Abstract
The outbreak of a novel coronavirus (SARS‐CoV‐2) since December 2019 in Wuhan, the major transportation hub in central China, became an emergency of major international concern. While several etiological studies have begun to reveal the specific biological features of this virus, the epidemic characteristics need to be elucidated. Notably, a long incubation time was reported to be associated with SARS‐CoV‐2 infection, leading to adjustments in screening and control policies. To avoid the risk of virus spread, all potentially exposed subjects are required to be isolated for 14 days, which is the longest predicted incubation time. However, based on our analysis of a larger dataset available so far, we find there is no observable difference between the incubation time for SARS‐CoV‐2, severe acute respiratory syndrome coronavirus (SARS‐CoV), and middle east respiratory syndrome coronavirus (MERS‐CoV), highlighting the need for larger and well‐annotated datasets.
Keywords: coronavirus, incubation, local infection/replication/spread, pandemic, virulence
Highlights
It is unclear whether there are statistically significant differences in incubation times amongst SARS‐CoV‐2, SARS‐CoV and MERS‐CoV. However, this is because: 1) Limited available data challenges investigation of the current coronavirus outbreak. 2) Lack of annotation also makes it difficult to identify and consolidate the datasets. Thus, care should be taken when selecting datasets for comparative analysis with other viruses or outbreaks.
Biological investigation of SARS‐CoV‐2 biology has revealed several general characteristics of the virus. SARS‐CoV‐2 is a novel coronavirus, which has a ~30 kb single‐stranded positive sense RNA genome with an organization typical of other coronaviruses such as SARS and MERS. 1 Although phylogenetic analysis indicate it belongs to the same β‐coronavirus genus as severe acute respiratory syndrome coronavirus (SARS‐CoV) and middle east respiratory syndrome coronavirus (MERS‐CoV), SARS‐CoV‐2 has a higher genome‐sequence similarity to several β‐coronaviruses detected in bats. It shows more than 96% identity to a known bat coronavirus, compared to 79.5% identity to SARS‐CoV BJ01. 1 , 2
Studies investigating the clinical characteristics, epidemic and treatment have also been carried out. According to clinical investigation of the pneumonia cases in China, SARS‐CoV‐2 infection causes SARS with major symptoms such as fever, cough, myalgia, or fatigue and minor symptoms such as sputum production, headache, hemoptysis, and diarrhea. 3
As more data become available, additional case features are also being revealed. More than half of the initial cases had visited the Wuhan Huanan seafood market. It is also apparent that the outcome of SARS‐CoV‐2 pneumonia is enormously destructive, despite a mortality rate less than 3% (according to the latest data on February 8th, 2020 in China 4 ) when compared with SARS‐CoV (mortality rate 9.6%) and MERS‐CoV (mortality rate 9.6% and 34%). 5 However, the spread of SARS‐CoV‐2 infection is much broader than SARS or MERS‐CoV and involves larger numbers of patients.
The symptom onset date of the first identified patient infected by SARS‐CoV‐2 was December 1st, 2019, which is about 14 days before the subsequent reported cases. 3 The first estimate of mean incubation time was based on the exposure information of 10 confirmed early SARS‐CoV‐2 infected cases in Wuhan, China and was predicted to be 5.2 days (95% confidence interval [CI]: 4.1‐7.0), 6 supporting the case for a longer incubation time compared to SARS‐CoV (mean incubation time 4.0 days, 95% CI: 3.6‐4.4) 7 and MERS‐CoV in Saudi Arabia/Middle east area (range of incubation times 4.5‐5.2 days, mean value/95% CI not reported). 8 A longer incubation time may lead to a high rate of asymptomatic and subclinical infection among immunocompetent individuals. A popular hypothesis circulating on social media is that the rapid spread of SARS‐CoV‐2 is a consequence of a longer incubation time, that is, while it can give the host the chance to develop immunity, it may also facilitate the spread of infection.
The reported estimate of the SARS‐CoV‐2 incubation time was based on limited case data. A subsequent unpublished study from 88 cases estimated a mean incubation time of 6.4 days (95% CI, 5.6‐7.7 days). 9 However, the data were taken from an online resource, 10 and only a subset of these data (25 patients) had both clearly defined start and stop dates for exposure, together with a date for onset of symptoms. The patients from Wuhan had extended exposure times by December 14th. As an alternative approach, we limited our dataset to the patients whose exposure periods were well‐defined. As of February 8th, 2020, this comprised 50 patients (Supporting Information Material). We also collected additional raw data from earlier reports on SARS (153 patients) and MERS (70 patients) outbreaks (Supporting Information Material). We then fitted “Weibull”, “lognormal,” and “gamma” functions to the respective datasets. These are shown for SARS‐CoV‐2, SARS, and MERS datasets in Figure 1A‐C, respectively. The corresponding mean and 95% CI were: SARS‐CoV‐2, 4.9 (4.4‐5.5) days; SARS, 4.7 (4.3‐5.1) days; and MERS 5.8 (5.0‐6.5) days (Figure 1E). A pairwise comparison between each of the three datasets (Mann–Whitney test) showed no significant differences (Supporting Information Material). The discrepancy between our results and other studies can be understood by examining the individual datasets. These are summarized as boxplots in Figure 1D and have notably varying distributions. For the MERS datasets, for example, we found only five reports published with accessible raw data, but one report had several patients with incubation times ranged from 0 to 21 days. It is unclear whether these data were included in the other datasets. If these data are included in the analysis, it significantly impacts the outcome for estimation of incubation times 7.5 (7.1‐7.9) days (Figure S1). Thus, any conclusions based on comparisons between SARS‐CoV‐2, SARS, and MERS incubation times are arbitrarily dependent on the selected data sources.
Figure 1.
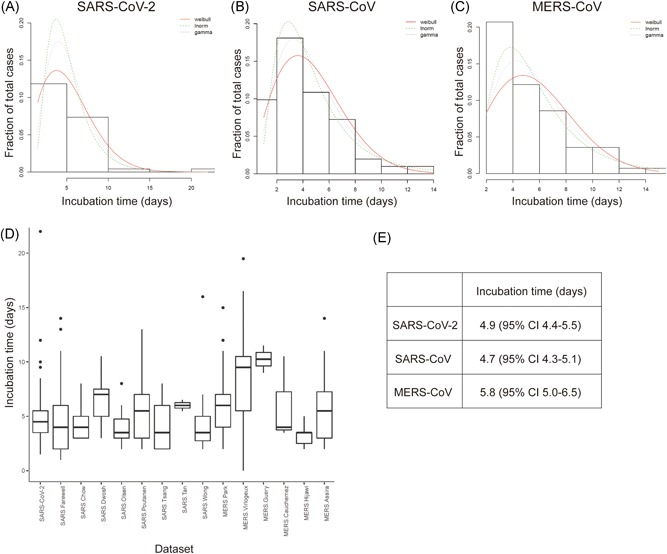
Estimated incubation times for SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV. A, Fitted Weibull, lognormal, and gamma distributions to data from 49 SARS‐CoV‐2 infected patients with defined exposure start date, exposure end date, and symptoms onset date. B, Corresponding analysis for 153 SARS‐CoV patients consolidated from seven different studies. C, Corresponding analysis for 70 MERS‐CoV patients consolidated from four different studies. D, Box and whisker plot showing distribution of incubation times for each of the individual studies used in (A), (B), and (C) and one additional MERS dataset that was not included in the analysis. E, Estimated incubation times for SARS‐CoV‐2, SARS‐CoV, and MERS‐CoV. See Table S1 for raw data and references
The challenge of finding suitable data extends to investigations of other important characteristics of the SARS‐CoV‐2 outbreak:
Reproduction number R 0. There has been significant variation in the reported values of R 0 according to the dataset used in the analysis.
Clinical symptoms. Diverse clinical symptoms have been described, but this information is only available for a limited number of reported cases.
Transmission routes. There are still debates on possible routes besides aerosol transmission for human‐to‐human infection. There is now evidence of viral presence in feces and on the object surface, indicating possibilities of waterborne and contact transmission, which may account for the infection from asymptomatic patients.
Thus, access to well‐annotated data related to these topics from clinical patients and subclinical subjects will help our understanding for each of these factors. Our results indicate that the current 14 days isolation period should be continued until more comprehensive data are available. To this end, we make the following suggestions:
-
1.
Data should be ideally annotated using standard metadata tags, for example, from the Disease Ontology (disease‐ontology.org) to aid data standardization, integration, and analysis.
-
2.
Study subjects should include not only patients and suspected infected individuals, but also the samples from the “normal” population.
Clinicians can help support these efforts by carefully collecting and capturing as much relevant patient data where possible. In this way, more complete datasets can be constructed, allowing for more in‐depth analyses to better determine optimal intervention strategies and patient treatment.
Supporting information
Supplementary information
Supplementary information
Jiang X, Rayner S, Luo M‐H. Does SARS‐CoV‐2 has a longer incubation period than SARS and MERS? J Med Virol. 2020;92:476–478. 10.1002/jmv.25708
REFERENCES
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020. https://doi.org/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. National Health Commission of the People's Republic of China , 2020. http://www.nhc.gov.cn/xcs/yqtb/202002/6c305f6d70f545d59548ba17d79b8229.shtml [DOI] [PMC free article] [PubMed]
- 5. Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E. A novel coronavirus emerging in China—Key questions for impact assessment. N Engl J Med. 2020. 10.1056/NEJMp2000929 [DOI] [PubMed] [Google Scholar]
- 6. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lessler J, Reich NG, Brookmeyer R, Perl TM, Nelson KE, Cummings DA. Incubation periods of acute respiratory viral infections: a systematic review. Lancet Infect Dis. 2009;9(5):291‐300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Park JE, Jung S, Kim A, Park JE. MERS transmission and risk factors: a systematic review. BMC Public Health. 2018;18(1):574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Backer JA, Klinkenberg D, Wallinga J. The incubation period of 2019‐nCoV infections among travellers from Wuhan, China. medRxiv. 2020. 10.1101/2020.01.27.20018986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun K, Online repository, 2020. https://docs.google.com/spreadsheets/d/1jS24DjSPVWa4iuxuD4OAXrE3QeI8c9BC1hSlqrNMiU/edit#gid=1449891965
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Supplementary information


