Abstract
Autophagy is an important survival pathway and participates in the host response to infection. Beneficial effects of melatonin have been previously reported in an animal model of acute liver failure (ALF) induced by the rabbit hemorrhagic disease virus (RHDV). This study was aimed to investigate whether melatonin protection against liver injury induced by the RHDV associates to modulation of autophagy. Rabbits were infected with 2 × 104 hemagglutination units of a RHDV isolate and received 20 mg/kg melatonin at 0, 12, and 24 hr postinfection. RHDV induced autophagy, with increased expression of beclin‐1, ubiquitin‐like autophagy‐related (Atg)5, Atg12, Atg16L1 and sequestrosome 1 (p62/SQSTM1), protein 1 light chain 3 (LC3) staining, and conversion of LC3‐I to autophagosome‐associated LC3‐II. These effects reached a maximum at 24 hr postinfection, in parallel to extensive colocalization of LC3 and lysosome‐associated membrane protein (LAMP)‐1. The autophagic response induced by RHDV infection was significantly inhibited by melatonin administration. Melatonin treatment also resulted in decreased immunoreactivity for RHDV viral VP60 antigen and a significantly reduction in RHDV VP60 mRNA levels, oxidized to reduced glutathione ratio (GSSG/GSH), caspase‐3 activity, and immunoglobulin‐heavy‐chain‐binding protein (BiP) and CCAAT/enhancer‐binding protein homologous protein (CHOP) expression. Results indicate that, in addition to its antioxidant and antiapoptotic effects, and the suppression of ER stress, melatonin induces a decrease in autophagy associated with RHDV infection and inhibits RHDV RNA replication. Results obtained reveal novel molecular pathways accounting for the protective effect of melatonin in this animal model of ALF.
Keywords: acute liver failure, autophagy, melatonin, rabbit hemorrhagic disease
Introduction
Acute liver failure (ALF) (sometimes referred to as fulminant hepatic failure) is a clinical syndrome resulting from rapid loss in hepatocyte function, typically associated with coagulopathy and encephalopathy in patients without pre‐existing liver disease. Liver transplantation is the only therapy of proven benefit, but the rapidity of progression and the variable course of ALF limit its use 1. As a result, reproducible experimental animal models resembling the clinical conditions are still needed to test new therapeutic approaches, and ALF presents many challenging opportunities in basic research. Most common models are based on surgical techniques or the use of hepatotoxic drugs and very few satisfactory viral models are available 2. We have developed a viral model of ALF by means of the inoculation of rabbits with the virus of rabbit hemorrhagic disease (RHDV), which displays biochemical data, histological characteristics, and clinical features that resemble those in human ALF 3. In rabbits infected with RHDV melatonin inhibits oxidative damage, apoptotic mechanisms and endoplasmic reticulum (ER) stress 4, 5, 6, 7, which supports its potential therapeutic use in patients with ALF. A number of reports published in the last few years indicate that properties of the indol as a potent antioxidant, modulator of apoptosis and positive regulator of immune functions contribute to melatonin's antiviral actions 8.
Autophagy is a catabolic process involving self‐degradation of cellular components via the lysosomal pathway which acts as survival mechanism and can participate in the host response to infection 9, 10. Autophagy is known to participate in both sensing oxidative stress and removing oxidatively damaged proteins and organelles 11, and plays a multifunctional role in host defense, by promoting pathogen clearance and modulating innate and adaptative immune responses 12. The process begins with the engulfment of bulk amounts of cytoplasm by double membrane vesicles, termed autophagosomes, which ultimately fuse with lysosomes, generating an autolysosome in which the constituent is subsequently degraded 13. At the molecular level the nucleation of the autophagosomal membrane is controlled by a molecular complex containing Bcl‐2‐interacting protein (beclin)‐1, which allows the production of phosphatidylinositol 3‐phosphate to occur 14. Conjugation of ubiquitin‐like autophagy‐related (Atg)12 to Atg5 and formation of a complex with Atg16L1 is required in the elongation of the autophagosome membrane. Autophagosome formation also requires the conversion of cytosolic‐associated protein light chain 3 (LC3)‐I to the membrane‐bound LC3‐II form. LC3 then binds to the adaptator protein p62 sequestrosome 1 (p62/SQSTM1) which, in addition to its role in cell proliferation, inflammation or oxidative stress, facilitates the autophagic degradation of ubiquitinated protein aggregates in lysosomes 13, 15. The interplay between autophagy and programmed cell death is complex, and recent data suggest the existence of a cross talk between autophagic and apoptotic pathways 16. Some studies have demonstrated that autophagy may have an active contribution to cell death in virus infected cells 17. However, there are also reports that autophagy can prolong survival of virus‐infected cells by counteracting the apoptotic response 18, 19. Given the well‐known regulatory effect that melatonin has on apoptotic cell death, and the dual capabilities of autophagy to induce survival or death depending on cellular stress 20, melatonin modulation of autophagy is currently under consideration 21, and the regulatory role of the indol on the process of autophagy deserves further careful studies.
In this research, we analyzed whether melatonin administration modulates the autophagic response in RHDV‐infected rabbits. Our findings provide evidence that reduction in liver damage by melatonin is associated with a lower RHDV RNA replication and a decrease in the incomplete autophagy induced by RHDV. Further elucidation of the role of autophagy and its relationship to oxidative stress and apoptosis in the pathogenesis of RHDV infection will help to a better knowledge of molecular mechanisms accounting for the protective effect of melatonin in this animal model of ALF.
Materials and methods
Virus and experimental model
Nine‐week‐old male New Zealand white rabbits were kept in the animal facility of the University of León with 12‐hr light cycle at 21–22°C and 50% relative humidity. They were given a standard dry rabbit food and water ad libitum. Effects of melatonin were studied by sacrificing control rabbits and batches of infected animals at 18, 24, 30 and 36 hr post infection (hpi). Infection was induced by i.m. injection of 2 × 104 hemagglutination units of an RHDV isolate 3, 4. Melatonin was given (20 mg/kg body weight i.p.) at 0, 12 and 24 hpi; untreated animals received 4 mL of vehicle at 0, 12 and 24 hpi. Melatonin (Sigma, St Louis, MO, USA) was dissolved into absolute ethanol and further dilutions were made in saline. The final concentration of ethanol was 5%.
The study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and was specifically approved by the Ethics Committee of the University of León.
Real‐time RT‐PCR
Total RNA was extracted from frozen rabbit liver using a Trizol reagent (Life Technologies, Madrid, Spain) and quantified using a Nano Drop1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Residual genomic DNA was removed by incubating RNA with RQ1 RNase‐free DNase (Promega, Madison, WI, USA). RNA integrity was confirmed by formaldehyde gel electrophoresis. Total RNA (1 μg) was reverse transcribed as described 7 and mRNA was determined by real‐time PCR analysis using SYBR Green I Master (Roche Diagnostics GmbH, Mannheim, Germany) and the appropriate primers (Table 1). Relative changes in gene expression levels were determined using the method 22. The cycle number at which the transcripts were detectable (Ct) was normalized to the cycle number of β‐Actin gene detection, referred to as ΔCt.
Table 1.
Primers used in this study
| Gene | Sense primer (5'‐3') | Antisense primer (5'‐3') |
|---|---|---|
| Beclin‐1 | CATGCAATGGTGGCTTTCC | TCTCGCCCTTTTCAACCTCTT |
| Atg5 | CGTCCTGTGGCTGCAGATG | AAGGACACACTTCTTTGAGGAGATC |
| Atg12 | TGCTGAAGGCTGTGGGAGAT | TGTTCGCTCTACAGCCCATTT |
| Atg16L1 | CCACCAAACCGGCATGAG | CTTGCAGCTGGCTGTCATTC |
| p62/SQSTM1 | AACAGAGGTGACCACCCTTCA | AGCACAGACTGGCTGGAAGTC |
| RHDV | TAGCCCAACAGAAGCACAAG | AAACAAGTCGTCAACCTCCC |
| BiP | ATTGACAATGGTGTCTTCGAAGTC | CCCCGCCCAGGTGAGT |
| CHOP | ATACATCACCACACCTGAAAGCA | GCACTCGGCTGCCATCTC |
| β‐Actin | TGGCATCCTGACGCTCAA | TCGTCCCAGTTGGTCACGAT |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Western blot analysis
For Western blot analysis, liver tissue (25 mg) was homogenized in 1 mL RIPA buffer containing protease and phosphatase inhibitor cocktails (Roche Diagnostics GmbH), maintaining temperature at 4°C throughout all procedures. Then the homogenate was incubated on ice for 30 min and finally the samples were centrifuged at 13,000 g for 30 min at 4°C. The supernatant fraction was stored at −80°C in aliquots until use. Nuclear extracts were prepared from liver homogenates as described previously 23. Briefly, 100 mg of liver from all animals was homogenized in 5 × 10−4 L of buffer A (0.01 m Hepes‐KOH pH 7.9, 250 g/L glycerol, 0.420 m NaCl, 0.0015 m MgCl2, 2 × 10−4 m EDTA, 5 × 10−4 m DTT, 2 × 10−4 m PMSF) and a phosphatase inhibitor cocktail (Roche Diagnostics GmbH) to disrupt extracellular matrix and cellular membranes. Homogenates were centrifuged at 1000 g for 10 min at 4°C. The pellet was resuspended in 2.5 × 10−4 L of buffer B (0.02 m NaCl Hepes‐ KOH pH 7.9, 250 g/L glycerol, 0.042 m NaCl, 15 × 10−4 m PMSF), homogenized, and incubated at 4°C for 30 min. Cellular debris was removed by centrifugation at 14,000 g for 15 min at 4°C. The supernatant fraction containing DNA binding proteins was recollected and stored at −80°C in aliquots until use. Protein concentration was measured by Bradford assay. Equal amounts of protein extracts (10–50 μg) were separated by 7–12% sodium dodecyl sulfate (SDS)‐polyacrylamide gel electrophoresis and transferred electrically to polyvinyllidene difluoride membranes (Millipore, Bedford, MA). The membranes were then blocked with 5% nonfat dry milk in Tris‐buffered saline containing 0.05% Tween 20 (TBST) for 30 min at 37°C and probed overnight at 4°C with polyclonal anti‐p62/SQSTM1, poly(ADP‐ribose)polymerase‐1 (PARP‐1) and IRF7 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), phospho‐IRF7 (Cell Signaling Technology, Danvers, MA, USA) and LC3‐I/II (Abcam, Cambridge, UK) antibodies at 1:200–1:1000 dilution with PBST containing 2.5% nonfat dry milk. Equal loading of protein was demonstrated by probing the membranes with a rabbit anti‐β‐Actin polyclonal antibody (1:2000; Sigma) and a rabbit anti‐Lamin‐B polyclonal antibody (1:200; Santa Cruz Biotechnology). After washing with TBST, the membranes were incubated for 1 hr at room temperature with secondary HRP conjugated antibody (1:5000; Dako, Glostrup, Denmark), and visualized using ECL detection kit (Amersham Pharmacia, Uppsala, Sweden) 7. The density of the specific bands was quantified with an imaging densitometer (Scion Image J Software 1.46a, Bethesda, MD, USA).
Immunohistochemistry
Tissue samples were recovered, fixed in 10% buffered formalin and embedded in paraffin. Sections (4 μm) were dewaxed and hydrated through graded ethanol, cooked in 25 mm citrate buffer, pH 6.0, in a pressure cooker for 10 min, transferred into boiling deionized water and let to cool for 20 min. Tissue sections were then treated with 3% hydrogen peroxide to inactivate endogenous peroxidase activity. The slides were incubated with rabbit anti‐VP60 antibodies (Ingenasa, Madrid, Spain) overnight at 4°C. Subsequently, the sections were incubated for 30 min using the EnVision+ system and developed with a solution of 3‐3‐diaminobenzidine (DAB) (Vector Lab, Burlingame, CA, USA). The slides were stained with hematoxylin for 10 s and mounted. The specificity of the technique was evaluated by negative controls (omitting the incubation with the primary antibody and incubating it with nonimmune sera).
Double immunofluorescence
For immunofluorescent double staining sections were dewaxed in xylene and rehydrated in graded ethanol to distilled water, do not allowing slides to dry at any time during this process. Heat mediated antigen was performed in a cooker filled with 1 mm EDTA (pH = 8). Sections were brought to a boil and then maintain at a sub‐boiling temperature for 15 min. All subsequent incubations with immunochemicals were performed in a humidified chamber. After unmasking and after blocking the nonspecific binding the sections were co‐incubated with the LAMP‐1 antibody (Santa Cruz Biotechnology) and LC3 (Abcam) (at 1:50 and 1:200 dilution, respectively) overnight at 4°C. After incubation with primary antibodies, samples were washed twice in PBS for 10 min at room temperature. Thereafter, the secondary antibodies donkey anti‐rabbit conjugated with FITC and donkey anti‐mouse conjugated with Dylight™549 (Jackson Immuno‐Research, Baltimore, PA, USA) were applied for 2 hr at 21°C. After washing in TBS, the coverslips were mounted on Dako Cytomation Fluorescent Mounting Medium (Dako). In sections from each experimental group, the primary antibody was replaced by antibody diluent to assess for nonspecific binding of the secondary antibody 7. The preparations were analyzed with an inverted fluorescent microscope (Nikon Eclipse Ti, Tokyo, Japan).
Measurement of oxidized and reduced glutathione
Oxidized and reduced glutathione (GSSG and GSH, respectively) analysis was performed by the method of Hissin and Hilf 24, homogenizing 250 mg of tissue in 0.1 m sodium phosphate 5 mm EDTA buffer (pH 8.0) with 25% phosphoric acid at a proportion of 1:20.
Caspase‐3 activity
Lysates were prepared by homogenizing liver tissue in 0.25 mM sucrose, 1 mM EDTA, 10 mM Tris and a protease inhibitor cocktail (Roche Diagnostics GmbH). The lysates were then centrifuged at 14,000 g for 10 min at 4°C, and supernatants (50 μg protein) were incubated for 1 hr at 37°C in HEPES buffer containing 100 μM concentrations of the specific fluorogenic substrate 7‐amino‐4‐methylcoumarin N‐acetyl‐L‐aspartyl‐Lglutamyl‐L‐valyl‐l‐aspartic acid amide (DEVD‐AMC). Cleavage of the caspase substrate was monitored using a spectrofluorimeter (Hitachi F‐2000 fluorimeter, Hitachi LTD, Tokyo, Japan) at excitation/emission wavelengths of 380/460 nm. Activity was expressed as fluorescence units per milligram of protein per minute of incubation.
Statistical analysis
Results are expressed as mean values ± standard error of the mean (S.E.M.). Data were compared by analysis of variance (ANOVA); when the analysis indicated the presence of a significant difference, the means were compared with the Newman‐Keul's test. Significance was accepted when P was <0.05. Values were analyzed using the statistical package SPSS 19.0 (IBM Corporation, Armonk, NY, USA).
Results
To determine the presence of the virus in the liver of rabbits infected with the RHDV viral VP60 antigen was examined by immunohistochemical techniques. Data obtained indicate a progressive increase in the extent of labeling in RHDV‐infected animals which reached a maximum at 36 hpi (Fig. 1A,B) in parallel to the increase in RHDV mRNA level (Fig. 1C). Melatonin treatment resulted in decreased immunoreactivity and significantly reduced RHDV mRNA expression (Fig. 1A–C), revealing an inhibitory role of the indol on RHDV replication. Viral replication associated to increased expression of the phosphorylated form of IRF7, a master regulator of type I‐interferon‐dependent innate immunity, and this effect was not modified by melatonin administration (Fig. 1D).
Figure 1.
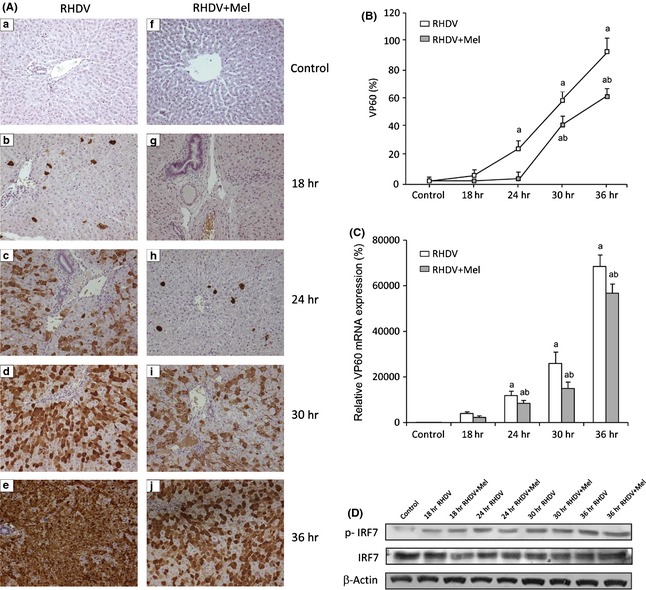
Expression of the capsid protein VP60 and IRF7 in rabbit hemorrhagic disease virus (RHDV)‐infected and melatonin‐treated rabbits. (A,B) VP60 immunohistochemical labeling in hepatocytes: (a,f) Control, (b,g) 18 hpi; (c,h) 24 hpi; (d,i) 30 hpi; (e,j) 36 hpi. Paraffin‐embedded sections were immunostained with a VP60 antibody. Original magnification: 200×. Data are presented as percentage change from the control group. Image analysis was performed using the ImageJ software v3.91 (http://rsbweb.nih.gov/ij/). (C) Levels of VP60 mRNA analyzed by real‐time PCR assay and normalized against β‐Actin. Values are expressed as means ± S.E.M. (n = 6). a P < 0.05, compared with Control, b P < 0.05, compared with RHDV, same period. (D) Western blot of IRF7. Proteins from liver extracts were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, followed by immunoblotting. Equal loading of proteins is illustrated by β‐Actin bands.
Autophagy activation by RHDV infection was confirmed by the significant increase of beclin‐1 expression and the upregulation of components of the Atg12‐Atg5‐Atg16L1 complex at 18–24 hpi (Table 2). Another frequently used autophagy marker is LC3, which is converted from the cytosolic form LC3‐I to the lapidated form LC3‐II. Western blot data indicate that LC3‐II expression increased markedly at 18–24 hpi, with a maximum rise of LC3‐II/LC3‐I ratio at 24 hpi, which suggests that RHDV could be blocking the autophagic process in liver cells at early postinfection periods (Fig. 2A). Effects on LC3 expression can reflect aggregated effects of induced autophagosome synthesis, decreased fusion with lysosomes and/or suppression of proteolytic turnover in the autophagolysosomes. Immunoflourescence analysis showed spots of green fluorescence which reached a maximum at 24 hpi, suggesting an elevated turnover of autophagosomes (Fig. 2B). At 18 hpi, there was little colocalization between LC3 and the lysosomal marker LAMP‐1, but extensive overlap was observed at 24 hpi, presumably due to increased accumulation of autophagolysosomes. However, the parallel increase in p62/SQSTM1 mRNA levels (Table 2) may reflect a dysfunctional autophagy with impairment of the autophagic flux. The autophagic response in RHDV‐infected rabbits was partially inhibited by melatonin treatment, as shown by the reduced LC3 immunofluorescent staining (Fig. 2B), the decrease in LC3‐II protein expression (Fig. 2A), and the diminished mRNA expression of beclin‐1, Atg5, Atg12 and Atg16L1 (Table 2).
Table 2.
Effect of rabbit hemorrhagic disease virus (RHDV) infection and melatonin on mRNA levels of genes related to autophagy and ER stress
| Control | RHDV18 | RHDV18 + Mel | RHDV24 | RHDV24 + Mel | RHDV30 | RHDV30 + Mel | RHDV36 | RHDV36 + Mel | |
|---|---|---|---|---|---|---|---|---|---|
| Beclin‐1 | 100 ± 8 | 222 ± 12 a | 177 ± 16 a , b | 234 ± 16 a | 94 ± 7 b | 143 ± 7 a | 87 ± 7 b | 87 ± 7 | 68 ± 7 a , b |
| Atg5 | 100 ± 4 | 170 ± 20 a | 141 ± 6 a | 149 ± 24 a | 70 ± 8 a , b | 129 ± 5 a | 78 ± 5 a , b | 95 ± 5 | 49 ± 5 a , b |
| Atg12 | 100 ± 12 | 193 ± 10 a | 177 ± 9 a | 135 ± 7 a | 94 ± 8 b | 89 ± 6 | 43 ± 7 a , b | 91 ± 7 | 53 ± 7 a , b |
| Atg16L1 | 100 ± 5 | 177 ± 11 a | 103 ± 12 b | 114 ± 9 | 57 ± 9 a , b | 86 ± 6 | 49 ± 9 a , b | 80 ± 10 | 38 ± 9 a , b |
| p62/SQSTM1 | 100 ± 4 | 134 ± 7 a | 92 ± 13 b | 158 ± 12 a | 108 ± 6 b | 130 ± 10 a | 75 ± 6 a , b | 81 ± 4 a | 53 ± 5 a , b |
| BiP | 100 ± 6 | 264 ± 8 a | 231 ± 9 a | 302 ± 6 a | 234 ± 7 a , b | 216 ± 12 a | 136 ± 15 a , b | 129 ± 10 a | 79 ± 9 a , b |
| CHOP | 100 ± 7 | 389 ± 13 a | 312 ± 24 a , b | 672 ± 9 a | 264 ± 4 a , b | 244 ± 9 a | 136 ± 18 a , b | 114 ± 4 | 90 ± 7 a |
Levels of mRNA were analyzed by real‐time PCR assays. Data, normalized against β‐Actin, are presented as percentage change from the control group. Values are expressed as means± S.E.M. (n = 6). a P < 0.05, compared with Control, b P < 0.05 compared with RHDV, same period.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 2.
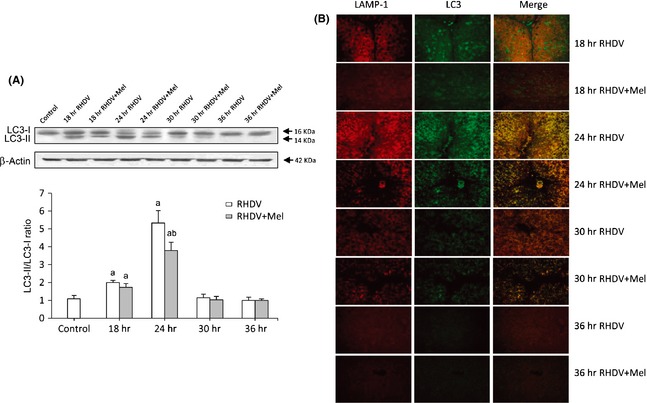
Markers of autophagy in rabbit hemorrhagic disease virus (RHDV)‐infected and melatonin‐treated rabbits. (A) Western blot of LC3. Proteins from liver extracts were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, followed by immunoblotting. Equal loading of proteins is illustrated by β‐Actin bands. The graph shows the LC3‐II/LC3‐I ratio. Values are expressed as means ± S.E.M. (n = 6). a P < 0.05, compared with Control, b P < 0.05, compared with RHDV, same period. (B) Colocalization of LC3 and LAMP‐1 by immunofluorescence analysis. In the left panels LAMP‐1 positive staining (red fluorescence); in middle panels positive staining of LC3 (green fluorescence); right panels show the merge of two fluorescences.
Endoplasmic reticulum (ER) stress has been reported to induce the accumulation of autophagosomes, and autophagy may be an important part of normal ER function 25. We have previously reported that melatonin treatment inhibits ER stress at 36 hpi in RHDV‐infected rabbits 7. During ER stress different transcription factors regulate the expression of ER chaperones that enhance the folding capacity of the ER, including CCAAT/enhancer‐binding protein homologous protein (CHOP) and immunoglobulin‐heavy‐chain‐binding protein (BiP/GRP78). As expected, mRNA levels for both chaperones increased following RHDV infection, with a maximum at 24 hpi, and this effect was significantly inhibited by melatonin administration (Table 2).
As previously reported 26, melatonin prevented the significant increase in the GSSG/GSH ratio observed in RHDV‐infected rabbits (Fig. 3A). Data obtained also confirmed the already described effect of melatonin on apoptotic death induced by RHDV 6; both activation of caspase‐3, the common event initiated by multiple different stimuli that induces apoptosis, and proteolysis of PARP‐1, a nuclear enzyme cleavaged into a 85‐kDA fragment by caspase‐3 27, increased significantly at 30 and 36 hpi, and these effects were significantly prevented by melatonin treatment (Fig. 3B,C).
Figure 3.
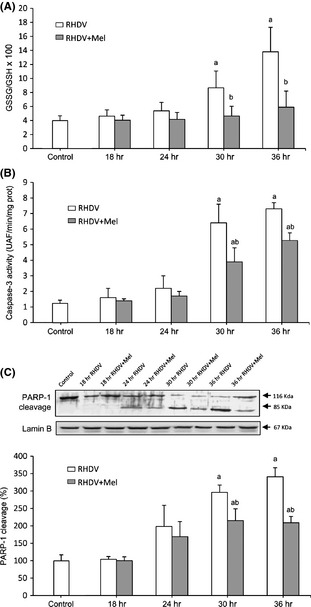
Markers of oxidative stress and apoptosis in rabbit hemorrhagic disease virus (RHDV)‐infected and melatonin‐treated rabbits. (A) Oxidized to reduced glutathione ratio (GSSG/GSH) in hepatic tissue. (B) Liver activity of caspase‐3. Samples were incubated with the specific fluorogenic substrate Ac‐DEVD‐AMC. (C) Western blot of PARP‐1. Proteins from liver extracts were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, followed by immunoblotting. Equal loading of proteins is illustrated by Lamin B bands. The graph shows densitometric quantification. Values are expressed as means ± S.E.M. (n = 6). a P < 0.05, compared with Control, b P < 0.05, compared with RHDV, same period.
Discussion
Results from this study demonstrate that RHDV induced autophagosome and autophagolysosome formation, with increased expression of beclin‐1, LC3‐II/LC‐I ratio and Atg5‐Atg12‐Atg16L1, and colocalization of LC3 and LAMP‐1. The parallel increase in p62/SQSTM1 expression may reflect a dysfunctional autophagy similar to the previous reports following infection of liver cells by other RNA viruses 28, 29. Data obtained also indicate that the RHDV‐induced autophagic response was inhibited by melatonin administration. Mechanisms through which viruses induce autophagy remain unclear. One of the typical stress responses initiated by viral infection is ER stress, which down‐regulates protein synthesis through the three branches of the unfolded protein response (UPR). During ER stress, different transcription factors regulate the expression of ER chaperones, such as CHOP and BiP, which enhance the folding capacity of the ER 7. It is known that sensors of the UPR control signal‐transduction pathways leading to the transcription of autophagic genes 30, and viruses capable of inducing ER stress have the potential to induce an autophagic response. Thus, previous studies have demonstrated that autophagy is associated to ER stress in HCV infection, and that persistent induction of ER stress and incomplete activation of autophagy play an important role in HCV pathogenesis 28, 31. In addition, a newer hypothesis that is gaining support is that autophagic activities are sensitive to oxidative stress and to the generation of reactive species 11. Indeed, a recent research has shown that oxidative stress triggers the accumulation of autophagic compartments without increasing the degradation of long‐lived proteins in human neuroblastoma cells infected by herpes simplex virus type 1 infection 32. Moreover, it has been reported that in MEF cells infected with the Chikungunya virus, independent induction of ER and oxidative stress pathways triggers autophagy through the inhibition of the mammalian target of rapamycin (mTOR) 18. Therefore, the previously reported reduction of oxidative stress and ER stress by melatonin 7, 26, confirmed in this research through the decrease in CHOP and BiP expression and the increase in the GSSG/GSH ratio, could contribute to the inhibitory effect of the indol on the autophagic response induced by viral infection.
In our research, the autophagic activity induced by the RHDV declined in late periods of infection (30–36 hpi) in parallel to the increase in apoptosis. Different studies have previously shown that virally induced autophagy is capable of preventing the early apoptotic death of cells 33, which suggests that autophagy might contribute to limit the cytopathic effect of viruses and the pathological consequences associated with cell death triggered by viral infection. This regulation of programmed cell death by autophagy could also be present in RHDV‐infected hepatocytes. However, mechanisms responsible for the regulation of apoptosis by autophagy and for the overhelming of the apoptotic response by the inhibitory effect of autophagy remain unknown. Melatonin is able to induce or inhibit autophagy based on cellular necessities and oxidative stress levels. There are reports that autophagy and survival are significantly enhanced in ischemic N2a cells treated with melatonin 34, and melatonin‐induced autophagy protects against human prion protein‐mediated neurotoxicity 35. Moreover, melatonin induces autophagy and results in the effective removal of mutant‐TGFBIp from granular corneal dystroph type 2 fibroblasts 36. In contrast, melatonin is known to protect HeLa cells from rotenone‐induced cell injury via inhibition of autophagy 37, to suppress cyclosporine A‐induced autophagy in rat pituitary GH3 cells 38, to inhibit kainic acid‐induce neurotoxicity in mouse hippocampus via inhibition of autophagy 39, and to attenuate methamphetamine‐induced autophagy 40. Consistent with studies which report parallel inhibition of autophagic processes and programmed cell death, in the present experiments both autophagy and apoptosis were significantly inhibited by melatonin. These effects of the indol could be the consequence of its antioxidant action, which may directly antagonize mechanisms contributing both to the intrinsic and extrinsic pathways of apoptosis 6, but also result in the inhibition of ER stress and, in turn, in diminished autophagic and apoptotic responses.
An additional interesting fact concerns the inhibition of RHDV induced by melatonin. The role of autophagy in relation to viral replication is dual and it may be beneficial to virus or host. Thus, although autophagy may be a cellular mechanism to clear viral infection, and its inhibition results in an increased replication and virulence of herpex simplex virus 1 (HSV1) or vesicular stomatitis virus (VSV), different viruses appear to use this response to enhance their own replication. It has been reported that the double membrane of the autophagosome supports poliovirus replication 41, the autophagic machinery is used for coronaviruses replication 42, HCV uses autophagy for the early protein translation 43, and Dengue virus activate autophagy to elevate viral replication 44. Moreover, it is known that ER stress may play a positive role in HCV RNA replication through activation of autophagy, because siRNAs directed against sensors of the different branches of the UPR suppress HCV‐induced lipidation of LC3, required for formation of autophagosomes, and reduce HCV RNA levels 28. It has also been reported that inhibitors of X‐box binding protein‐1 (XBP‐1), one of the ER stress mediators, inhibit in parallel HCV viral replication and autophagy in hepatoma cells 45. Therefore, data obtained in this study could indicate that the decrease in RHDV RNA replication in melatonin‐treated rabbits is a consequence, at least in part, of its negative effect on ER stress and on the autophagic response. Both HCV and Dengue virus are known to exploit the UPR‐autophagy pathway to promote replication by suppressing innate antiviral immunity 46, 47, and a number of in vitro and in vivo studies have documented that melatonin plays a role in innate immunity 48. However, in our experiments expression of the phosphorylated form of IRF7, a potent type I interferon inducer capable of modulating viral propagation in infected hepatocytes 49, increased in RHDV‐infected rabbits and was not modified by melatonin. This suggests that RHDV is not able to abrogate type I interferon‐dependent immune responses, and inhibition of viral replication by melatonin is probably unrelated to induction of innate immunity.
In conclusion, results from this study indicate that in addition to its anti‐oxidant and anti‐apoptotic effects, and the suppression of ER stress, melatonin induces a decrease in the autophagy associated with RHDV infection and inhibits RHDV RNA replication. Understanding the mechanisms behind the interplay of RHDV‐induced autophagy with oxidative stress, ER stress and apoptosis may help to identify molecular pathways accounting for the protective effect of melatonin in this animal model of ALF.
Acknowledgements
CIBERehd is funded by Instituto de Salud Carlos III, Spain.
References
- 1. Lee WM. Recent developments in acute liver failure. Best Pract Res Clin Gastroenterol 2012; 26:3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tuñon MJ, Álvarez M, Culebras JM et al. An overview of animal models for investigating the pathogenesis and therapeutic strategies in acute hepatic failure. World J Gastroenterol 2009; 15:3086–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tuñón MJ, Sánchez‐Campos S, García‐Ferreras J et al. Rabbit hemorrhagic viral disease: characterization of a new animal model of fulminant liver failure. J Lab Clin Med 2003; 141:272–278. [DOI] [PubMed] [Google Scholar]
- 4. Sánchez‐Campos S, Álvarez M, Culebras JM et al. Pathogenic molecular mechanisms in an animal model of fulminant hepatic failure: rabbit hemorrhagic viral disease. J Lab Clin Med 2004; 144:215–222. [DOI] [PubMed] [Google Scholar]
- 5. San‐Miguel B, Álvarez M, Culebras JM et al. N‐acetyl‐cysteine protects liver from apoptotic death in an animal model of fulminant hepatic failure. Apoptosis 2006; 11:1945–1957. [DOI] [PubMed] [Google Scholar]
- 6. Tuñón MJ, San Miguel B, Crespo I et al. Melatonin attenuates apoptotic liver damage in fulminant hepatic failure induced by the rabbit hemorrhagic disease virus. J Pineal Res 2011; 50:38–45. [DOI] [PubMed] [Google Scholar]
- 7. Tuñón MJ, San‐Miguel B, Crespo I et al. Melatonin treatment reduces endoplasmic reticulum stress and modulates the unfolded protein response in rabbits with lethal fulminant hepatitis of viral origin. J Pineal Res 2013; 55:221–228. [DOI] [PubMed] [Google Scholar]
- 8. Boga JA, Coto‐Montes A, Rosales‐Corral SA et al. Beneficial actions of melatonin in the management of viral infections: a new use for this “molecular handyman”. Rev Med Virol 2012; 22:23–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 2009; 5:527–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dreux M, Chisari FV. Viruses and the autophagy machinery. Cell Cycle 2010; 9:1295–1307. [DOI] [PubMed] [Google Scholar]
- 11. Lee J, Giordano S, Zhang J. Autophagy, mitochondria and oxidative stress: cross‐talk and redox signaling. Biochem J 2012; 441:523–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakahira K, Cloonan SM, Mizumura K et al. Autophagy: a crucial moderator of redox balance, inflammation, and apoptosis in lung disease. Antioxid Redox Signal 2013; 20:474–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol 2010; 22:124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wirawan E, Lippens S, Vanden Berghe T et al. Beclin: a role in membrane dynamics and beyond. Autophagy 2012; 8:6–17. [DOI] [PubMed] [Google Scholar]
- 15. Pankiv S, Clausen TH, Lamark T et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilite degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 28:24131–24145. [DOI] [PubMed] [Google Scholar]
- 16. Tovilovic G, Ristic B, Milenkovic M et al. The role and therapeutic potential of autophagy modulation in controlling virus‐induced cell death. Med Res Rev 2013; doi: 10.1002/med.21303 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 17. Xi X, Zhang X, Wang B et al. The interplays between autophagy and apoptosis induced by enterovirus 71. PLoS ONE 2013; 8:e56966. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Joubert PE, Werneke SW, de la Calle C et al. Chikungunya virus‐induced autophagy delays caspase‐dependent cell death. J Exp Med 2012; 209:1029–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tovilovic G, Ristic B, Siljic M et al. mTOR‐independent autophagy counteracts apoptosis in herpes simplex virus type 1‐infected U251 glioma cells. Microbes Infect 2013; 15:615–624. [DOI] [PubMed] [Google Scholar]
- 20. Carbajo‐Pescador S, García‐Palomo A, Martín‐Renedo J et al. Melatonin modulation of intracellular signaling pathways in hepatocarcinoma HepG2 cell line: role of the MT1 receptor. J Pineal Res 2011; 51:463–471. [DOI] [PubMed] [Google Scholar]
- 21. Coto‐Montes A, Boga JA, Rosales‐Corral S et al. Role of melatonin in the regulation of autophagy and mitophagy: a review. Mol Cell Endocrinol 2012; 361:12–23. [DOI] [PubMed] [Google Scholar]
- 22. García‐Mediavilla MV, Sánchez‐Campos S, González‐Pérez P et al. Differential contribution of hepatitis C virus NS5A and core proteins to the induction of oxidative and nitrosative stress in human hepatocyte‐derived cells. J Hepatol 2005; 43:606–613. [DOI] [PubMed] [Google Scholar]
- 23. Kretzmann NA, Fillmann H, Mauriz JL et al. Effects of glutamine on proinflammatory gene expression and activation of nuclear factor kappa B and signal transducers and activators of transcription in TNBS‐induced colitis. Inflamm Bowel Dis 2008; 14:1504–1513. [DOI] [PubMed] [Google Scholar]
- 24. Hissin PJ, Hilf R. A fluorometric method for determination of oxidized and reduced glutathione in tissues. Anal Biochem 1976; 74:214–226. [DOI] [PubMed] [Google Scholar]
- 25. Ogata M, Hino S, Saito A et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 2006; 26:9220–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crespo I, Miguel BS, Laliena A et al. Melatonin prevents the decreased activity of antioxidant enzymes and activates nuclear erythroid 2‐related factor 2 signaling in an animal model of fulminant hepatic failure of viral origin. J Pineal Res 2010; 49:193–200. [DOI] [PubMed] [Google Scholar]
- 27. Mauriz JL, González P, Jorquera F et al. Caspase inhibition does not protect against liver damage in hemorrhagic shock. Shock 2003; 19:33–37. [DOI] [PubMed] [Google Scholar]
- 28. Sir D, Chen WL, Choi J et al. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 2008; 48:1054–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Air‐Goughoulte M, Kanda T, Meyer K et al. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol 2008; 82:2241–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang J, Lam GY, Brumell JH. Autophagy signaling through reactive oxygen species. Antioxid Redox Signal 2011; 14:2215–2231. [DOI] [PubMed] [Google Scholar]
- 31. Huang H, Kang R, Wang J et al. Hepatitis C virus inhibits AKT‐tuberous sclerosis complex (TSC), the mechanistic target of rapamycin (MTOR) pathway, through endoplasmic reticulum stress to induce autophagy. Autophagy 2013; 9:175–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Santana S, Sastre I, Recuero M et al. Oxidative stress enhances neurodegeneration markers induced by herpes simplex virus type 1 infection in human neuroblastoma cells. PLoS ONE 2013; 8:e75482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joubert PE, Werneke S, de la Calle C et al. Chikungunya‐induced cell death is limited by ER stress and oxidative stress‐induced autophagy. Autophagy 2012; 8:1261–1263. [DOI] [PubMed] [Google Scholar]
- 34. Guo Y, Wang J, Wang Z et al. Melatonin protects N2a against ischemia/reperfusion injury through autophagy enhancement. J Huazhong Univ Sci Technolog Med Sci 2010; 30:1–7. [DOI] [PubMed] [Google Scholar]
- 35. Jeong JK, Moon MH, Lee YJ et al. Melatonin‐induced autophagy protects against human prion protein‐mediated neurotoxicity. J Pineal Res 2012; 53:138–146. [DOI] [PubMed] [Google Scholar]
- 36. Choi SI, Kim KS, Oh JY et al. Melatonin induces autophagy via an mTOR‐dependent pathway and enhances clearance of mutant‐TGFBIp. J Pineal Res 2012; 54:361–372. [DOI] [PubMed] [Google Scholar]
- 37. Zhou H, Chen J, Lu X et al. Melatonin protects against rotenone‐induced cell injury via inhibition of Omi and Bax‐mediated autophagy in Hela cells. J Pineal Res 2012; 52:120–127. [DOI] [PubMed] [Google Scholar]
- 38. Yoo YM, Jeung EBM. Melatonin suppresses cyclosporine A‐induced autophagy in rat pituitary GH3 cells. J Pineal Res 2010; 48:204–211. [DOI] [PubMed] [Google Scholar]
- 39. Chang CF, Huang HJ, Lee HC et al. Melatonin attenuates kainic acid‐induced neurotoxicity in mouse hippocampus via inhibition of autophagy and α‐synuclein aggregation. J Pineal Res 2012; 52:312–321. [DOI] [PubMed] [Google Scholar]
- 40. Nopparat C, Porter JE, Ebadi M et al. The mechanism for the neuroprotective effect of melatonin against methamphetamine‐induced autophagy. J Pineal Res 2010; 49:383–389. [DOI] [PubMed] [Google Scholar]
- 41. Taylor MP, Kierkegaard M. Modification of cellular autophagy protein LC3 by poliovirus. J Virol 2007; 81:12543–12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maier HJ, Britton P. Involvement of autophagy in coronavirus replication. Viruses 2012; 4:3440–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dreux M, Gastaminza P, Wieland SF et al. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA 2009; 106:14046–14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee YR, Hu HY, Kuo SH et al. Dengue virus infection induces autophagy: an in vivo study. J Biomed Sci 2013; 20:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shinohara Y, Imajo K, Yoneda M et al. Unfolded protein response pathways regulate hepatitis C virus replication via modulation of autophagy. Biochem Biophys Res Commun 2013; 432:326–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest 2011; 121:37–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang T, Lin RT, Li Y et al. Hepatitis C virus inhbitis intracellular interpheron alpha expression in human hepatic cell lines. Hepatology 2005; 42:819–827. [DOI] [PubMed] [Google Scholar]
- 48. Calvo JR, González‐Yanes C, Maldonado MD. The role of melatonin in the cells of the innate immunity: a review. J Pineal Res 2013; 55:103–120. [DOI] [PubMed] [Google Scholar]
- 49. Wen X, Abe T, Kukihara H et al. Elimination of hepatitis C virus from hepatocytes by a selective activation of therapeutic molecules. PLoS ONE 2011; 6:e15967. [DOI] [PMC free article] [PubMed] [Google Scholar]


