Abstract
Human metapneumovirus (hMPV) has been recognized as an important cause of respiratory tract infections in all age groups and in all geographical area. The role of hMPV in causing respiratory tract infections in Kuwait was not yet investigated. The aim of this study was to determine the prevalence of hMPV infection in Kuwait among patients with respiratory tract infection with respect to other respiratory viruses. During January–December 2009, 460 respiratory samples from 388 patients with respiratory tract infection were collected from different hospitals. They were tested for hMPV RNA by real‐time PCR, and for other respiratory viruses by conventional PCR. Out of 388 patients, 110 (28%) were positive for viral respiratory infections; 21 (5.4%) were positive for hMPV, 29 (7.5%) were positive for rhinovirus, 13 (4%) were positive for respiratory syncytial virus, and 10 (3%) were positive for adenovirus. Most (n = 19, 90.5%) of hMPV‐positive patients were admitted to the intensive care unit, 76% of them were of age 2 years and below, and 24% of age 59 years and above. All hMPV‐positive elderly patients had pneumonia while 50% of hMPV‐positive infants had bronchopneumonia. Children with hMPV/rhinovirus co‐infection (n = 3, 1%) had recurrent chest infection and frequent intensive care unit admission. The hMPV infection was mostly detected between December and May, and genotype B was more prevalent than genotype A. This is the first study demonstrating the prevalence of hMPV infection in Kuwait, and suggests that hMPV infection is prevalent in infants and elderly patients with lower respiratory tract infection. J. Med. Virol. 83:1811–1817, 2011. © 2011 Wiley‐Liss, Inc.
Keywords: human metapneumovirus (hMPV), prevalence, respiratory tract infections, viral load, co‐infection
INTRODUCTION
Human metapneumovirus (hMPV) was discovered in 2001 in the Netherlands from a nasopharyngeal aspirate of young children with acute respiratory tract infection [Van den Hoogen et al., 2001]. Since then, hMPV has been identified frequently in respiratory tract infections worldwide [Kahn, 2006], and lately recognized as a major cause of lower respiratory tract infections in infants, elderly, and immunocompromised patients [Bao et al., 2008]. Around 12% of all respiratory tract infections in children are caused by hMPV [Kahn, 2006], and almost all children are infected by the age of 5 [Van den Hoogen et al., 2001]. Human metapneumovirus also accounts for 10% of all hospitalizations of elderly patients with respiratory tract infections [Falsey et al., 2003], and is a significant pathogen in immunocompromised patients [Muir and Pillay, 1998; Williams et al., 2005].
The clinical symptoms associated with hMPV infections are very similar to those symptoms observed in respiratory syncytial virus (RSV) infection. Human metapneumovirus can cause both upper and lower respiratory tract infections, such as bronchitis, bronchiolitis, and pneumonia [Boivin et al., 2002], as well as asthma exacerbation [Kahn, 2003]. Symptoms such as fever, cough, tachypnea, wheezing, and hypoxia are frequently observed in infected children, and in children with a clinical syndrome consistent with bronchiolitis, hyperinflation, as well as focal infiltrates and peribronchial cuffing [Williams et al., 2004]. Some reports have shown that hMPV infection is associated with encephalitis, and there is at least one example where hMPV was detected in a patient who died of complications associated with encephalitis [Schildgen et al., 2005]. In addition, some studies have reported a hospital‐acquired hMPV infection in infants, children, and elderly patients in long‐term care facilities [Chano et al., 2005; Honda et al., 2006; Boivin et al., 2007; Kim et al., 2009].
Genetic analysis revealed a relatively high degree of sequence variability between different hMPV isolates, and two major subgroups A and B were identified. Subsequent genetic analysis led to a further subdivision of the hMPV A and B subgroups into the subtypes 1A, 2A, 1B, and 2B [Biacchesi et al., 2003]. Moreover, subgroup A2 was further subdivided into two minor subgroups A2a and A2b [Huck et al., 2006]. Although viral respiratory tract infections are very common in Kuwait and responsible of acute respiratory diseases and pneumonia [Hijazi et al., 1996, 1997], the prevalence of hMPV infection is not yet documented in Kuwait. The aims of this study were therefore to determine the prevalence of hMPV infection in Kuwait in patients with respiratory tract infections with respect to the other respiratory viruses, to identify the type of circulating hMPV, and to determine the seasonal distribution of hMPV infection.
MATERIALS AND METHODS
Specimen Collection
All respiratory samples were collected during January–December, 2009 from different hospitals in Kuwait; 236 (51%) were from the Mubarak Al‐Kabir Hospital, 136 (30%) from the Al‐Sabah Hospital, 31 (7%) from the Farwanya Hospital, 23 (5%) from the Amiri Hospital, 11 (2.5%) from the Abdul‐Aziz Al‐Rashed Allergy Center, 9 (2%) from the Organ Transplant Center, 4 (1%) from the Adan Hospital, 4 (1%) from the Ibn‐Sina Hospital, 4 (1%) from the Jahra Hospital, 1 (0.3%) from the Infectious Diseases Hospital, and 1 (0.3%) from the Kuwait Cancer Center. Since the year of 2009 had witnessed the H1N1 pandemic, respiratory samples that were H1N1 positive starting from August to December have been excluded. The 460 respiratory samples were obtained from 388 patients, 210 (54%) were males and 178 (46%) were females, with either upper (n = 94, 24%) or lower (n = 294, 76%) respiratory tract infections of all age groups, from 0 day up to 80 years, and from different nationalities. The respiratory samples included bronchoalveolar lavage, endotracheal tube secretions, tracheal aspirate, throat swab, nasopharyngeal aspirate, nasopharyngeal swab, nasal swab, nasal and throat swabs, and pleural fluid. All respiratory samples were collected after obtaining written informed consent from patients. The ethical permission on this research study was granted by the Ethical Decision Committee of the Research Administration, Faculty of Medicine, Kuwait University.
Viral RNA Extraction
The hMPV RNA extraction was done using the automated nucleic acid extraction method, MagNA Pure LC 2.0 (Roche Diagnostics Ltd, Rotkreuz, Switzerland). All 460 respiratory samples were extracted using the MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instruction.
Real‐Time RT‐PCR for Detection of hMPV RNA
The hMPV RNA was detected using a commercial real‐time PCR detection assay that amplifies the hMPV nucleoprotein gene (PrimerDesign, PrimerDesignTM Ltd, Southampton, Hants, UK), in combination with the AgPath‐IDTM one‐step RT‐PCR mix reagent (Ambion, Applied Biosystems, Austin, TX). RT‐PCR master mix was prepared on ice by adding 12.5 µl of 2× RT‐PCR buffer, 1.5 µl of the pathogen specific primer/probe mix, 1 µl of 25× RT‐PCR enzyme mix, 4 µl nuclease‐free water, and 6 µl of the extracted RNA sample. The Applied Biosystem 7500 Fast Real‐Time PCR System (Applied Biosystems, Foster City, CA) was used for hMPV detection according to the following cycling: 45°C for 10 min, 95°C for 10 min, 50 cycles of 95°C for 15 sec, and 60°C for 60 sec.
Detection of Other Respiratory Viruses by PCR
In addition to the hMPV detection, all samples were also tested for other respiratory viruses as described elsewhere [Khadadah et al., 2010]. Briefly, a single PCR was used to detect adenovirus and parainfluenza virus‐2 (PIV‐2); duplex PCR was carried out to detect influenza A and B viruses; triplex PCR was carried out to detect respiratory syncytial virus (RSV), parainfluenza viruses (PIV) ‐1 and ‐3, and another triplex PCR was performed to detect human rhinovirus, human coronaviruses‐229E and ‐OC43 [Khadadah et al., 2010].
Genotyping of Detected hMPV RNA
A set of degenerate primers was selected on the basis of sequences published by Mackay et al. [2003] (forward primer hMPVF; 5′‐AAYMGWGTRYTAAGTGATGCRCTC‐3′, nucleotide position 601–624, and reverse primer hMPVR; 5′‐CAKTGTYTGRCCRGCHCCRTAA‐3′, nucleotide position 792–813) to amplify the N gene (hMPV isolate 00‐1, GenBank accession number under AF371337). The RT‐PCR was performed using the Qiagen One‐Step RT‐PCR Kit (Qiagen, Hilden, Germany). The reaction mixture was exposed to a 30 min, 45°C reverse transcription incubation, and then 40 cycles of 94°C for 20 sec, 50°C for 20 sec, and 68°C for 20 sec, with a final 68°C extension for 7 min using the GeneAmp® PCR System 9700 (Applied Biosystems). The RT‐PCR amplification reaction resulted in a 213‐bp PCR product when separated by 2% agarose gel electrophoresis and visualized by ethidium bromide staining.
The PCR fragments were purified with QIAquick PCR purification columns as described by the manufacturer (Qiagen, Suite, Valencia), and both strands were directly sequenced with both hMPVF and hMPVR primers using the ABI PRISM® BigDye® Terminator Cycle Sequencing v3.1 Ready Reaction kit (Applied Biosystems). Labeled DNA fragments were separated and detected using ABI genetic analyzer 3130 (Applied Biosystems). The obtained sequences were then analyzed for genotype identification using Basic Local Alignment Search Tool (BLAST) alignment from GenBank (http://www.ncbi.nlm.nih.gov/Blast.cgi) and compared to sequences available from the GenBank database (GenBank accession numbers were as follows: Subgroup A1, AF371337; subgroup A2a, NC_004148, FJ168779, and AB503857; subgroup A2b, DQ843659, and GQ153651; subgroup B1, AY525843; and subgroup B2, DQ843658, AY297748, and FJ168778).
Statistical Analysis
The difference in the hMPV viral load between different groups of patients was assessed using Mann–Whitney and Kruskal–Wallis tests. The significance of the difference in categorical variables among various groups was tested using the χ2‐test and Fisher exact test, as appropriate. The statistical analysis was performed using SPSS v17.0 software (SPSS Inc., Chicago, IL).
RESULTS
The clinical data of the study population is shown in Table I. All H1N1‐negative respiratory samples were screened for hMPV and other respiratory viruses. Table II shows that rhinovirus was the most detected virus in clinical respiratory specimens of patients with respiratory symptoms (6.3%), followed by hMPV (5%), respiratory syncytial virus (3%), and adenovirus (2.8%). Coronavirus‐OC43 and parainfluenza virus type 2 were not detected. The prevalence of hMPV infection in patients with respiratory tract infections was 5.4%. Human metapneumovirus was also found in the presence of rhinovirus in three patients (1%), and adenovirus in one patient (0.3%), with a co‐infection rate of 1%.
Table I.
Clinical Data of the Study Population
| Respiratory symptoms | Patients, n (%) |
|---|---|
| Lower respiratory tract infection | |
| Pneumonia | 137 (35%) |
| Bronchopneumonia | 36 (9%) |
| Bronchiolitis | 38 (10%) |
| Acute exacerbations of bronchial asthma | 16 (4%) |
| COPD | 6 (1.6%) |
| Pulmonary edema | 1 (0.3%) |
| Pulmonary hypertension | 1 (0.3%) |
| Respiratory distress | 1 (0.3%) |
| Respiratory failure | 7 (2%) |
| Wheezy chest | 2 (0.5%) |
| Unspecified LRTI symptoms | 49 (13%) |
| Total | 294 (76%) |
| Upper respiratory tract infection | 94 (24%) |
| Total | 388 (100%) |
COPD, chronic obstructive pulmonary disease.
Upper respiratory tract infections include: Cough, Rhinitis, Fever, Tonsillitis, and Flu‐like Illnesses.
Table II.
Nucleic Acid Detection of Respiratory Viruses in Clinical Specimens From Patients With Respiratory Symptoms
| Respiratory viruses | Patients, n (%) |
|---|---|
| hMPV | 21 (5.4%) |
| RSV | 13 (4%) |
| Adenoviruses | 10 (3%) |
| Influenza‐A | 5 (1.3%) |
| Influenza‐B | 1 (0.3%) |
| Rhinoviruses | 29 (7.5%) |
| PIV‐1 | 10 (3%) |
| PIV‐2 | 0 |
| PIV‐3 | 6 (1.5%) |
| Corona‐229E | 1 (0.3%) |
| Corona‐OC43 | 0 |
| hMPV/Rhinovirus | 3 (1%) |
| hMPV/Adenovirus | 1 (0.3%) |
| Rhinovirus/RSV | 2 (0.5%) |
| Rhinovirus/Adenovirus | 1 (0.3%) |
| Rhinovirus/PIV‐1 | 1 (0.3%) |
| Rhinovirus/PIV‐3 | 2 (0.5%) |
| PIV‐1/PIV‐3 | 4 (1%) |
| Total | 110 (28%) |
hMPV, human metapneumovirus; RSV, respiratory Syncytial virus; PIV, parainfluenza virus.
The hMPV‐positive patients were segregated according to their sex, age, and intensive care unit/ward admission (Table III). The overall proportion of hMPV infection in females was 7% (n = 13) and that in males was 4% (n = 8) (P = 0.13). Most of the hMPV‐positive patients (76%) were infants having less than 2 years, while 24% of them were patients having 59 years and above, and the overall prevalence of hMPV infection in infants and elderly was 11 and 8%, respectively. The hMPV infection was not detected among children aged more than 2 years and adults aged less than 59 years. Most of the hMPV‐positive patients (90%) were admitted to the intensive care unit; the overall proportion of hMPV infection in patients admitted to intensive care unit was 8%. The hMPV was only detected in the following types of respiratory samples: Endotracheal tube secretions (30%), tracheal aspirate (30%), nasopharyngeal aspirate (30%), and nasopharyngeal swab (10%). The difference in the hMPV RNA load among different types of respiratory samples was not significant.
Table III.
Characteristics of hMPV‐Positive Patients
| Patient, n (%) | |
|---|---|
| Sex | |
| Male | 8 (4%) |
| Female | 13 (7%) |
| Total | 21 (5.4%) |
| Age | |
| <2 years | 16 (11%) |
| >59 years | 5 (8%) |
| Hospital admission | |
| Ward | 2 (1%) |
| Intensive Care Unit | 19 (8%) |
All hMPV‐positive patients had lower respiratory tract infections with high detectable RNA load. All elderly patients (n = 5) had pneumonia, while 50% (n = 8) of infants less than 2 years had bronchopneumonia, 25% (n = 4) had pneumonia, and 13% (n = 2) had acute exacerbation of bronchial asthma. The median hMPV RNA load for each group of patients according to their clinical data was as the following: 1.76 × 106 copies/ml for patients with pneumonia, 4.08 × 107 copies/ml for patients with bronchopneumonia, and 2.31 × 108 copies/ml for patients presented with acute exacerbation of bronchial asthma. In addition, one patient with bronchiolitis had hMPV RNA load of 7.68 × 107 copies/ml, and one with respiratory distress had 4.48 × 106 copies/ml. The difference in the viral load between each group of patients with lower respiratory tract infection was not significant (P = 0.55). The median hMPV RNA load in infants aged 2 years and below (9.48 × 106 copies/ml) was not statistically different from that in old patients aged 59 years and above (1.31 × 107 copies/ml) (P = 0.89). The genotype of hMPV RNA detected was identified by direct sequencing. Eleven hMPV RT‐PCR products could be sequenced showing high similarity with hMPV sequences available in the GenBank database. Two hMPV genotypes were detected, A subgroup 2b and B subgroup 2; although genotype B2 was more prevalent than A2b (Table IV). The hMPV infection was detected during winter and spring seasons (Fig. 1), with high incidence rate in spring, and with very low incidence during summer (P = 0.005).
Table IV.
Genotypes of Detected hMPV in Patients With Respiratory Tract Infections
| Patient | Age | Sex | Clinical Diagnosis | hMPV RNA Load copies/ml | hMPV Genotype (Homology %)a |
|---|---|---|---|---|---|
| A | 2 months | Female | Bronchiolitis | 7.68 × 107 | B2 (99%) |
| B | 5 months | Female | Bronchopneumonia | 6.78 × 108 | A2b (97%) |
| C | 5 months | Female | Respiratory Distress | 4.48 × 106 | B2 (97%) |
| D | 6 months | Male | Bronchopneumonia | 8.97 × 106 | B2 (97%) |
| E | 9 months | Female | Bronchopneumonia | 1.61 × 106 | B2 (97%) |
| F | 10 months | Female | Bronchopneumonia | 1.44 × 109 | B2 (92%) |
| G | 1 year | Female | Pneumonia | 5.73 × 108 | B2 (99%) |
| H | 2 years | Female | Acute Exacerbation of Bronchial Asthma | 4.62 × 108 | B2 (97%) |
| I | 59 years | Male | Pneumonia | 9.39 × 105 | A2b (95%) |
| J | 67 years | Male | Pneumonia | 1.47 × 1010 | B2 (99%) |
| K | 70 years | Female | Pneumonia | 4.21 × 109 | B2 (99%) |
Homology with reference genome from GenBank database.
Figure 1.
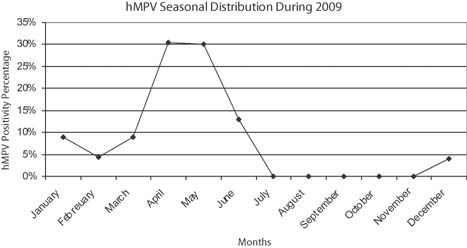
The seasonal distribution of hMPV infection in Kuwait.
DISSCUSSION
This is the first study in Kuwait exploring the importance of hMPV infection among patients with respiratory tract infections in hospitals. Previous studies on hMPV infection have been published in the Middle East area however, most of them have focused on one age group, children less than 5 years [Al‐Sonboli et al., 2005; Regev et al., 2006; Arabpour et al., 2008; Kaplan et al., 2008; Ali et al., 2010; Moattari et al., 2010]. Not surprising, most of respiratory samples collected in this study were from infants less than 2 years, showing that the most affected age group vulnerable for respiratory infections is infants. Infants are considered to be at high risk for respiratory infections, since their immunity system is being progressively developed during their first 2 years [File, 2000]. Respiratory infections are considered as a major cause of morbidity and mortality in developing countries [Campbell, 1995], and this agrees with our observation that most of the respiratory samples (65%) were collected from patients admitted to the intensive care unit for acute lower respiratory tract infections.
In the current study, rhinovirus was found to be the major cause of acute respiratory infections followed by hMPV and then respiratory syncytial virus. These results are in agreement with a previous study done in Kuwait showing that respiratory syncytial virus and human rhinovirus were the major cause of severe lower respiratory tract infections in Kuwait [Khadadah et al., 2010]. In addition, the best respiratory samples for hMPV detection were endotracheal tube secretions, tracheal aspirate, nasopharyngeal aspirate followed by nasopharyngeal swab, whereas hMPV RNA was not detected in throat swab, nasal swab, bronchoalveolar lavage and pleural fluid. This support earlier studies which recommended nasopharyngeal aspirate and swabs specimens for hMPV detection [Van den Hoogen et al., 2001, 2004a]. The prevalence of hMPV infection among the study population was 5.4%. Previous studies have reported that it accounts for at least 5 to 7% of the respiratory tract infections in hospitalized children, and at least 3% in the general community of patients who visit a general practitioner for respiratory tract infections [Van den Hoogen et al., 2004b].
Most of the hMPV‐positive patients were of age 2 years and less (76%), and of age 59 years and above (24%), and these results parallel most if not all studies that have shown that hMPV is a common disease in children and can cause severe infections in the elderly [Falsey et al., 2003; Wolf et al., 2003; Principi et al., 2006; Hermos et al., 2010]. In Qatar the hMPV virus was the most commonly detected virus, accounting for 74% of all respiratory infections among pediatric patients aged 1 month to 5 years, and this result is very similar to the current study finding that most of hMPV‐positive patients were of age 2 years and less than 76% [Al‐Thani et al., 2010]. Our results highlight the importance to introduce a screening test for hMPV infection in routine diagnosis, and to have appropriate medical management for hMPV‐positive cases as other studies have recommended previously [Deffrasnes et al., 2007]. Moreover, the clinical data of hMPV‐positive patients have shown that all elderly people presented with pneumonia mainly, while infants presented with a variety of clinical syndromes such as bronchopneumonia, pneumonia, acute exacerbation of bronchial asthma and bronchiolitis, a finding which has also been observed in previous studies [Boivin et al., 2002; Williams et al., 2004]. In this context, a study conducted on 10 elderly patients with underlying illness, has shown that four patients had developed pneumonitis and two have died due to hMPV infection [Boivin et al., 2002].
Of the 23 hMPV‐positive patients, three (1%) were also positive for rhinovirus and one (0.3%) was positive for adenovirus, with total co‐infection rate of 1%. Some previous reports have shown a co‐infection of hMPV/respiratory syncytial virus, which correlated with the severity of infection [Greensill et al., 2003; Semple et al., 2005]. On the other hand, other studies have shown that hMPV/respiratory syncytial virus co‐infection may normally be rare in most populations [Mackay et al., 2006], while some publications have failed to detect this co‐infection [Esper et al., 2003; Al‐Sonboli et al., 2006; Van Woensel et al., 2006]. Furthermore, some recent studies have found co‐infection of hMPV with SARS‐coronavirus and human bocavirus [Choi et al., 2006; Lee et al., 2007]. The co‐infection of hMPV/rhinovirus might be responsible for the severe symptoms presented by our patients. Indeed, one 2‐years‐old female had high hMPV RNA load (4.62 × 108 copies/ml) and presented with acute exacerbation of bronchial asthma. A male infant aged 2 months had recurrent chest infection with frequent intensive care unit admission, and his hMPV viral load was high (1.76 × 106 copies/ml). The last patient was a 1‐year‐old female with recurrent chest infection and frequent intensive care unit admission, and her hMPV RNA load was high (5.73 × 108 copies/ml). This suggests that the presence of hMPV infection with other respiratory virus may complicate the clinical condition and prolong patient stay in the hospital, as it has been reported earlier [Greensill et al., 2003; Semple et al., 2005; Xiao et al., 2010]. The high viral load of hMPV detected in our patients with lower respiratory tract infection may suggest a correlation between the severity of clinical symptoms and the virus replication rate, as described elsewhere [Boivin et al., 2002; Gerna et al., 2007; Bosis et al., 2008; Peng et al., 2010].
The detection of hMPV in the respiratory samples was confirmed by RT‐PCR and sequencing of the hMPV N gene as described earlier [Mackay et al., 2003]. Sequence analysis has shown that both genotypes A and B circulate in Kuwait, with predominance of B2 subgroup. Genotype A has been reported as the predominant genotype in respiratory samples [Bastien et al., 2003; Boivin et al., 2003; Maggi et al., 2003; Peiris et al., 2003], but other studies have documented the presence of both genotypes A and B [Huck et al., 2006; Regev et al., 2006; Matsuzaki et al., 2008].
The seasonal distribution of hMPV infection during the year 2009 indicates that hMPV infection incidence is high in winter and peaks during spring with almost no infection detected during summer. These results are not different from those of previous observations that have found high level of hMPV detection in the spring and winter months [Sugrue et al., 2008; Ali et al., 2010].
Acknowledgements
We thank Dr. Gyorgy Szucs for his helpful comments.
REFERENCES
- Ali SA, Williams JV, Chen Q, Faori S, Shehabi A, Jundi EA, Khuri‐Bulos N, Halasa N. 2010. Human metapneumovirus in hospitalized children in Amman. Jordan J Med Virol 82: 1012–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Sonboli N, Hart CA, Al‐Aeryani A, Banajeh SM, Al‐Aghbari N, Dove W, Cuevas LE. 2005. Respiratory syncytial virus and human metapneumovirus in children with acute respiratory infections in Yemen. Pediatr Infect Dis J 24: 34–736. [DOI] [PubMed] [Google Scholar]
- Al‐Sonboli N, Hart CA, Al‐Aghbari N, Al‐Ansi A, Ashoor O, Cuevas LE. 2006. Human metapneumovirus and respiratory syncytial virus disease in children. Yemen Emerg Infect Dis 12: 1437–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Thani A, Azzam S, Al‐Sheik abboubaker H, Abdel‐Hadi F, ELsheikh M, Janahi I. 2010. The role of human metapneumovirus in pediatric respiratory tract infection in Qatar. Future Virol 5: 355–360. [Google Scholar]
- Arabpour M, Samarbafzadeh AR, Makvandi M, Shamsizadeh A, Percivalle E, Englud J, Latifi SM. 2008. The highest prevalence of human metapneumovirus in Ahwaz children accompanied by acute respiratory infections. Indian J Med Microbiol 26: 123–126. [DOI] [PubMed] [Google Scholar]
- Bao X, Sinha M, Liu T, Hong C, Luxon BA, Garofalo RP, Casola A. 2008. Identification of human metapneumovirus‐induced gene networks in airway epithelial cells by microarray analysis. Virology 374: 114–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien N, Normand S, Taylor T, Ward D, Peret TC, Boivin G, Anderson LJ, Li. Y. 2003. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res 93: 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biacchesi S, Skiadopoulos MH, Boivin G, Hanson CT, Murphy BR, Collins PL, Buchholz UJ. 2003. Genetic diversity between human metapneumovirus subgroups. Virology 315: 1–9. [DOI] [PubMed] [Google Scholar]
- Boivin G, Abed Y, Pelletier G, Ruel L, Moisan D, Cote S, Peret TC, Erdman DD, Anderson LJ. 2002. Virological features and clinical manifestations associated with human metapneumovirus: A new paramyxovirus responsible for acute respiratory‐tract infections in all age groups. J Infect Dis 186: 1330–1334. [DOI] [PubMed] [Google Scholar]
- Boivin G, De Serres G, Cote S, Gilca R, Abed Y, Rochette L, Bergeron MG, Dery. P. 2003. Human metapneumovirus infections in hospitalized children. Emerg Infect Dis 9: 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin G, De Serres G, Hamelin M, Cote S, Argouin M, Tremblay G, Maranda‐Aubut R, Sauvageau C, Ouakki M, Boulianne N, Couture C. 2007. An outbreak of severe respiratory tract infection due to human metapneumovirus in a long term care facility. Clin Infect Dis 44: 1152–1158. [DOI] [PubMed] [Google Scholar]
- Bosis S, Esposito S, Osterhaus A, Tremolati E, Begliatti E, Tagliabue C, Corti F, Principi N, Niesters H. 2008. Association between high nasopharyngeal viral load and disease severity in children with human metapneumovirus infection. J Clin Virol 42: 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell H. 1995. Acute respiratory infections: A global challenge. Arch Dis Child 73: 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chano F, Rousseau C, Laferriere C, Couillard M, Charest H. 2005. Epidemiological survey of human metapneumovirus infection in a large pediatric tertiary care center. J Clin Microbiol 43: 5520–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EH, Lee HJ, Kim SJ, Eun BW, Kim NH, Lee JA, Lee JH, Song EK, Kim SH, Park JY, Sung JY. 2006. The association of newly identified respiratory viruses with lower respiratory tract infections in Korean children, 2000‐2005. Clin Infect Dis 43: 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffrasnes C, Hamelin ME, Boivin G. 2007. Human metapneumovirus. Semin Respir Care Med 28: 213–221. [DOI] [PubMed] [Google Scholar]
- Esper F, Boucher D, Weibel C, Martinello RA, Kahn JS. 2003. Human metapneumovirus infection in the United States: Clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics 111: 1407–1410. [DOI] [PubMed] [Google Scholar]
- Falsey AR, Erdman D, Anderson LJ, Walsh EE. 2003. Human metapneumovirus infections in young and elderly adults. J Infect Dis 187: 785–790. [DOI] [PubMed] [Google Scholar]
- File TM. 2000. The epidemiology of respiratory tract infections. Semin Respir Infect 15: 184–194. [DOI] [PubMed] [Google Scholar]
- Greensill J, McNamara PS, Dove W, Flanagan B, Smyth RL, Hart CA. 2003. Human metapneumovirus in severe respiratory syncytial virus bronchiolitis. Emerg Infect Dis 9: 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna G, Sarasini A, Percivalle E, Campanini G, Rovida F, Marchi A, Baldanti F. 2007. Prospective study of human metapneumovirus infection: Diagnosis, typing and virus quantification in nasopharyngeal secretions from pediatric patients. J Clin Virol 40: 236–240. [DOI] [PubMed] [Google Scholar]
- Hermos C, Vargas S, McAdam A. 2010. Human metapneumovirus. Clin Lab Med 30: 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijazi Z, Pacsa A, Eisa S, el Shazli A, Abd el‐Salam RA. 1996. Laboratory diagnosis of acute lower respiratory tract viral infections in children. J Trop Pediatr 42: 276–280. [DOI] [PubMed] [Google Scholar]
- Hijazi Z, Pacsa A, el‐Gharbawy F, Chugh TD, Essa S, el Shazli A, Abd el‐Salam RA. 1997. Acute lower respiratory tract infections in children in Kuwait. Ann Trop Paediatr 17: 127–134. [DOI] [PubMed] [Google Scholar]
- Honda H, Iwahashi J, Kashiwagi T, Imamura Y, Hamada N, Anraku T, Ueda S, Kanda T, Takahashi T, Morimoto S. 2006. Outbreak of human metapneumovirus infection in elderly inpatients in Japan. J Am Geriatr Soc 54: 177–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck B, Scharf G, Neumann‐Haefelin D, Puppe W, Weigl J, Falcone V. 2006. Novel human metapneumovirus sublineage. Emerg Infect Dis 12: 147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn JS. 2003. Human metapneumovirus: A newly emerging respiratory pathogen. Curr Opin Infect Dis 16: 255–258. [DOI] [PubMed] [Google Scholar]
- Kahn JS. 2006. Epidemiology of human metapneumovirus. Clin Microbiol Rev 19: 546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan NM, Dove W, Abd‐Eldayem SA, Abu‐Zeid AF, Shamoon HE, Hart CA. 2008. Molecular epidemiology and disease severity of respiratory syncytial virus in relation to other potential pathogens in children hospitalized with acute respiratory infection in Jordan. J Med Virol 80: 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadadah M, Essa S, Higazi Z, Behbehani N, Al‐Nakib W. 2010. Respiratory syncytial virus and human rhinoviruses are the major causes of severe lower respiratory tract infections in Kuwait. J Med Virol 82: 1462–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Sung H, Im H, Hong S, Kim M. 2009. Molecular epidemiological investigation of a nosocomial outbreak of human metapneumovirus infection in a pediatric hemato‐oncology patient population. J Clin Microbiol 47: 1221–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Chan PK, Yu IT, Tsoia KK, Lui G, Sung JJ, Cockram CS. 2007. Co‐circulation of human metapneumovirus and SARS‐associated coronavirus during a major nosocomial SARS outbreak in Hong Kong. J Clin Virol 40: 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IM, Jacob KC, Woolhouse D, Waller K, Syrmis MW, Whiley DM, Siebert DJ, Nissen M, Sloots TP. 2003. Molecular assays for detection of human metapneumovirus. J Clin Microbiol 41: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IM, Bialasiewicz S, Jacob KC, McQueen E, Arden KE, Nissen MD, Sloots TP. 2006. Genetic diversity of human metapneumovirus over 4 consecutive years in Australia. J Infect Dis 193: 1630–1633. [DOI] [PubMed] [Google Scholar]
- Maggi F, Pifferi M, Vatteroni M, Fornai C, Tempestini E, Anzilotti S, Lanini L, Andreoli E, Ragazzo V, Pistello M, Specter S, Bendinelli M. 2003. Human metapneumovirus associated with respiratory tract infections in a 3‐year study of nasal swabs from infants in Italy. J Clin Microbiol 41: 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Itagaki T, Abiko C, Aoki Y, Suto A, Mizuta K. 2008. Clinical impact of human metapneumovirus genotypes and genotype‐specific seroprevalence in Yamagata, Japan. J Med Virol 80: 1084–1089. [DOI] [PubMed] [Google Scholar]
- Moattari A, Aleyasin S, Arabpour M, Sadeghi S. 2010. Prevalence of human metapneumovirus (hMPV) in children with wheezing in Shiraz‐Iran. Iran J Allergy Asthma Immunol 9: 250–254. [PubMed] [Google Scholar]
- Muir D, Pillay D. 1998. Respiratory virus infections in immunocompromised patients. J Med Microbiol 47: 561–562. [DOI] [PubMed] [Google Scholar]
- Peiris JS, Tang WH, Chan KH, Khong PL, Guan Y, Lau YL, Chiu. SS. 2003. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis 9: 628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D, Zhao X, Liu E, Huang Y, Yang X, Zhao Y, Chen X, Zhang Z. 2010. Analysis of viral load in children infected with human metapneumovirus. Iran J Pediatr 20: 393–400. [PMC free article] [PubMed] [Google Scholar]
- Principi N, Bosis S, Esposito S. 2006. Human metapneumovirus in pediatric patients. Clin Microbiol Infect 12: 301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev L, Hindiyeh M, Shulman LM, Barak A, Levy V, Azar R, Shalev Y, Grossman Z, Mendelson E. 2006. Characterization of human metapneumovirus infections in Israel. J Clin Microbiol 44: 1484–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildgen O, Glatzel T, Geikowski T, Scheibner B, Matz B, Bindl L, Born M, Viazov S, Wilkesmann A, Knöpfle G, Roggendorf M, Simon A. 2005. A human metapneumovirus RNA in encephalitis patient. Emerg Infect Dis 11: 467–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple MG, Cowell A, Dove W, Greensill J, McNamara PS, Halfhide C, Shears P, Smyth RL, Hart CA. 2005. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J Infect Dis 191: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugrue R, Tan B‐H, Loo L‐H. 2008. The emergence of human metapneumovirus: HMPV detection and epidemiology. Future Virol 3: 363–371. [Google Scholar]
- Van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Hoogen BG, Herfst S, Sprong L, Cane PA, Forleo‐Naeto E, DeSwart RL, Osterhaus AD, Fouchier RA. 2004a. Antigenic and genetic variability of human metapneumovirus. Emerg Infect Dis 10: 658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Hoogen BG, Osterhaus DM, Fouchier RA. 2004b. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr Infect Dis J 23: S25–32. [DOI] [PubMed] [Google Scholar]
- Van Woensel JB, Bos AP, Lutter R, Rossen JW, Schuurman R. 2006. Absence of human metapneumovirus co‐infection in cases of severe respiratory syncytial virus infection. Pediatr Pulmonol 41: 872–874. [DOI] [PubMed] [Google Scholar]
- Williams JV, Harris PA, Tollefson SJ, Halburnt‐Rush LL, Pingsterhaus JM, Edwards KM, Wright PF, Crowe JE Jr. 2004. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med 350: 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JV, Martino R, Rabella N, Otegui M, Parody R, Heck JM, Crowe JEA Jr. 2005. A prospective study comparing human metapneumovirus with other respiratory viruses in adults with hematologic malignancies and respiratory tract infections. J Infect Dis 192: 1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Zakay‐Rones Z, Fadeela A, Greenberg D, Dagan R. 2003. High seroprevalence of human metapneumovirus among young children in Israel. J Infect Dis 188: 1865–1867. [DOI] [PubMed] [Google Scholar]
- Xiao NG, Xie ZP, Zhang B, Yuan XH, Song JR, Gao HC, Zhang RF, Hou YD, Duan ZJ. 2010. Prevalence and clinical and molecular characterization of human metapneumovirus in children with acute respiratory infection in China. Pediatr Infect Dis J 29: 131–134. [DOI] [PubMed] [Google Scholar]


