by the spike glycoprotein trimmer (S). The membrane (M) protein (a type III transmembrane glycoprotein) and the envelope (E) protein are located among the S proteins in the virus envelope
The coronavirus subfamily is genotypically and serologically divided into four genera, the α, β, ɣ, and δ coronaviruses. The β‐coronavirus can be further classified into four viral lineages, namely lineage A‐D. There are nearly 30 recognized CoVs that infect humans, mammals, fowl, and other animals. Human CoV infections are caused by α‐ and β‐CoVs. CoVs are common human pathogens, and 30% to 60% of the Chinese population is positive for anti‐CoV antibodies. The viral infections are generally associated with upper respiratory tract infections, of which the signs and symptoms commonly include fever, headache, and cough; some patients may have lower respiratory tract infections. In contrast, SARS‐CoV and MERS‐CoV infections may remain asymptomatic in the early stage until severe pneumonia, dyspnea, renal insufficiency, and even death (Figure 2).
Figure 2.
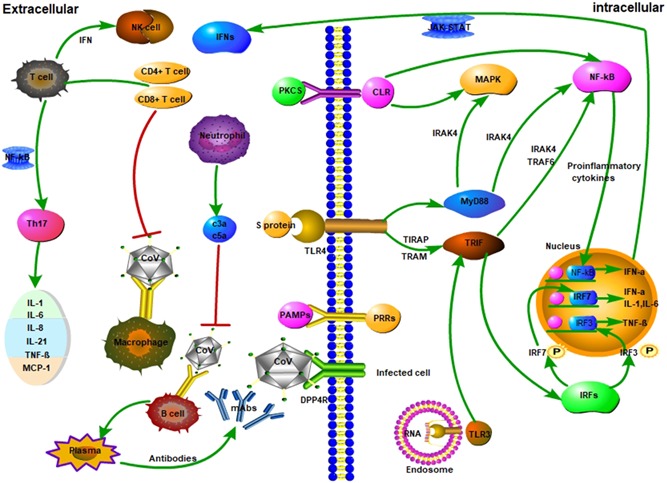
The innate immune response and adaptive immune responses of Coronaviruses (CoV) infection during an infection. A, CoV infects macrophages, and then macrophages present CoV antigens to T cells. This process leads to T cell activation and differentiation, including the production of cytokines associated with the different T cell subsets (ie, Th17), followed by a massive release of cytokines for immune response amplification. The continued production of these mediators due to viral persistence has a negative effect on NK, and CD8 T cell activation. However, CD8 T cells produce very effective mediators to clear CoV. B, Attachment of CoV to DPP4R on the host cell through S protein leads to the appearance of genomic RNA in the cytoplasm. An immune response to dsRNA can be partially generated during CoV replication. TLR‐3 sensitized by dsRNA and cascades of signaling pathways (IRFs and NF‐κB activation, respectively) are activated to produce type I IFNs and proinflammatory cytokines. The production of type I IFNs is important to enhance the release of antiviral proteins for the protection of uninfected cells. Sometimes, accessory proteins of CoV can interfere with TLR‐3 signaling and bind the dsRNA of CoV during replication to prevent TLR‐3 activation and evade the immune response. TLR‐4 might recognize S protein and lead to the activation of proinflammatory cytokines through the MyD88‐dependent signaling pathway. Virus‐cell interactions lead to the strong production of immune mediators. The secretion of large quantities of chemokines and cytokines (IL‐1, IL‐6, IL‐8, IL‐21, TNF‐β, and MCP‐1) is promoted in infected cells in response to CoV infection. These chemokines and cytokines, in turn, recruit lymphocytes and leukocytes to the site of infection. Red lines refer to inhibitory effects. Green lines refer to activating effects
Histopathological observations of pulmonary lesions in SARS cases not only show nonspecific inflammatory responses such as edema and inflammatory cell infiltration but also exhibit severe exfoliation of alveolar epithelial cells, alveolar septal widening, damage to alveolar septa, and alveolar space infiltration in a distinctly organized manner. Pathologically, inflammation includes degeneration (necrosis), infiltration, and hyperplasia. Thus, SARS‐CoV infection can cause pathological changes, degeneration, infiltration, and hyperplasia. Damage to the pulmonary interstitial arteriolar walls indicates that inflammatory response plays an important role throughout the course of disease in spites of the pathogenic effect of CoVs.
Although the pathologies of SARS and MERS are not yet fully understood, viral and host factors play a key role in SARS‐CoV and MERS‐CoV infections. During virus infection, host factors trigger an immune response against the virus. However, it should be noted that immunopathogenesis is associated with an immune response out of control, which may result in pulmonary tissue damage, functional impairment, and reduced lung capacity. Chemotactic factors are essential to the immune responses against the virus infections, given their regulatory effect on dilations and positions of leukocytes in the host lungs. Therefore, spectral changes in chemotactic factors may lead to severely maladjusted immune responses. Immune insufficiency or misdirection may increase viral replication and cause tissue damages. In contrast, overactive immune responses may induce immunopathological conditions. In this review article, we provide an analysis of the role of cytokines secreted upon CoV infections and their potentially detrimental contribution to the damages of the respiratory tract and other tissues.
2. INNATE IMMUNE RESPONSE
2.1. Pathogen‐recognition receptors
The host innate immune system detects viral infections by using pattern recognition receptors (PRRs) to recognize pathogen‐associated molecular patterns (PAMPs). At present, the known PRRs mainly include toll‐like receptor (TLR), RIG‐I‐like receptor (RLR), NOD‐like receptor (NLR), C‐type lectin‐like receptors (CLmin), and free‐molecule receptors in the cytoplasm, such as cGAS, IFI16, STING, DAI, and so on.
2.2. Toll‐like receptors
PAMPs recognized by Toll‐like receptors (TLRs) include lipids, lipoproteins, proteins, and nucleic acids of the bacterial, viral, parasite, and fungal origins. 6 The recognition of PAMPs by TLRs also occurs in cell membranes, endosomes, lysosomes, and endocytolysosomes and other locations in cells. 6 Different TLRs can induce different biological responses via subsequent activation of varied adapter proteins, such as MyD88, TIRAP, TRIP, and TRAM, but these adapter proteins all share the Toll/Interleukin‐1 receptor (TIR) structure. 7 MyD88 is the first identified TIR family member, which acts as an adapter protein by almost all TLRs except TLR3. It mainly activates the transcription factors NF‐kB and mitogen‐activated protein kinases (MAPKs) pathways to induce inflammatory factors' expression. 6 Unlike MyD88, TRIF is an adapter protein of TLR3 and TLR4, which activates the transcription factors IRF3 and NF‐kB to induce the expression of type I interferon and immune‐inflammatory factors. The function of TRAM and TIRAP is to recruit TRIF molecules to the TLR4 receptor and MyD88 to the TLR2 and TLR4 receptors. Therefore, the TLR signaling pathways are classified as the MyD88‐dependent pathway, which functions to activate immune‐inflammatory factors, and the TRIF‐dependent pathway, which functions to activate the type I interferons and inflammatory factors. 6 After a TLR is activated by the corresponding PAMP, MyD88 recruits the busy‐1 receptor‐related kinases IRAK4, IRAKI, IRAK2, and IRAK‐M. IRAK4 plays an important role in activating NF‐kB and MAPKs downstream of MyD88. IRAK interacts with TRAF6, which causes its K‐63 ubiquitination, and facilitates NEMO ubiquitination to activate NF‐kB. TRIF‐dependent pathways activate IRF3 and NF‐kB. 8 , 9 In addition to activating NF‐kB, TRIF‐dependent pathways, they also activate IRF3 and interferon‐β. 10 , 11
2.3. RIG‐I‐like receptors
RIG‐I‐like receptors (RLRs), including the H family members RIG‐I (DDX58), MDA5 (IFIH), and LGP2, primarily recognize nucleic acids of RNA viruses. 12 , 13 They have a DExD/H‐box RNA helicase structure and a C‐terminal termination structure (CTD), while RIG‐I and MDA5 have an N‐terminal caspase recruitment structure (CARD), to interact with the downstream adapter MAVS. The C‐terminal RNA helicase and CTD structure are considered to recognize RNA, and its conformational change requires ATP to make the CARD structure interact with MAVS. 14
RIG‐I is activated by a variety of RNA viruses' infections, including Influenza A virus (IAV), Newcastle disease virus (NDV), Sendai virus (SeV), and Vesicular stomatitis virus (VSV), Measles virus (MV), and Hepatitis C virus (HCV). 15 , 16 The common features of the viral RNAs are short double‐stranded with a triphosphate structure, and complementary ends and/or poly‐U/UC‐rich structure. The viral nucleocapsid proteins containing triphosphine RNA at the 5′‐end can be recognized by RIG‐I. 17 The double‐stranded RNA with double‐basic acid at the 5′‐end can be recognized by RIG‐I. 18 When the viral 5′‐terminal triphosphate end is recognized by the CTD structure, the ATP‐dependent conformational change brings the CTD structure to form a complex with the double‐stranded RNA, and the CARD structure is then released from its self‐inhibition and interacts with MAVS. 19
MDA5 recognizes RNAs of picornaviruses, including poliovirus (PV) and Encephalomyocarditis virus (EMCV). MDAS‐recognized RNA is characterized by long double‐stranded RNA more than 1 kbp. Crystal structure analysis shows that the helicase and CTD structure of MDA5 are also surrounded by double‐stranded RNA, the same as RIG‐I. However, the CTD structure of MDA5 does not have a hat structure, 20 and the hat structure is necessary to have a triphosphate RNA interaction at the 5′‐end. The CTD structure of MDA5 directly interacts with the double‐stranded RNA, so that the 5′‐end RNA can be freely released. 21
2.4. Nucleotide‐binding and oligomerization domain‐like receptors
Nucleotide‐binding and oligomerization domain (NOD)‐like receptors (NLRs) are a class of pattern recognition receptors, 22 which recognize components of pathogens and contain a conserved NOD structure. 23 NLR receptor family members are divided into three subclasses according to their functions. The first NLR subclass forms complexes with a variety of proteins and these complexes are defined as inflammasome that contains at least eight NLR proteins, including NLRP1, NLRP3, NLRP6, NLRC4, NLRC5W, and AY2. 24 , 25 , 26 The second subclass is essential to reproduction and embryo regeneration. 27 The third subclass is comprised of regulatory NLRs. These NLRs are positive or negative conditioned inflammatory signaling cascade pathways.
2.5. C‐type lectin‐like receptor
C‐type lectin‐like receptor (CLRs) are a large family of soluble, transmembrane pattern recognition receptors with more than 1000 members, which are widely expressed in myeloid cells. Due to its motif structure in the intracellular region with multiple signaling pathways, the CLR receptor has a wide range of functions, including cell adhesion, induction of endocytosis, phage, tissue repair, platelet activation, and natural immune responses. There are two main ways of CLR receptor activation in the cells. The first type is direct activation, such as macrophage‐induced Mincle and CLEC4E receptors, and Dectin‐2 (CLEC6A) receptors. The second type is to activate the receptor by activating HAM‐like motifs in the intracellular tail of the receptor, such as Dectin‐1 (CLEC7A) and DNGR‐1 (CLEC9A). 28 , 29 Both mechanisms involve the recruitment of acidified spleen tyrosine kinases, which in turn promotes CARD9, B‐cell lymphoid tissue 10 (BcL10), and Maltl complex formation. At the same time, apoptosis‐related granule‐like proteins, including ASC, is acidified by SyK and JNK, 30 and PKCS is also a key essential element in the pathway. 31 The signaling pathways activate downstream molecules, including NF‐kB and MAPKs, and trigger a variety of cellular responses, including cell phagocytosis, maturation of DC cells, and chemotaxis of cells. 31
2.6. Cytoplasmic DNA receptor
Exogenous microbial DNAs are recognized by host DNA receptors. In addition to TLR9 in the TLR family, Cytoplasmic DNA receptor (CDR) can recognizes DNA CpG islands. 32 More than 10 CDRs distributed in the cytoplasm have been identified, including AIM2‐like receptors (ALRs), DNA‐dependent activator of IFN‐regulatory factor (DAI), interference stimulator of interferon gene (STING), leucine‐rich repeat flightless‐interacting protein 1 (LRRFIP1), DExD/H‐box RNA helicase (DDX), Meiotic recombinant protein 11 Homolog A (MRE11), RNA polymerase III (Pol III), DNA dependent protein kinase (DNA‐PK), DNA repair‐related proteins Rad50, cyclic GMP‐AMP synthase (cGAS), and Sry‐related HMG box 2 (Sox2). 4 , 32 , 33 DAI recognizes Z‐type DNA and B‐type DNA, which does not depend on sequence specificity, but on the length of DNA. 34 , 35 AIM2 is mainly involved in the recognition of double‐stranded DNA. Exogenous microbial DNAs are also recognized by host DNA receptors. IFI16 and cGAS are reported to be novel DNA receptors that mediate cytosolic DNA recognition and induce type I interferon. 36
2.7. Type I interferons
When a virus invades the host, PRRs initially recognize the viral nucleic acid, collect the specific signal adapter protein, activate IRF3 and IRF7 before being translocated to the nucleus and promote the synthesis of type I interferons (IFNs). Type I IFNs subsequently activate the downstream JAK‐STAT signal pathway, promote the expression of IFN‐stimulated genes (ISGs). 37 , 38
As the host's major antiviral molecules, IFNs limit virus spread, and play an immunomodulatory role to promote macrophage phagocytosis of antigens, as well as NK cells restriction of infected target cells and T/B cells. Thus, blocking the production of IFNs has a direct effect on the survival of the virus in the host. 39 , 40 So far, PRRs are divided into three types according to their forms of existence 41 : the membrane type includes TLR2, TLR4, mannose receptor (MR), scavenger receptor (SR); the secretory type comprises mannose‐binding lectin (MBL) and C‐reactive protein (CRP); the cytoplasmic type consists of TLR3, TLR7/8, and NLRs. Among them, the IFN production‐related PRRs mainly include TLRs, RLRs, and NLRs. The signaling pathways induce downstream IFNs production. 42 Upon infecting plasma‐like dendritic cells (pDCs), the viral nucleic acids are recognized by TLR7/TLR9 to induce the production of inflammatory cytokines and type I IFNs mediated by NF‐κB and IRF7. 43 , 44 VSV infection induces miR‐146a expression in macrophages through the RIG‐I/NF‐κB‐dependent pathway 45 and the disorder of the JAK‐STAT signaling pathway directly affects the spread of virus. 46
Although SARS‐CoV and other coronaviruses are sensitive to IFN‐a/b, these viruses remain highly pathogenic. Reportedly, the N protein of SARS‐CoV acts as an antagonist of immune escape protein and host interferon response. 47 , 48 , 49 It is reported that EV71 infection downregulates JAK1, p‐JAK1, and p‐TYK2, inhibits p‐STAT1/2, and blocks the JAK‐STAT signaling pathway mediated by type I IFNs, thereby hindering the function of IFNs and promoting EV71 replication and proliferation in host cells. 50 Ebola virus (EBOV) promotes cytokine signal inhibitory factor‐1 (SOCS1) and blocks the JAK‐STAT signal pathway by directly binding to phosphorylated JAK, resulting in the inhibition of JAK activation. 51 In addition, influenza A virus can inhibit the IFN‐I downstream pathway by inducing the expression of SOCS3. 52
2.8. Dendritic cells
Dendritic cells (DCs) play a key role in innate immune and adaptive immune responses. As the strongest antigen‐presenting cells in the organism, they effectively stimulate the activation of T‐lymphocytes and B‐lymphocytes, thus combining innate and adaptive immunity. Immature DCs have strong migration ability, and mature DCs can effectively activate T cells in the central link of start‐up, regulation, and maintenance of immune responses. Thus, once the maturation process of DCs is blocked, it directly affects the initiation of subsequent adaptive immune responses. 53 , 54 , 55 DC precursor cells differentiate into DCs by adding such inducers as GM‐CSF, IL‐4, and TNF‐α. 56 However, DC precursor cells cannot differentiate into DCs if transfected with HIV‐1 Nef protein in the presence of the inducers, indicating that Nef blocks the differentiation of DC precursor cells into mature DCs. Both the core protein and NS3 protein of HCV inhibit the expression of CD1a, CD1b, and DC‐SIGN molecules on human peripheral blood mononuclear precursor cells (PBMCs), which play an important role in the development of peripheral blood mononuclear precursor cells to DCs. 57 In addition, HIV‐1 attenuates the major histocompatibility antigen I (MHC I) on the surface of DCs, thereby reducing the ability of DCs to present the viral antigens. HIV‐1 infection enhances the expression of DC‐specific intercellular adhesion molecule‐3‐grabbing nonintegrity (DC‐SIGN), thus inhibiting CC chemokine receptor 7 (CCR7) and MHC‐II, which are important receptors of DC homing. 58 , 59 These results indicate that virus infection interferes with the differentiation and function of DCs, hinders the subsequent adaptive immune response mediated by DCs, and makes the virus evade the adaptive immune response of the host successfully.
2.9. Defensins
Defensins are a family of endogenous antibiotic peptide molecules, which widely exist in human, animals, and plants, and are important for the host's innate defense system. Defensins have broad‐spectrum antimicrobial activities. In vitro inhibition experiments show that defensins have killing effects on bacteria, fungi, mycoplasma, chlamydia, spirochetes, tumor cells, and viruses. 60 , 61
Defensins of human and rabbit neutrophils are mainly found in the eosinophilic granules of neutrophils. They are small molecular cationic polypeptides composed of 29 to 34 amino acid residues, with a relative molecular weight of 3500 to 4000 dolt and three intramolecular disulfide bonds. They are main components of the neutrophils independent of oxygen sterilization. 62 , 63 Human α‐defensin HNP‐1 inactivates herpes simplex virus type I and type II (HSV‐1 and HSV‐2), cytomegalovirus (CMV), VSV, and IAV. 64 , 65 Purified defensins of guinea pigs, rabbits, and rats have weak anti‐HIV‐1 activity. 66 , 67
However, some studies showed that purified human neutrophil defensin (HNP1‐3) and rabbit neutrophil defensins (RNP1–5) could neither inhibit nor kill SARS‐CoV. 68 , 69
3. ADAPTIVE IMMUNE RESPONSES
3.1. Immune response of T cells
MERS‐CoV and SARS‐CoV are β‐coronaviruses that can cause fatal lower respiratory tract infections and extrapulmonary manifestations. 70 , 71 , 72 T cells, CD4+ T cells, and CD8+ T cells particularly play a significant antiviral role by balancing the combat against pathogens and the risk of developing autoimmunity or overwhelming inflammation. 73 CD4+ T cells promote the production of virus‐specific antibodies by activating T‐dependent B cells. However, CD8+ T cells are cytotoxic and can kill viral infected cells. CD8+ T cells account for about 80% of total infiltrative inflammatory cells in the pulmonary interstitium in SARS‐CoV‐infected patients and play a vital role in clearing CoVs in infected cells and inducing immune injury. 74 In addition, by comparing T‐cell‐deficient BALB/c mice (transduced by ad5‐hdp4) with controls and B‐cell‐deficient mice, some researchers determined that T cells could survive in the infected lungs and destroy the infected cells. 75 It emphasizes the important role of T cells rather than B cells in the control of pathogenesis of MERS‐CoV infection. A cross‐reactive T cell response leads to a decrease in MERS‐CoV. 76 However, CD4+ T cells are more susceptible to MERS‐CoV infection. The depletion of CD8+ T cells do not affect and delay viral replication at the time of infection with SARS‐CoV. 77 , 78 Depletion of CD4+ T cells is associated with reduced pulmonary recruitment of lymphocytes and neutralizing antibody and cytokine production, resulting in a strong immune‐mediated interstitial pneumonitis and delayed clearance of SARS‐CoV from lungs. 79 Additionally, T helper cells produce proinflammatory cytokines via the NF‐kB signaling pathway. 80 IL‐17 cytokines recruit monocytes and neutrophils to the site of infection with inflammation and activate other downstream cytokine and chemokine cascades, such as IL‐1, LL‐6, IL‐8, IL‐21, TNF‐β, and MCP‐1. 81 , 82
On the other hand, MERS‐CoV induces T cell apoptosis by activating the intrinsic and extrinsic apoptosis pathways. A novel BH3‐like region located in the C‐terminal cytosolic domain of SARS‐CoV protein mediates its binding to Bcl‐xL and induced T‐cell apoptosis. 83 During the later stage of infection, depletion of T cells having antiviral effects may prolong the infection and promote viral survival. 84
The reappearance of SARS‐CoV is still a noteworthy problem. SARS‐CoV‐specific T cells have been screened in SARS convalescent patients. All the detected memory T cell responses are directed at SARS‐CoV structural proteins. Two CD8+T cell responses to SARS‐CoV membrane (M) and Nucleocapsid (N) protein are characterized by measuring their HLA restriction and minimal T cell epitope regions. Further, these reactions are found to last up to 11 years after infection. Absence of cross‐reactivity of these CD8+T cell responses against the MERS‐CoV is also demonstrated. 78
Results of the current research show that the T cell response to S protein and other structural proteins (including the M and N proteins) is long‐lasting and persistent. This provides evidence for the design of the SARS vaccine composed of viral structural proteins, which can induce dominant, effective, and long‐term memory cell responses against the virus.
3.2. Humoral immune responses
B cell subsets with phenotypes characteristic of naive, non‐isotype‐switched, memory cells and antibody‐secreting cells accumulate in CoVs. 85 The antigen stimulation of MERS‐CoV infection was clarified by using the specific 9‐mer peptide “CYSSLILDY”, which located at position 437 to 445 within the region of the S glycoprotein. 85 The sequence has the highest B cell antigenicity plot and has the ability to form the greatest number of interactions with MHCI alleles in a computerized simulation. 86 Reports show that humoral immunity is essential to control the persistent phase of CoV infection. More antibodies isolated from patients who have survived MERS‐CoV infection have been described, including MCA1, CDC‐C2, CSC‐C5, CDC‐A2, CDC‐A10, MERS‐GD27, and MERS‐GD33. 87 , 88 , 89
The complement system plays a vital role in the host immune response to CoV infection. Primitively identified as a host‐sensitive and nonspecific complement to adaptive immune pathways, the complement system provides a way for the innate immune system to detect and respond to foreign antigens. 90 Given its potential to damage the host tissues, the complement system is tightly controlled by inhibiting proteins in the serum. Virus encoded proteins help them evade the detection of the complement system, suggesting that complements are vital to the antiviral response. C3a and C5a have potent proinflammatory properties and can trigger inflammatory cell recruitment and neutrophil activation. C3a and C5a blockade acts as a treatment for acute lung injury, and anti‐C5a antibody shows to protect mice from infection with MERS‐CoV. 91 SARA‐CoV infection activates the complement pathway and complement signaling contributes to disease. 92
3.3. Antibody responses to coronaviruses' infections
The antibody response in vivo is a dynamic and complex mixture of monoclonal antibodies (mAbs), which work together to target different antigenic domains on the envelope glycoprotein of the virus. It is important to determine whether the antibodies are powerful in the adaptive immune responses to MERS‐CoV infection. Research from all over the world have described more than 20 kinds of monoclonal antibodies, most of which are human or humanized antibodies. The virus uses its spike proteins as an adhesion factor to facilitate host entry through a special receptor called dipeptidyl peptidase‐4 (DPP4). This receptor is considered a key factor in the signal transmission and activation of acquired and innate immune responses in infected patients. Thus, compared with the time‐consuming vaccine preparation, the design of monoclonal antibodies against these proteins has a better protective effect.
Human monoclonal antibody (m336) isolated from the phage display library interacts with the receptor‐binding region of MES coronavirus spike protein and displays strong neutralization activity to MES‐CoV in vitro. 93 Human monoclonal antibody m336 shows high neutralization activity to MERS‐CoV in vitro. m336 reduces the RNA titer of lung by 40 000 to 90 000 folds. 94 After infection with MERS‐CoV, monkeys were treated with high‐titer hyperimmune plasma or monoclonal antibody m336. Both groups had relieved symptoms of clinical diseases, but the reduction of respiratory viral load was only found in the hyperimmune plasma group. Although both super immune plasma and m336 therapy show to mitigate the disease of the common marmoset, neither has the ability to prevent the disease completely. 95 Yet, HMab m336 is found to significantly reduce the viral RNA titers and viral‐associated pathological changes in rabbit lung tissue. 94 Mice inoculated with S nanoparticles produced high‐level neutralizing antibodies against homologous viruses, and these antibodies have no cross‐protection with heteroviruses. 96 After being stimulated by SARS‐CoV, immunized ferrets produced more rapid and stronger neutralizing antibody reaction than the control animals; however, the strong inflammatory reaction is observed in liver tissue. All this suggests that the expression of SARS‐CoV S protein is associated with enhanced hepatitis. 97 On the other hand, the time course of SARS‐CoV viremia and antibody response has been studied. 98 SARS‐CoV viremia is not detected in the blood samples of convalescent patients. In the peak period of viremia, 75% of the blood samples of patients diagnosed as SARS in the first 1 to 2 weeks before detection can detect virus RNA. The prolongation of IgG production may indicate the significance of IgG in both humoral immune response to acute SARS‐CoV infection and clearance of the remaining virus sources during recovery. This is an important subject that needs further study.
4. CONCLUSIONS
Since the emergence of SARS‐CoV in 2002 and its spread throughout 32 countries and areas, the world has experienced the outbreak of MERS‐CoV and now, the 2019‐nCoV. All these viruses belong to the subfamily Coronavirinae in the family Coronaviridae. Since CoVs emerge periodically and unpredictably, spread rapidly, and induce serious infectious diseases, they become a continuous threat to human health. This is especially true when there are no approved vaccines or drugs for the treatment of CoV infections and there exists a range of animal reservoirs for CoVs and recombinant CoVs. In recent years, profound understandings of the innate immune response to viruses have been made. This type of immune response inhibits virus replication, promotes virus clearance, induces tissue repair, and triggers a prolonged adaptive immune response against the viruses. In most cases, pulmonary and systemic inflammatory responses associated with CoVs are triggered by the innate immune system when it recognizes the viruses. Although a broadly protective, universal vaccine is considered the ultimate protection against the virus spread, vaccine development can be time‐consuming. To fulfill the pressing need, we should propose effective therapeutic measures using the accumulated knowledge of the innate immune response system. Targeted immunotherapy is a good alternative to some antivirals that have narrow treatment windows and meet with drug resistance easily. In 2003, glucocorticoid was widely used in SARS treatment to control pulmonary infection by regulating inflammatory responses. Except for viral pathogenicity, the inflammatory response of the body also plays a crucial role in SARS‐induced lung injury cases. Therefore, in CoV pneumonia cases, it is important to control cytokine production and inflammatory response, given that they are responsible for the accumulation of cells and fluids. This strategy is challenging as we have not yet clearly identified any features in an immune response that can be inhibited specifically without compromising the beneficial host defense.
However, accomplishing this is not impossible. Notable achievements have been made in analyzing detrimental and protective mechanisms. For instance, completely blocking a proximal event in the immune response (eg, activation of IFN response‐related PRRs) seems unwise considering its general role in regulating the host defense. In contrast, more limited and specific effector arms, such as controlled production of oxygen radicals, NET formation, IL‐1, IL‐4, IL‐6, IL‐8, and IL‐21 production, are probably practicable targets. At last, further research is needed to improve the understanding of the temporal features of CoV‐induced inflammatory response in relation to the timing of therapeutic interventions.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
ACKNOWLEDGMENTS
This work was supported by research grants from the National Natural Science Foundation of China (81902066, 81730061, and 81471942).
Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92:424–432. 10.1002/jmv.25685
REFERENCES
- 1. World Health Organization . Laboratory testing of human suspected cases of novel coronavirus (nCoV) infection [published online ahead of print January 21, 2020]. https://apps.who.int/iris/bitstream/handle/10665/330374/WHO-2019-nCoV-laboratory-2020.1-eng.pdf
- 2. World Health Organization . Novel Coronavirus (2019‐nCoV) situation report‐2 [published online ahead of print January 21, 2020]. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200122-sitrep-2-2019-ncov.pdf
- 3. World Health Organization . Middle East respiratory syndrome coronavirus (MERS‐CoV) [published online ahead of print January 21, 2020]. https://www.who.int/emergencies/mers-cov/en/
- 4. World Health Organization . WHO MERS global summary and assessment of risk [published online ahead of print January 21, 2020]. https://www.who.int/csr/disease/coronavirus_infections/risk-assessment-august-2018.pdf?ua=1
- 5. Koh D, Sng J. Lessons from the past: perspectives on severe acute respiratory syndrome. Asia Pac J Public Health. 2010;22(3 Suppl):132s‐136s. [DOI] [PubMed] [Google Scholar]
- 6. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783‐801. [DOI] [PubMed] [Google Scholar]
- 7. Kawai T, Akira S. The role of pattern‐recognition receptors in innate immunity: update on Toll‐like receptors. Nature Immunol. 2010;11(5):373‐384. [DOI] [PubMed] [Google Scholar]
- 8. Pobezinskaya YL, Kim YS, Choksi S, et al. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF‐dependent Toll‐like receptors. Nature Immunol. 2008;9(9):1047‐1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ermolaeva MA, Michallet MC, Papadopoulou N, et al. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF‐dependent inflammatory responses. Nature Immunol. 2008;9(9):1037‐1046. [DOI] [PubMed] [Google Scholar]
- 10. Hacker H, Karin M. Regulation and function of IKK and IKK‐related kinases. Sci STKE. 2006;2006(357):re13. [DOI] [PubMed] [Google Scholar]
- 11. Häcker H, Redecke V, Blagoev B, et al. Specificity in Toll‐like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439(7073):204‐207. [DOI] [PubMed] [Google Scholar]
- 12. Kell AM, Gale M Jr. RIG‐I in RNA virus recognition. Virology. 2015;479‐480:110‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoneyama M, Fujita T. RNA recognition and signal transduction by RIG‐I‐like receptors. Immunol Rev. 2009;227(1):54‐65. [DOI] [PubMed] [Google Scholar]
- 14. Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG‐I‐like receptors. Curr Opin Immunol. 2015;32:48‐53. [DOI] [PubMed] [Google Scholar]
- 15. Yoneyama M, Fujita T. Recognition of viral nucleic acids in innate immunity. Rev Med Virol. 2010;20(1):4‐22. [DOI] [PubMed] [Google Scholar]
- 16. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805‐820. [DOI] [PubMed] [Google Scholar]
- 17. Weber M, Gawanbacht A, Habjan M, et al. Incoming RNA virus nucleocapsids containing a 5′‐triphosphorylated genome activate RIG‐I and antiviral signaling. Cell Host Microbe. 2013;13(3):336‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goubau D, Schlee M, Deddouche S, et al. Antiviral immunity via RIG‐I‐mediated recognition of RNA bearing 5'‐diphosphates. Nature. 2014;514(7522):372‐375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Civril F, Bennett M, Moldt M, et al. The RIG‐I ATPase domain structure reveals insights into ATP‐dependent antiviral signalling. EMBO Rep. 2011;12(11):1127‐1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takahasi K, Kumeta H, Tsuduki N, et al. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C‐terminal domains: identification of the RNA recognition loop in RIG‐I‐like receptors. J Biol Chem. 2009;284(26):17465‐17474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu B, Peisley A, Richards C, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell. 2013;152(1‐2):276‐289. [DOI] [PubMed] [Google Scholar]
- 22. Inohara Chamaillard, McDonald C, Nunez G. NOD‐LRR proteins: role in host‐microbial interactions and inflammatory disease. Annu Rev Biochem. 2005;74:355‐383. [DOI] [PubMed] [Google Scholar]
- 23. Inohara N, Nunez G. The NOD: a signaling module that regulates apoptosis and host defense against pathogens. Oncogene. 2001;20(44):6473‐6481. [DOI] [PubMed] [Google Scholar]
- 24. Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL‐1beta‐processing inflammasome with increased activity in Muckle‐Wells autoinflammatory disorder. Immunity. 2004;20(3):319‐325. [DOI] [PubMed] [Google Scholar]
- 25. Davis BK, Roberts RA, Huang MT, et al. Cutting edge: NLRC5‐dependent activation of the inflammasome. J Immunol. 2011;186(3):1333‐1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hornung V, Ablasser A, Charrel‐Dennis M, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase‐1‐activating inflammasome with ASC. Nature. 2009;458(7237):514‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van Gorp H, Kuchmiy A, Van Hauwermeiren F, Lamkanfi M. NOD‐like receptors interfacing the immune and reproductive systems. FEBS J. 2014;281(20):4568‐4582. [DOI] [PubMed] [Google Scholar]
- 28. Rogers NC, Slack EC, Edwards AD, et al. Syk‐dependent cytokine induction by Dectin‐1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22(4):507‐517. [DOI] [PubMed] [Google Scholar]
- 29. Geijtenbeek TB, Gringhuis SI. Signalling through C‐type lectin receptors: shaping immune responses. Nat Rev Immunol. 2009;9(7):465‐479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hara H, Tsuchiya K, Kawamura I, et al. Phosphorylation of the adaptor ASC acts as a molecular switch that controls the formation of speck‐like aggregates and inflammasome activity. Nature Immunol. 2013;14(12):1247‐1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strasser D, Neumann K, Bergmann H, et al. Syk kinase‐coupled C‐type lectin receptors engage protein kinase C‐sigma to elicit Card9 adaptor‐mediated innate immunity. Immunity. 2012;36(1):32‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hemmi H, Takeuchi O, Kawai T, et al. A Toll‐like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740‐745. [DOI] [PubMed] [Google Scholar]
- 33. Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG‐I pathway. Cell. 2009;138(3):576‐591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP‐AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science (New York, NY). 2013;339(6121):786‐791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gallego‐Marin C, Schrum JE, Andrade WA, et al. Cyclic GMP‐AMP synthase is the cytosolic sensor of plasmodium falciparum genomic DNA and activates type I IFN in malaria. J Immunol. 2018;200(2):768‐774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012;86(6):2900‐2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ma DY, Suthar MS. Mechanisms of innate immune evasion in re‐emerging RNA viruses. Current opinion in virology. 2015;12:26‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nelemans T, Kikkert M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses. 2019;11(10):961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14(1):36‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cao L, Ji Y, Zeng L, et al. P200 family protein IFI204 negatively regulates type I interferon responses by targeting IRF7 in nucleus. PLOS Pathog. 2019;15(10):e1008079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gordon S. Pattern recognition receptors: doubling up for the innate immune response. Cell. 2002;111(7):927‐930. [DOI] [PubMed] [Google Scholar]
- 42. Baccala R, Gonzalez‐Quintial R, Lawson BR, et al. Sensors of the innate immune system: their mode of action. Nat Rev Rheumatol. 2009;5(8):448‐456. [DOI] [PubMed] [Google Scholar]
- 43. Phadwal K, Alegre‐Abarrategui J, Watson AS, et al. A novel method for autophagy detection in primary cells: impaired levels of macroautophagy in immunosenescent T cells. Autophagy. 2012;8(4):677‐689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seranova E, Ward C, Chipara M, Rosenstock TR, Sarkar S. In vitro screening platforms for identifying autophagy modulators in mammalian cells. Methods Mol Biol. 2019;1880:389‐428. [DOI] [PubMed] [Google Scholar]
- 45. Hou J, Wang P, Lin L, et al. MicroRNA‐146a feedback inhibits RIG‐I‐dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J Immunol. 2009;183(3):2150‐2158. [DOI] [PubMed] [Google Scholar]
- 46. Schneider WM, Chevillotte MD, Rice CM. Interferon‐stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513‐545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Spiegel M, Pichlmair A, Martinez‐Sobrido L, et al. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two‐step model for activation of interferon regulatory factor 3. J Virol. 2005;79(4):2079‐2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kopecky‐Bromberg SA, Martinez‐Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol. 2007;81(2):548‐557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lu X, Pan J, Tao J, Guo D. SARS‐CoV nucleocapsid protein antagonizes IFN‐beta response by targeting initial step of IFN‐beta induction pathway, and its C‐terminal region is critical for the antagonism. Virus Genes. 2011;42(1):37‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Y, Zhang Z, Zhao X, et al. Enterovirus 71 inhibits cellular type I interferon signaling by downregulating JAK1 protein expression. Viral Immunol. 2014;27(6):267‐276. [DOI] [PubMed] [Google Scholar]
- 51. Okumura A, Pitha PM, Yoshimura A, Harty RN. Interaction between Ebola virus glycoprotein and host toll‐like receptor 4 leads to induction of proinflammatory cytokines and SOCS1. J Virol. 2010;84(1):27‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pauli EK, Schmolke M, Wolff T, et al. Influenza A virus inhibits type I IFN signaling via NF‐kappaB‐dependent induction of SOCS‐3 expression. PLOS Pathog. 2008;4(11):e1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kaewraemruaen C, Ritprajak P, Hirankarn N. Dendritic cells as key players in systemic lupus erythematosus. Asian Pac J Allergy Immunol. 2019. 10.12932/AP-070919-0639 [DOI] [PubMed] [Google Scholar]
- 54. Wu L, Dakic A. Development of dendritic cell system. Cell Mol Immunol. 2004;1(2):112‐118. [PubMed] [Google Scholar]
- 55. Nouri‐Shirazi M, Guinet E. Exposure to nicotine adversely affects the dendritic cell system and compromises host response to vaccination. J Immunol. 2012;188(5):2359‐2370. [DOI] [PubMed] [Google Scholar]
- 56. Guo Y, Xu WW, Song J, Deng W, Liu DQ, Zhang HT. Intracellular overexpression of HIV‐1 Nef impairs differentiation and maturation of monocytic precursors towards dendritic cells. PLOS One. 2012;7(7):e40179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tu Z, Hamalainen‐Laanaya HK, Nishitani C, Kuroki Y, Crispe IN, Orloff MS. HCV core and NS3 proteins manipulate human blood‐derived dendritic cell development and promote Th 17 differentiation. Int Immunol. 2012;24(2):97‐106. [DOI] [PubMed] [Google Scholar]
- 58. Fairman P, Angel JB. The effect of human immunodeficiency virus‐1 on monocyte‐derived dendritic cell maturation and function. Clin Exp Immunol. 2012;170(1):101‐113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cardone M, Ikeda KN, Varano B, Gessani S, Conti L. HIV‐1‐induced impairment of dendritic cell cross talk with gammadelta T lymphocytes. J Virol. 2015;89(9):4798‐4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. White MR, Tecle T, Crouch EC, Hartshorn KL. Impact of neutrophils on antiviral activity of human bronchoalveolar lavage fluid. Am J Physiol. 2007;293(5):L1293‐L1299. [DOI] [PubMed] [Google Scholar]
- 61. Campbell O, Gagnon J, Rubin JE. Antibacterial activity of chemotherapeutic drugs against Escherichia coli and Staphylococcus pseudintermedius. Lett Appl Microbiol. 2019;69(5):353‐357. [DOI] [PubMed] [Google Scholar]
- 62. Zharkova MS, Orlov DS, Golubeva OY, et al. Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics‐A novel way to combat antibiotic resistance? Front Cell Infect Microbiol. 2019;9:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lehrer RI, Lichtenstein AK, Ganz T. Defensins: antimicrobial and cytotoxic peptides of mammalian cells. Annu Rev Immunol. 1993;11:105‐128. [DOI] [PubMed] [Google Scholar]
- 64. Li SZ, Shu QP, Song Y, et al. Phosphorylation of MAVS/VISA by Nemo‐like kinase (NLK) for degradation regulates the antiviral innate immune response. Nat Commun. 2019;10(1):3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Daher KA, Selsted ME, Lehrer RI. Direct inactivation of viruses by human granulocyte defensins. J Virol. 1986;60(3):1068‐1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nakashima H, Yamamoto N, Masuda M, Fujii N. Defensins inhibit HIV replication in vitro. AIDS. 1993;7(8):1129. [DOI] [PubMed] [Google Scholar]
- 67. Zapata W, Aguilar‐Jiménez W, Feng Z, et al. Identification of innate immune antiretroviral factors during in vivo and in vitro exposure to HIV‐1. Microb Infect. 2016;18(3):211‐219. [DOI] [PubMed] [Google Scholar]
- 68. Yasui F, Kai C, Kitabatake M, et al. Prior immunization with severe acute respiratory syndrome (SARS)‐associated coronavirus (SARS‐CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS‐CoV. J Immunol. 2008;181(9):6337‐6348. [DOI] [PubMed] [Google Scholar]
- 69. Zhu X, Wang Y, Zhang H, et al. Genetic variation of the human alpha‐2‐Heremans‐Schmid glycoprotein (AHSG) gene associated with the risk of SARS‐CoV infection. PLOS One. 2011;6(8):e23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS‐like disease. Clin Microbiol Rev. 2015;28(2):465‐522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20(4):660‐694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sato K, Misawa N, Takeuchi JS, et al. Experimental adaptive evolution of simian immunodeficiency virus sivcpz to pandemic human immunodeficiency virus type 1 by using a humanized mouse model. J Virol. 2018;92(4):e01905‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cecere TE, Todd SM, Leroith T. Regulatory T cells in arterivirus and coronavirus infections: do they protect against disease or enhance it? Viruses. 2012;4(5):833‐846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Maloir Q, Ghysen K, von Frenckell C, Louis R, Guiot J. [Acute respiratory distress revealing antisynthetase syndrome]. Rev Med Liege. 2018;73(7‐8):370‐375. [PubMed] [Google Scholar]
- 75. Zhao J, Li K, Wohlford‐Lenane C, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA. 2014;111(13):4970‐4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pascal KE, Coleman CM, Mujica AO, et al. Pre‐ and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS‐CoV infection. Proc Natl Acad Sci USA. 2015;112(28):8738‐8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Channappanavar R, Fett C, Zhao J, Meyerholz DK, Perlman S. Virus‐specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol. 2014;88(19):11034‐11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ng OW, Chia A, Tan AT, et al. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post‐infection. Vaccine. 2016;34(17):2008‐2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen J, Lau YF, Lamirande EW, et al. Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS‐CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS‐CoV infection. J Virol. 2010;84(3):1289‐1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Manni ML, Robinson KM, Alcorn JF. A tale of two cytokines: IL‐17 and IL‐22 in asthma and infection. Expert review of respiratory medicine. 2014;8(1):25‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bunte K, Beikler T. Th17 Cells and the IL‐23/IL‐17 axis in the pathogenesis of periodontitis and immune‐mediated inflammatory diseases. Int J Mol Sci. 2019;20(14):3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dutzan N, Abusleme L. T helper 17 cells as pathogenic drivers of periodontitis. Adv Exp Med Biol. 2019;1197:107‐117. [DOI] [PubMed] [Google Scholar]
- 83. Yang Y, Xiong Z, Zhang S, et al. Bcl‐xL inhibits T‐cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem J. 2005;392(Pt 1):135‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mubarak A, Alturaiki W, Hemida MG. Middle East respiratory syndrome coronavirus (MERS‐CoV): infection, immunological response, and vaccine development. J Immunol Res. 2019;2019:6491738‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ababneh M, Alrwashdeh M, Khalifeh M. Recombinant adenoviral vaccine encoding the spike 1 subunit of the Middle East Respiratory Syndrome Coronavirus elicits strong humoral and cellular immune responses in mice. Vet World. 2019;12(10):1554‐1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tuhin ali M, Morshed MM, Musa MA, et al. Computer aided prediction and identification of potential epitopes in the receptor binding domain (RBD) of spike (S) glycoprotein of MERS‐CoV. Bioinformation. 2014;10(8):533‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Niu P, Zhang S, Zhou P, et al. Ultrapotent human neutralizing antibody repertoires against Middle East Respiratory syndrome coronavirus from a recovered patient. J Infect Dis. 2018;218(8):1249‐1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chen Z, Bao L, Chen C, et al. Human neutralizing monoclonal antibody inhibition of Middle East Respiratory Syndrome coronavirus replication in the common marmoset. J Infect Dis. 2017;215(12):1807‐1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Niu P, Zhao G, Deng Y, et al. A novel human mAb (MERS‐GD27) provides prophylactic and postexposure efficacy in MERS‐CoV susceptible mice. Science China Life sciences. 2018;61(10):1280‐1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Baker S, Kessler E, Darville‐Bowleg L, Merchant M. Different mechanisms of serum complement activation in the plasma of common (Chelydra serpentina) and alligator (Macrochelys temminckii) snapping turtles. PLOS One. 2019;14(6):e0217626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sun S, Zhao G, Liu C, et al. Inhibition of complement activation alleviates acute lung injury induced by highly pathogenic avian influenza H5N1 virus infection. Am J Respir Cell Mol Biol. 2013;49(2):221‐230. [DOI] [PubMed] [Google Scholar]
- 92. Gralinski LE, Sheahan TP, Morrison TE, et al. Complement activation contributes to severe acute respiratory syndrome coronavirus pathogenesis. mBio. 2018;9(5):e01753‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ying T, Du L, Ju TW, et al. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol. 2014;88(14):7796‐7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Houser KV, Gretebeck L, Ying T, et al. Prophylaxis with a Middle East Respiratory Syndrome Coronavirus (MERS‐CoV)‐specific human monoclonal antibody protects rabbits from MERS‐CoV Infection. J Infect Dis. 2016;213(10):1557‐1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. van Doremalen N, Falzarano D, Ying T, et al. Efficacy of antibody‐based therapies against Middle East respiratory syndrome coronavirus (MERS‐CoV) in common marmosets. Antiviral Res. 2017;143:30‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Coleman CM, Liu YV, Mu H, et al. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014;32(26):3169‐3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Weingartl H, Czub M, Czub S, et al. Immunization with modified vaccinia virus Ankara‐based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78(22):12672‐12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chen W. Antibody response and viraemia during the course of severe acute respiratory syndrome (SARS)‐associated coronavirus infection. J Med Microbiol. 2004;53(Pt 5):435‐438. [DOI] [PubMed] [Google Scholar]


