Abstract
Coronavirus (CoV) infections are commonly associated with respiratory and enteric disease in humans and animals. In 2012, a new human disease called Middle East respiratory syndrome (MERS) emerged in the Middle East. MERS was caused by a virus that was originally called human coronavirus‐Erasmus Medical Center/2012 but was later renamed as Middle East respiratory syndrome coronavirus (MERS‐CoV). MERS‐CoV causes high fever, cough, acute respiratory tract infection, and multiorgan dysfunction that may eventually lead to the death of the infected individuals. The exact origin of MERS‐CoV remains unknown, but the transmission pattern and evidence from virological studies suggest that dromedary camels are the major reservoir host, from which human infections may sporadically occur through the zoonotic transmission. Human to human transmission also occurs in healthcare facilities and communities. Recent studies on Middle Eastern respiratory continue to highlight the need for further understanding the virus‐host interactions that govern disease severity and infection outcome. In this review, we have highlighted the major mechanisms of immune evasion strategies of MERS‐CoV. We have demonstrated that M, 4a, 4b proteins and Plppro of MERS‐CoV inhibit the type I interferon (IFN) and nuclear factor‐κB signaling pathways and therefore facilitate innate immune evasion. In addition, nonstructural protein 4a (NSP4a), NSP4b, and NSP15 inhibit double‐stranded RNA sensors. Therefore, the mentioned proteins limit early induction of IFN and cause rapid apoptosis of macrophages. MERS‐CoV strongly inhibits the activation of T cells with downregulation of antigen presentation. In addition, uncontrolled secretion of interferon ɣ‐induced protein 10 and monocyte chemoattractant protein‐1 can suppress proliferation of human myeloid progenitor cells.
Keywords: immune evasion, immune response, Middle East respiratory syndrome coronavirus
This study was conducted to highlight the major mechanisms of immune evasion strategies of MERS‐CoV. In this review, we have demonstrated that M, 4a, 4b proteins and Plppro of MERS‐CoV inhibit the type I interferon (IFN) and nuclear factor‐κB signalling pathways and therefore facilitate innate immune evasion. In addition, NSP4a, NSP4b, and NSP15 inhibit dsRNA sensors. Therefore, the mentioned proteins limit early induction of IFN and cause rapid apoptosis of macrophages. MERS‐CoV strongly inhibits the activation of T cells with downregulation of antigen presentation. In addition, uncontrolled secretion of interferon ɣ‐induced protein 10 and monocyte chemoattractant protein‐1 can suppress proliferation of human myeloid progenitor cells.

Abbreviations
- CARD
caspase activation and recruitment domains
- CREBBP
cAMP‐response element binding protein
- CXCL10
C‐X‐C motif chemokine 10
- DPP4
dipeptidyl peptidase 4
- dsRNA
double‐stranded RNA
- IKKε
IκB kinase ε
- IL‐10
interleukin 10
- IP‐10
interferon ɣ‐induced protein 10
- IRF3
interferon regulatory factor 3
- ISG15
interferon‐stimulated gene 15
- ISG56
IFN‐stimulated gene 56
- ISGs
interferon‐stimulated genes
- MAVS
mitochondrial antiviral signaling
- MCP
monocyte chemoattractant protein‐1
- MDA5
melanoma differentiation associated protein 5
- MERS
Middle East respiratory syndrome
- NEMO
NF‐κB essential modulator
- NF‐κB
nuclear factor‐κB
- NSP15
nonstructural protein 15
- OAS
oligoadenylate synthetase
- ORFs
open reading frames
- PDE
phosphodiesterase
- PKR
protein kinase R
- PLpro
papain‐like protease
- PRR
pattern recognition receptors
- RIG‐I
cytoplasmic retinoic acid‐inducible gene I
- ssRNA
single‐stranded RNA
- TBK1
TANK‐binding kinase 1
- TLR3
toll‐like receptor 3
- TLRs
toll‐like receptors
- TRAF3
TNF receptor‐associated factor 3
1. INTRODUCTION
During summer of 2012, in Jeddah, Saudi Arabia, a novel coronavirus (CoV; Middle East respiratory syndrome coronavirus [MERS‐CoV]) was isolated from the sputum of a patient with acute pneumonia and renal failure (de Groot et al., 2013). Since September 2012, 27 countries in the Middle East, North Africa, Europe, the United States of America and Asia have reported cases of MERS‐CoV. Globally, 2143 laboratory‐confirmed cases of MERS‐CoV and at least 750 deaths have been reported to the World Health Organization (WHO), 82% of whom were from the kingdom of Saudi Arabia. According to the latest WHO report (26 January 2018) related to the kingdom of Saudi Arabia, 20 cases including nine deaths have been reported between December 9, 2017 and January 17, 2018 (Organization, 2017). The largest MERS outbreak outside of Saudi Arabia has occurred in South Korea with 186 cases and 39 deaths (CFR: 21%) from May to July 2015 (Chen, Chughtai, Dyda, & MacIntyre, 2017). Most people infected with MERS‐CoV developed a severe respiratory illness with clinical symptoms of fever, cough, and shortness of breath. Others may develop gastrointestinal symptoms such as diarrhea and nausea or vomiting, and kidney failure. MERS can be life‐threatening. Many deaths due to MERS‐CoV have been reported (Al‐Tawfiq & Memish, 2016). The highest virus loads are found in the lower respiratory tract samples, although low concentrations of viral RNA can also be found in stool, urine, and blood samples (Drosten et al., 2013).
MERS‐CoV has been classified into lineage C of Betacoronavirus and is most phylogenetically related to two bat coronaviruses, Tylonycteris bat coronavirus HKU4 (Ty‐BatCoV HKU4) and Pipistrellus bat coronavirus HKU5 (Pi‐BatCoV HKU5), providing insight on its evolutionary origin (Lau et al., 2013; Zumla, Hui, & Perlman, 2015). Table 1 shows the amino acid (aa) sequences identities between HKU4, HKU5, and MERS‐CoV (Corman et al., 2014).
Table 1.
Comparison of amino acid identities between HKU4, HKU5, and MERS‐CoV
| ORF | Nucleotide positions (start–end) | No. of amino acids | Amino acid identity (%) | ||
|---|---|---|---|---|---|
| MERS‐CoV | HKU4 | HKU5 | |||
| ORF 1ab | 281–21,528 | 7,082 | 92.7 | 73.7 | 76.0 |
| Spike | 21,470–25,504 | 1,344 | 64.3–64.6 | 60.5–60.8 | 61.5 |
| ORF3 | 25,519–25,830 | 103 | 76.5–78.4 | 40.7–44.0 | 47.9 |
| ORF4a | 25,839–26,168 | 109 | 87.0–88.0 | 37.4–38.3 | 41.9–42.9 |
| ORF4b | 26,044–26,820 | 258 | 83.7–85.4 | 83.7–85.4 | 26.8–26.8 |
| ORF5 | 26,827–27,501 | 224 | 87.1–88.4 | 47.1 | 54.8–55.7 |
| E | 27,577–27,825 | 82 | 89.0 | 73.2–74.4 | 72.0 |
| M | 27,840–28,499 | 219 | 93.6–94.5 | 81.7–82.2 | 82.6–83.1 |
| N | 28,603–29,202 | 199 | 81.1–83.9 | 48.7–50.8 | 52.6–55.8 |
Note. MERS‐CoV: Middle East respiratory syndrome coronavirus; ORF: open reading frame.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
The genome structure is a polycistronic positive‐sense single‐stranded RNA with ~30 kb in size and encoding 20 proteins. The 5' end of the genome contains two open reading frames (ORFs), ORF1a and ORF1b, which encode two polyproteins (pp), pp1a and pp1ab; production of pp1ab requires a ribosomal frameshift to transcribe the portion encoded by ORF1b. ORF1a encodes viral proteases, main protease (Mpro, also called 3CLpro), and papain‐like protease (PLpro), which are responsible for cleavage of the ORF1a and ORF1b. These polyproteins are further cleaved into 16 nonstructural proteins, whereas the 3′ end of the viral genome encodes four structural proteins (E, N, S, and M; Mustafa, Balkhy, & Gabere, 2017). Upon infection, these proteins are expressed to facilitate viral replication and propagation in the host (Cho, Lin, Chuang, & Hsu, 2016; Durai, Batool, Shah, & Choi, 2015).
Each coronavirus has a specific group of genes, which is responsible for encoding accessory proteins (Y. Yang et al., 2013). These accessory proteins do not participate in the structure of MERS‐CoV particles but have an essential role in viral replication and evasion of the host immune response (de Haan, Masters, Shen, Weiss, & Rottier, 2002; Haijema, Volders, & Rottier, 2004; D. X. Liu, Fung, Chong, Shukla, & Hilgenfeld, 2014; Matthews, Coleman, van der Meer, Snijder, & Frieman, 2014; Niemeyer et al., 2013). They are difficult to study because of their low expression level as well as their low molecular weight. In addition, they are not conserved in the coronavirus subfamilies. Although the accessory proteins can be targeted by antiviral therapeutics, the biological function of these proteins is still not well understood. MERS‐CoV has five accessory proteins: 3, 4a, 4b, 5, and 8b, encoded by various ORFs (Durai et al., 2015). Figure 1 provides details on the organization of the MERS‐CoV genome.
Figure 1.

Genome organization of the MERS‐CoV is illustrated by boxes in this genome scheme. The genome contains two large 5′‐proximal ORFs (ORF1a and 1b) that encode two replicase polyproteins, whose mature products assemble into the viral replication and transcription complex. The 3′ end of the genome encodes structural and accessory proteins. The 3, 4a, 4b, 5, and 8b accessory proteins are located between the structural proteins S and E, whereas 8b resides downstream of the N protein. MERS‐CoV: Middle East respiratory syndrome coronavirus; MPRO: main protease; ORF: open reading frame; PLPRO: papain‐like protease; PP: polyprotein [Color figure can be viewed at wileyonlinelibrary.com]
A functional receptor of MERS‐CoV is dipeptidyl peptidase 4 (DPP4) from both human and bat (Raj et al., 2013). DPP4, a 766‐aa‐long type‐II transmembrane glycoprotein, is mainly expressed on epithelial cells and controls the activity of hormones and chemokines (Bosch, Raj, & Haagmans, 2013; Chan, Lau, & Woo, 2013; Zhang, Jiang, & Du, 2014). This receptor binds to a 231 residue region in the spike (S) protein of MERS‐CoV, a domain different from the receptor‐binding site of other Betacoronaviruses. The S protein and the receptor‐binding site within this protein induce neutralizing antibodies and, in principle, could serve as a subunit vaccine (Lu et al., 2013; Mou et al., 2013). Small molecules or peptides that prevent the binding of DPP4 are potential MERS‐CoV entry inhibitors, and a few have been identified. Adenosine deaminase, a DPP4 binding protein, acts as a competitive inhibitor for MERS‐CoV S protein (Raj et al., 2014). An anti‐cluster of differentiation 26 (CD26) polyclonal antibody has also shown inhibitory effects on MERS‐CoV infection in vitro (Ohnuma et al., 2013).
The case fatality rate (CFR) in MERS‐CoV (CFR: 35%) is higher than that of the severe acute respiratory syndrome (SARS; CFR: 9.6%; Al Hajjar, Memish, & Mcintosh, 2013). The virulence of these human infections is undeniably linked to immune evasion mechanisms. Viruses have learned how to manipulate host immune control mechanisms. Our knowledge of viral gene functions could lead to new antiviral strategies and the ability to exploit viral functions as tools in medicine (Alcami & Koszinowski, 2000). Here, we provide an overview of the different mechanisms that MERS‐CoV use to evade host immune responses.
2. HOST IMMUNE SYSTEM OVERVIEW
The immune system is canonically divided into two major branches, innate and adaptive immunity. The innate response elicited by an invading pathogen involves the rapid recognition of general molecular patterns by nonimmune cells or cells of the innate immune system, such as monocytes or macrophages, dendritic cells, and natural killer cells (Nicholson, 2016). Hence, the first barrier to overcome for successful viral infection is the rapid innate immune response of the host, which are involved in several effector mechanisms, including complement cascade, type I Interferons (IFNs), inflammatory cytokine, NK cell immunity, apoptosis, autophagy, and toll‐like receptors (TLRs) pathway. When the innate immune system confronts a pathogen, it becomes activated and prepares the adaptive arm of the immune system to respond appropriately (Takeuchi & Akira, 2009).
The adaptive immune system consists of two branches: the humoral immune response arm (production of antibodies by B cells) and the cellular immune response arm (activities carried out by cytotoxic CD4+ and CD8+ T cells). Both typically require antigen presentation in conjunction with major histocompatibility complex (MHC) and a costimulatory signal for full activation (Lee, Lee, Chaudhary, Gill, & Jung, 2010). There are three possible outcomes of viral infection: Early clearance of the pathogen either directly or by phagocytosis, overwhelming infection with failure to control, persistent infection where a balance between the pathogen and the host is achieved (Simmons, Willberg, & Paul, 2001).
Viruses use diverse mechanisms to avoid and antagonize the immune response of their hosts. These mechanisms include: enable the virus to avoid recognition by the humoral immune response, interfere with the functioning of the cellular immune response, and interfere with immune effector functions (Vossen, Westerhout, Söderberg‐Nauclér, & Wiertz, 2002).
3. INNATE IMMUNE
3.1. IFN responses
Viral infection of mammalian cells prompts the innate immune system to mount the first line of defense (Takeuchi & Akira, 2009). Type I IFNs (mainly IFN‐α and IFN‐β) are major effector cytokines in innate antiviral response (González‐Navajas, Lee, David, & Raz, 2012). These IFNs activate the JAK–STAT pathway to stimulate the expression of interferon‐stimulated genes (ISGs), which collectively inhibit viral replication and assembly. The genes encoding IFNs are regulated by the assembly of an enhanceosome containing several transcription factors including nuclear factor‐κB (NF‐κB) and interferon regulatory factor 3 (IRF3), both of which are regulated by subcellular localization (Seth, Sun, Ea, & Chen, 2005).
In tissue cells, transcription of the IFN‐β gene represents the primary response to virus infection (Haagmans et al., 2004). Induction of the IFN‐β gene requires the constitutively expressed transcription factor IRF3. IRF3 is the key transcription factor for IFN‐β, ISG56, interferon ɣ‐induced protein 10 (IP‐10), and other antiviral genes (Hiscott, 2007). In uninfected cells, IRF‐3 is inactive and resides in the cytoplasm but transported to the nucleus.
Upon infection, IRF‐3 is phosphorylated and dimerized before it enters the nucleus to upregulate IFN‐α and IFN‐β gene transcription, where it recruits the transcriptional coactivator cAMP‐response element binding protein and p300 to initiate IFN‐β mRNA synthesis (Fitzgerald et al., 2003; Suhara, Yoneyama, Kitabayashi, & Fujita, 2002). Upon virus infection, to induce IFN production, pathogen‐associated molecular patterns such as viral double‐stranded RNA are sensed by host pattern recognition receptors such as endosomal TLR3, melanoma differentiation associated protein 5 (MDA5), and cytoplasmic retinoic acid‐inducible gene I (RIG‐I; Kato, Takahasi, & Fujita, 2011). RIG‐I and MDA5 are cytoplasmic sensors of virus‐derived RNAs (Goubau, Deddouche, & Reis e Sousa, 2013). RIG‐I contains two N‐terminal caspase activation and recruitment domains (CARD)‐like domains, and a C‐terminal RNA helicase domain that binds to double‐stranded RNA (dsRNA; Sumpter et al., 2005). Presumably, the binding of viral RNA to RIG‐I leads to a conformational change that exposes the CARD‐like domain, which then activates downstream signaling. Consistent with this model, overexpression of the N‐terminal CARD‐like domains of RIG‐I is sufficient to activate both NF‐κB and IRF3 (Yoneyama et al., 2004).
Optimal activity of RIG‐I and MDA5 also requires Protein kinase, interferon‐inducible double stranded RNA dependent activator (PACT), a cellular dsRNA‐binding protein which binds to RIG‐I and MDA5 to activate IFN production (Kok et al., 2011). The activation of these receptors transmits a signal to downstream kinases TANK‐binding kinase 1 (TBK1) and IκB kinase ε (IKKε) that form a functional complex with TNF receptor‐associated factor 3 (TRAF3) and TANK. Consequent phosphorylation of IRF3 transcription factors by these kinases leads ultimately to transcriptional activation of IFN promoters.
Once secreted, IFN‐β binds to its receptor on the cell surface and activates the synthesis of proteins with antiviral, antiproliferative, and immunomodulatory properties (De Veer et al., 2001). IFN‐β also participates in the induction of IFN‐αs, which further amplify the antiviral response (Haagmans et al., 2004).
Mitochondrial antiviral signaling (MAVS) is essential for NF‐κB and IRF3 activation by RNA viruses. This protein contains an N‐terminal caspase activation and recruitment domains (CARD)‐like domain and a C‐terminal transmembrane domain that targets the protein to the mitochondrial membrane. MAVS functions downstream of RIG‐I and upstream of IκB and IRF3 phosphorylation (Seth et al., 2005).
NF‐κB transcription factor also plays an important role in regulating the cellular response to IFNs. NF‐κB clearly plays a critical role in the signal transduction pathway that senses viral nucleic acids during pathogenic infection (Pfeffer, 2011). Similar to IRF3, NF‐κB is sequestered in the cytoplasm in association with an inhibitor of the IκB family. Stimulation of cells with cytokines leads to the activation of a large kinase complex consisting of the catalytic subunits IKKα and IKKβ and the essential regulatory subunit NF‐κB essential modulator. The activated IKK complex phosphorylates IκB and targets this inhibitor for degradation by the ubiquitin‐proteasome pathway. NF‐κB is then liberated to enter the nucleus to turn on a battery of genes essential for immune and inflammatory responses (T. Liu, Zhang, Joo, & Sun, 2017). The activation of NF‐κB is a double‐edged sword: While normal functions of NF‐κB are needed for proper innate and adaptive immune responses, dysregulation of NF‐κB can lead to inflammatory diseases and tumorigenesis (Karin, 2006).
Viruses have evolved effective strategies to evade the IFN system. They can inhibit IFN induction, IFN signal transduction, or the action of particular antiviral proteins by various mechanisms, including double‐stranded RNA binding and IRF‐3 sequestration. Coronaviruses encode multiple proteins to counteract the host innate antiviral response (Kindler & Thiel, 2014; Totura & Baric, 2012; Wong, Lui, & Jin, 2016). MERS‐CoV‐infected cells exhibited reduced IFN and cytokine expression (Ferran & Skuse, 2017). MERS‐CoV M, 4a, 4b, and PLP proteins were recently found to antagonize type I IFN production (Y. Yang et al., 2013). Schematic illustration of inhibition of IFN signalling is shown in Figure 2.
Figure 2.
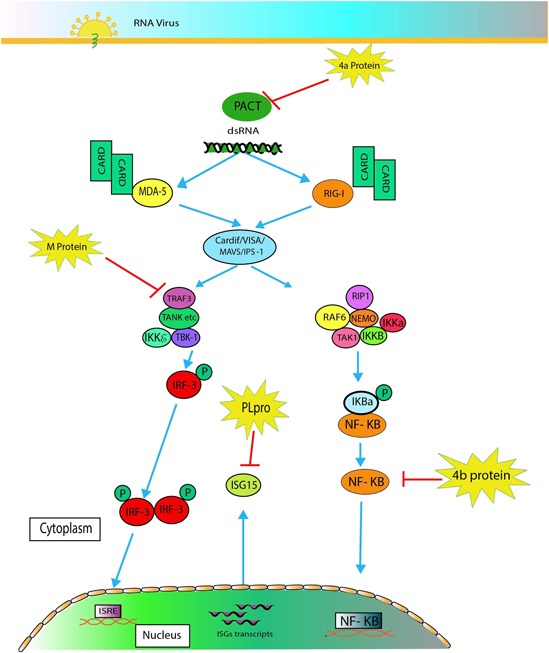
Illustration of virus activation of IFN‐I synthesis and its inhibition by the MERS‐CoV M, 4a, 4b and PLP proteins. M protein interacts with TRAF3 and suppresses IRF3 activity. 4a protein interacts with PACT and inhibits activation of RIG‐I and MDA5. 4b protein is unlikely to be a potent inhibitor of the NF‐κB signalling pathway. PLPs are known to have the ability to remove ISG15 conjugates from cellular substrates and reduce the levels of ubiquitinated and ISGylated host cell proteins. IFN: interferon; IRF: interferon regulatory factor; ISG: interferon‐stimulated gene; MDA: melanoma differentiation associated protein; MERS‐CoV: Middle East respiratory syndrome coronavirus; NF‐κB: nuclear factor‐κB; RIG‐I: cytoplasmic retinoic acid‐inducible gene I; TRAF: TNF receptor‐associated factor [Color figure can be viewed at wileyonlinelibrary.com]
3.1.1. M protein
M proteins from MERS‐CoV have three highly similar conserved N‐terminal transmembrane domains named TM1, TM2, and TM3 that localized predominantly to the Golgi complex and is required for virion assembly and a C‐terminal region (Neuman et al., 2011; Y. Yang et al., 2013). Using chimeric and truncation mutants, the N‐terminal transmembrane domains of the MERS‐CoV M protein were found to be sufficient for its inhibitory effect on IFN expression, whereas the C‐terminal domain was unable to induce this suppression (Lui et al., 2016). MERS‐CoV M protein suppresses IRF3 activity but not NF‐κB signaling. It is known that the activation of RIG‐I and MAVS results in activation of both IRF3 and NF‐κB (Seth et al., 2005; Yoneyama et al., 2004). MERS‐CoV M protein is capable of differentially suppressing the RIG‐I‐induced activation of IRF3. M protein interacts with TRAF3 and disrupts TRAF3–TBK1 association, leading to reduced activation of IRF3 (Lui et al., 2016). TRAF3 functions as an adapter that bridges the mitochondrial transducer MAVS with the downstream signaling complex containing TBK1 and IKK‐ɛ kinases that are essential for IRF3 activation (Guo & Cheng, 2007). SARS‐CoV M protein also prevents the formation of TRAF3·TANK·TBK1/IKK‐ɛ complex leading to reduced IRF3 activation and thereby inhibits IFN production (Siu et al., 2009). MERS‐CoV and SARS‐CoV are two highly pathogenic coronaviruses that have caused hundreds of deaths (Coleman et al., 2016). M proteins of MERS‐CoV and SARS‐CoV have high sequence similarity (>70%; Lui et al., 2016). In contrast, M protein encoded by human coronavirus HKU1, associated with common cold, has no influence on IFN production. HKU1 is commonly found in human population and is associated with less severe respiratory illnesses and has 35% identity with full M proteins of SARS coronavirus (Kanwar, Selvaraju, & Esper, 2017; Siu, Chan, Kok, Chiu‐Yat Woo, & Jin, 2014).
3.1.2. Nonstructural protein 4a (NSP4a)
ORF4a encoded one of the accessory proteins that block IFN induction and works as a strong inhibitor of type 1 IFN by inhibiting dsRNA recognition by cellular RIG‐I and MDA5 (Niemeyer et al., 2013; Siu et al., 2014). MERS‐CoV 4a protein has a suppression effect on the PACT‐induced activation of RIG‐I and MDA5, however, this double‐stranded RNA‐binding protein has no effect on the activity of downstream effectors such as RIG‐I, MDA5, MAVS, TBK1, and IRF3 (Siu, Yeung et al., 2014).
Protein 4a is 109 aa long and contains an RNA‐binding domain comprising 72 aa. The RNA‐binding domain of 4a binds dsRNA and does not allow it to bind to the RNA‐binding domain of RIG‐I, thereby inhibiting the antiviral signaling pathway. Thus, the virus blocks the innate immune response and continues infecting cells. The two key residues involved in the binding of RNA to the RNA‐binding domain in 4a are K63 and K67 (Siu, Yeung et al., 2014).
ORF4a is the most potent antagonist, because can inhibit multiple levels of the IFN response via the inhibition of both IFN production (IFN‐β promoter activity, IRF‐3 and NF‐κB activation) and the ISRE promoter element signaling pathway (Y. Yang et al., 2013).
3.1.3. NSP4b protein
ORF4b encodes an accessory protein of MERS‐CoV. NSP4b proteins with 246aa are multifunctional with activities in both the cytoplasm and nucleus, consistent with its localization in both cellular compartments. ORF4b protein is unlikely to be a potent inhibitor of the NF‐κB signalling pathway (Matthews et al., 2014). Replication of a recombinant MERS‐CoV with 4a and 4b proteins deletions was attenuated (Almazán et al., 2013).
3.1.4. Papain‐like protease ubiquitin‐like domain
Coronaviruses encode multifunctional enzymes with protease activity (PLpro). These enzymes are necessary for deubiquitinating (DUB)/deISGylating activity and processing the viral replicase polyproteins (Mielech, Kilianski, Baez‐Santos, Mesecar, & Baker, 2014). The ubiquitin pathway is important for regulating a number of innate immune pathways, and the ability of a viral protein to cleave ubiquitin from host cell proteins can contribute to virus pathogenesis. In addition to ubiquitination, modification of cellular proteins with ISG15 is known to have a broad spectrum antiviral activity. ISG15 is the ubiquitin‐like protein that can be conjugated to cellular targets via a mechanism called ISGylation, regulating innate immune responses. Coronavirus PLPs are known to have the ability to remove ISG15 conjugates from cellular substrates and reduces the levels of ubiquitinated and ISGylated host cell proteins (Clementz et al., 2010). MERS‐CoV PLpro acts as a potential IFN antagonist by interfering with the IRF3 and NF‐kB (X. Yang et al., 2014).
3.2. Modulation of apoptosis
Macrophages are immune cells equipped with multiple dsRNA sensors designed to detect viral infection and amplify innate antiviral immunity. Activation of dsRNA sensors results in an early induction of IFN, rapid apoptosis of macrophages and a protective immune response. However, many coronaviruses can infect and propagate in macrophages without activating dsRNA sensors.
Coronaviruses are positive‐sense RNA viruses that generate dsRNA intermediates during replication. MERS‐CoV, NSP4a, NSP4b, and NSP15 inhibit dsRNA sensors (Deng et al., 2017). MERS‐CoV NSP4a encodes a dsRNA‐binding protein that limits the activation of protein kinase R (PKR). PKR phosphorylates eIF2α, leading to the inhibition of cellular and viral translation. Expression of MERS‐CoV p4a impedes dsRNA‐mediated PKR activation, thereby rescuing translation inhibition (Rabouw et al., 2016). NSP4b encodes a phosphodiesterase, which inhibits RNase L activity. Limitation of RNase L activation can have a profound effect on enhancing virus replication and spread as well as leading to decreased IFN production in cells. The IFN‐inducible oligoadenylate synthetase (OAS)‐RNase L pathway activates upon sensing of viral dsRNA. Activated RNase L cleaves viral and hosts single‐stranded RNA, which leads to the translational arrest and subsequent cell death, preventing viral replication and spread. NSP4b protein antagonizes the OAS‐RNase L pathway by cleaving 2–5A and blocking the subsequent activation of RNase L (Thornbrough et al., 2016; L. Zhao et al., 2012).
NSP15 is an endoribonuclease. The mechanism by which NSP15 endoribonuclease activity suppresses the activation of dsRNA sensors is unknown. Deng et al. (2017) reported that macrophages infected with the NSP15 mutant virus had a significant dispersal of dsRNA as compared to the cells infected with wild‐type virus, where the majority of dsRNA has been accompanied by replication complexes. Figure 3 shows the MERS‐CoV escape from the dsRNA sensors.
Figure 3.

Activation of dsRNA sensors results in an early induction of interferon, rapid apoptosis of macrophages and a protective immune response. NSP4a limits the activation of PKR, thereby rescuing translation inhibition. NSP4b inhibits RNase L activity. Limitation of RNase L activation leads to decreased IFN production in cells. dsRNA: double‐stranded RNA; NSP: nonstructural protein; PKR: protein kinase R; OAS: oligoadenylate synthetase [Color figure can be viewed at wileyonlinelibrary.com]
4. ADAPTIVE IMMUNITY
In infection of MERS‐CoV, T cells play critical roles in controlling the pathogenesis (J. Zhao et al., 2014). Furthermore, T cells from human peripheral blood mononuclear cells, human lymphoid tissues, and the spleen of common marmosets were highly susceptible to MERS‐CoV. MERS‐CoV induces substantial apoptosis in the infected T cells that involve the activation of the intrinsic and extrinsic caspase‐dependent apoptosis pathways, resulting in high pathogenicity of the virus (Chu et al., 2015; Zhou, Chu, Chan, & Yuen, 2015).
MERS‐CoV can directly infect and replicate productively in macrophages and dendritic cells, which results in their malfunction and failure to present virus antigen to T cells (Chu et al., 2014; Zhou et al., 2014). Downregulation of antigen presentation pathways (decreased MHC I and II, costimulatory molecules) in macrophages and dendritic cells during MERS‐CoV infection would strongly inhibit the activation of T cells. (Faure et al., 2014; Josset et al., 2013).
The clinical course of MERS in patients shows that secretion of monocyte chemoattractant protein‐1 (MCP‐1), C‐X‐C motif chemokine 10 also known as IP‐10 and interleukin 10 is uncontrolled (Kim et al., 2016; Ying, Li, & Dimitrov, 2016). IP‐10 and MCP‐1 can suppress proliferation of human myeloid progenitor cells (Broxmeyer et al., 1993). The induction of these chemokines may inevitably aggravate the lymphopenia in patients with MERS (Zhou et al., 2015).
5. CONCLUSION
The relationship between a virus and its host is a complicated affair: a myriad of factors from the virus and host are involved in viral infection and consequential pathogenesis. The pathogenesis of human MERS‐CoV infection remains poorly understood. Multiple antagonistic mechanisms are developed by MERS‐CoV to evade the induction of proinflammatory cytokines and to attenuate the host defense. There is an urgent need for developing the most effective MERS therapy. More research findings on the pathogenesis and immune evasion mechanisms of MERS‐CoV may help to improve the treatment and control of MERS. Reliable vaccines have not yet been developed but antivirals inhibiting virus replication, such as mycophenolic acid, cyclosporine A, IFN‐α, IFN‐β, or ribavirin are available. The attenuated viruses generated might prove useful in vaccine development. Reverse genetics system is a tool to study the molecular biology of the virus, understanding novel gene functions, high‐throughput screening of antiviral drugs, and to develop attenuated viruses as vaccine candidates. Using this system a collection of MERS‐CoV deletion mutants has been generated. If deletion of genes that causing the virus to escape from the immune system presented growth kinetics similar to those of the wild‐type virus, indicating which is helpful to prevent MERS‐CoV infection.
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
Shokri S, Mahmoudvand S, Taherkhani R, Farshadpour F. Modulation of the immune response by Middle East respiratory syndrome coronavirus. J Cell Physiol. 2018;234:2143–2151. 10.1002/jcp.27155
References
REFERENCES
- Al Hajjar, S. , Memish, Z. A. , & Mcintosh, K. (2013). Middle East respiratory syndrome coronavirus (MERS‐CoV): A perpetual challenge. Annals of Saudi Medicine, 33(5), 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcami, A. , & Koszinowski, U. H. (2000). Viral mechanisms of immune evasion. Immunology Today, 21(9), 447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazan, F. , DeDiego, M. L. , Sola, I. , Zuniga, S. , Nieto‐Torres, J. L. , Marquez‐Jurado, S. , … Enjuanes, L. (2013). Engineering a replication‐competent, propagation‐defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio, 4(5), e00650–00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Tawfiq, J. , & Memish, Z. (2016). The middle east respiratory syndrome coronavirus respiratory infection: An emerging infection from the Arabian Peninsula, The Microbiology of Respiratory System Infections (pp. 55–63). Elsevier. [Google Scholar]
- Bosch, B. J. , Raj, V. S. , & Haagmans, B. L. (2013). Spiking the MERS‐coronavirus receptor. Cell Research, 23(9), 1069–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxmeyer, H. E. , Sherry, B. , Cooper, S. , Lu, L. , Maze, R. , Beckmann, M. P. , … Ralph, P. (1993). Comparative analysis of the human macrophage inflammatory protein family of cytokines (chemokines) on proliferation of human myeloid progenitor cells. Interacting effects involving suppression, synergistic suppression, and blocking of suppression. The Journal of Immunology, 150(8), 3448–3458. [PubMed] [Google Scholar]
- Chan, J. F.‐W. , Lau, S. K.‐P. , & Woo, P. C.‐Y. (2013). The emerging novel Middle East respiratory syndrome coronavirus: The “knowns” and “unknowns”. Journal of the Formosan Medical Association, 112(7), 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Chughtai, A. A. , Dyda, A. , & MacIntyre, C. R. (2017). Comparative epidemiology of Middle East respiratory syndrome coronavirus (MERS‐CoV) in Saudi Arabia and South Korea. Emerging Microbes & Infections, 6(6), e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, C.‐C. , Lin, M.‐H. , Chuang, C.‐Y. , & Hsu, C.‐H. (2016). Macro domain from Middle East respiratory syndrome coronavirus (MERS‐CoV) is an efficient ADP‐ribose binding module crystal structure and biochemical studies. Journal of Biological Chemistry, 291(10), 4894–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H. , Zhou, J. , Ho‐Yin Wong, B. , Li, C. , Cheng, Z.‐S. , Lin, X. , … Yuen, K.‐Y. (2014). Productive replication of Middle East respiratory syndrome coronavirus in monocyte‐derived dendritic cells modulates innate immune response. Virology, 454‐455, 197–205. [Supplement C]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H. , Zhou, J. , Wong, B. H.‐Y. , Li, C. , Chan, J. F.‐W. , Cheng, Z.‐S. , … Yuen, K. Y. (2015). Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. The Journal of Infectious Diseases, 213(6), 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz, M. A. , Chen, Z. , Banach, B. S. , Wang, Y. , Sun, L. , Ratia, K. , … Baker, S. C. (2010). Deubiquitinating and interferon antagonism activities of coronavirus papain‐like proteases. Journal of Virology, 84(9), 4619–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, C. M. , Sisk, J. M. , Mingo, R. M. , Nelson, E. A. , White, J. M. , & Frieman, M. B. (2016). Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. Journal of Virology, 90(19), 8924–8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman, V. M. , Ithete, N. L. , Richards, L. R. , Schoeman, M. C. , Preiser, W. , Drosten, C. , & Drexler, J. F. (2014). Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. Journal of Virology, 88(19), 11297–11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X. , Hackbart, M. , Mettelman, R. C. , O’Brien, A. , Mielech, A. M. , Yi, G. , … Baker, S. C. (2017). Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proceedings of the National Academy of Sciences, 114(21), E4251–E4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten, C. , Seilmaier, M. , Corman, V. M. , Hartmann, W. , Scheible, G. , Sack, S. , … Wendtner, C. M. (2013). Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. The Lancet Infectious Diseases, 13(9), 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai, P. , Batool, M. , Shah, M. , & Choi, S. (2015). Middle East respiratory syndrome coronavirus: Transmission, virology and therapeutic targeting to aid in outbreak control. Experimental & Molecular Medicine, 47(8), e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure, E. , Poissy, J. , Goffard, A. , Fournier, C. , Kipnis, E. , Titecat, M. , … Guery, B. (2014). Distinct immune response in two MERS‐CoV‐infected patients: Can we go from bench to bedside? PLoS One, 9(2), e88716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferran, M. C. , & Skuse, G. R. (2017). Evasion of host innate immunity by emerging viruses: Antagonizing host RIG‐I pathways. J Emerg Dis Virol, 3(3). [Google Scholar]
- Fitzgerald, K. A. , McWhirter, S. M. , Faia, K. L. , Rowe, D. C. , Latz, E. , Golenbock, D. T. , … Maniatis, T. (2003). IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nature Immunology, 4(5), 491–496. [DOI] [PubMed] [Google Scholar]
- González‐Navajas, J. M. , Lee, J. , David, M. , & Raz, E. (2012). Immunomodulatory functions of type I interferons. Nature Reviews Immunology, 12(2), 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau, D. , Deddouche, S. , & Reis e Sousa, C. (2013). Cytosolic sensing of viruses. Immunity, 38(5), 855–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot, R. J. , Baker, S. C. , Baric, R. S. , Brown, C. S. , Drosten, C. , Enjuanes, L. , … Ziebuhr, J. (2013). Middle East respiratory syndrome coronavirus (MERS‐CoV): Announcement of the coronavirus study group. Journal of Virology, 87(14), 7790–7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, B. , & Cheng, G. (2007). Modulation of the interferon antiviral response by the TBK1/IKKi adaptor protein TANK. Journal of Biological Chemistry, 282(16), 11817–11826. [DOI] [PubMed] [Google Scholar]
- Haagmans, B. L. , Kuiken, T. , Martina, B. E. , Fouchier, R. A. M. , Rimmelzwaan, G. F. , Van Amerongen, G. , … Osterhaus, A. D. M. E. (2004). Pegylated interferon‐α protects type 1 pneumocytes against SARS coronavirus infection in macaques. Nature Medicine, 10(3), 290–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan, C. A. M. , Masters, P. S. , Shen, X. , Weiss, S. , & Rottier, P. J. M. (2002). The group‐specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology, 296(1), 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema, B. J. , Volders, H. , & Rottier, P. J. M. (2004). Live, attenuated coronavirus vaccines through the directed deletion of group‐specific genes provide protection against feline infectious peritonitis. Journal of Virology, 78(8), 3863–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott, J. (2007). Triggering the innate antiviral response through IRF‐3 activation. Journal of Biological Chemistry, 282(21), 15325–15329. [DOI] [PubMed] [Google Scholar]
- Josset, L. , Menachery, V. D. , Gralinski, L. E. , Agnihothram, S. , Sova, P. , Carter, V. S. , … Katze, M. G. (2013). Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio, 4(3), e00165–00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar, A. , Selvaraju, S. , & Esper, F. (2017). Human coronavirus‐HKU1 infection among adults in Cleveland, Ohio. Open Forum Infectious Diseases, 4(2), ofx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin, M. (2006). Nuclear factor‐κB in cancer development and progression. Nature, 441(7092), 431–436. [DOI] [PubMed] [Google Scholar]
- Kato, H. , Takahasi, K. , & Fujita, T. (2011). RIG‐I‐like receptors: Cytoplasmic sensors for non‐self RNA. Immunological Reviews, 243(1), 91–98. [DOI] [PubMed] [Google Scholar]
- Kim, E. S. , Choe, P. G. , Park, W. B. , Oh, H. S. , Kim, E. J. , Nam, E. Y. , … Oh, M. (2016). Clinical progression and cytokine profiles of Middle East respiratory syndrome coronavirus infection. Journal of Korean Medical Science, 31(11), 1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler, E. , & Thiel, V. (2014). To sense or not to sense viral RNA—Essentials of coronavirus innate immune evasion. Current Opinion in Microbiology, 20, 69–75. [Supplement C]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok, K.‐H. , Lui, P.‐Y. , Ng, M. H. , Siu, K.‐L. , Au, S. W. , & Jin, D.‐Y. (2011). The double‐stranded RNA‐binding protein PACT functions as a cellular activator of RIG‐I to facilitate innate antiviral response. Cell Host and Microbe, 9(4), 299–309. [DOI] [PubMed] [Google Scholar]
- Lau, S. K. , Li, K. S. , Tsang, A. K. , Lam, C. S. , Ahmed, S. , Chen, H. , … Yuen, K.‐Y. (2013). Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: Implications for the origin of the novel Middle East respiratory syndrome coronavirus. Journal of Virology, 87(15), 8638–8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.‐R. , Lee, S. , Chaudhary, P. M. , Gill, P. , & Jung, J. U. (2010). Immune evasion by Kaposi’s sarcoma‐associated herpesvirus. Future Microbiology, 5(9), 1349–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. X. , Fung, T. S. , Chong, K. K.‐L. , Shukla, A. , & Hilgenfeld, R. (2014). Accessory proteins of SARS‐CoV and other coronaviruses. Antiviral Research, 109, 97–109. [Supplement C]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Zhang, L. , Joo, D. , & Sun, S.‐C. (2017). NF‐κB signaling in inflammation. Signal Transduction and Targeted Therapy, 2, 17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, G. , Hu, Y. , Wang, Q. , Qi, J. , Gao, F. , Li, Y. , … Gao, G. F. (2013). Molecular basis of binding between novel human coronavirus MERS‐CoV and its receptor CD26. Nature, 500(7461), 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui, P.‐Y. , Wong, L. Y. , Fung, C.‐L. , Siu, K.‐L. , Yeung, M.‐L. , Yuen, K.‐S. , … Jin, D.‐Y. (2016). Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1‐dependent phosphorylation of IRF3. Emerging Microbes & Infections, 5(4), e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, K. L. , Coleman, C. M. , van der Meer, Y. , Snijder, E. J. , & Frieman, M. B. (2014). The ORF4b‐encoded accessory proteins of Middle East respiratory syndrome coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signalling. The Journal of General Virology, 95(Pt 4), 874–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech, A. M. , Kilianski, A. , Baez‐Santos, Y. M. , Mesecar, A. D. , & Baker, S. C. (2014). MERS‐CoV papain‐like protease has deISGylating and deubiquitinating activities. Virology, 450‐451, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, H. , Raj, V. S. , van Kuppeveld, F. J. , Rottier, P. J. , Haagmans, B. L. , & Bosch, B. J. (2013). The receptor binding domain of the new MERS coronavirus maps to a 231‐residue region in the spike protein that efficiently elicits neutralizing antibodies. Journal of Virology, 87, 01277–01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa, S. , Balkhy, H. , & Gabere, M. N. (2017). Current treatment options and the role of peptides as potential therapeutic components for Middle East Respiratory Syndrome (MERS): A review. Journal of Infection and Public Health, 11, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman, B. W. , Kiss, G. , Kunding, A. H. , Bhella, D. , Baksh, M. F. , Connelly, S. , … Buchmeier, M. J. (2011). A structural analysis of M protein in coronavirus assembly and morphology. Journal of Structural Biology, 174(1), 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson, L. B. (2016). The immune system. Essays in Biochemistry, 60(3), 275–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer, D. , Zillinger, T. , Muth, D. , Zielecki, F. , Horvath, G. , Suliman, T. , … Muller, M. A. (2013). Middle East respiratory syndrome coronavirus accessory protein 4a Is a Type I interferon antagonist. Journal of Virology, 87(22), 12489–12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma, K. , Haagmans, B. L. , Hatano, R. , Raj, V. S. , Mou, H. , Iwata, S. , … Morimoto, C. (2013). Inhibition of Middle East respiratory syndrome coronavirus infection by anti‐CD26 monoclonal antibody. Journal of Virology, 87(24), 13892–13899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization, W. H. (2017). Middle East respiratory syndrome coronavirus (MERS‐CoV)–Saudi Arabia, Geneva: WHO; Accessed 21 July 2017. Available at http://wwwwhoint/emergencies/mers-cov/en/ [Google Scholar]
- Pfeffer, L. M. (2011). The Role of nuclear factor κB in the interferon response. Journal of Interferon & Cytokine Research, 31(7), 553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouw, H. H. , Langereis, M. A. , Knaap, R. C. M. , Dalebout, T. J. , Canton, J. , Sola, I. , … van Kuppeveld, F. J. M. (2016). Middle East respiratory coronavirus accessory protein 4a inhibits PKR‐mediated antiviral stress responses. PLoS Pathogens, 12(10), e1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, V. S. , Mou, H. , Smits, S. L. , Dekkers, D. H. W. , Müller, M. A. , Dijkman, R. , … Haagmans, B. L. (2013). Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus‐EMC. Nature, 495(7440), 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, V. S. , Smits, S. L. , Provacia, L. B. , van den Brand, J. M. A. , Wiersma, L. , Ouwendijk, W. J. D. , … Haagmans, B. L. (2014). Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4‐mediated entry of the Middle East respiratory syndrome coronavirus. Journal of Virology, 88(3), 1834–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth, R. B. , Sun, L. , Ea, C.‐K. , & Chen, Z. J. (2005). Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF‐κB and IRF3. Cell, 122(5), 669–682. [DOI] [PubMed] [Google Scholar]
- Simmons, R. A. , Willberg, C. B. , & Paul, K. (2001). Immune evasion by viruses. eLS John Wiley & Sons, Ltd. [Google Scholar]
- Siu, K.‐L. , Chan, C.‐P. , Kok, K.‐H. , Chiu‐Yat Woo, P. , & Jin, D.‐Y. (2014). Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cellular and Molecular Immunology, 11(2), 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu, K.‐L. , Kok, K.‐H. , Ng, M.‐H. J. , Poon, V. K. M. , Yuen, K.‐Y. , Zheng, B.‐J. , & Jin, D.‐Y. (2009). Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3·TANK·TBK1/IKKϵ Complex. The Journal of Biological Chemistry, 284(24), 16202–16209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu, K.‐L. , Yeung, M. L. , Kok, K.‐H. , Yuen, K.‐S. , Kew, C. , Lui, P.‐Y. , … Jin, D.‐Y. (2014). Middle East respiratory syndrome coronavirus 4a protein is a double‐stranded RNA‐binding protein that suppresses PACT‐induced activation of RIG‐I and MDA5 in the innate antiviral response. Journal of Virology, 88(9), 4866–4876. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Suhara, W. , Yoneyama, M. , Kitabayashi, I. , & Fujita, T. (2002). Direct involvement of CREB‐binding protein/p300 in sequence‐specific DNA binding of virus‐activated interferon regulatory factor‐3 holocomplex. Journal of Biological Chemistry, 277(25), 22304–22313. [DOI] [PubMed] [Google Scholar]
- Sumpter, R. , Loo, Y.‐M. , Foy, E. , Li, K. , Yoneyama, M. , Fujita, T. , … Gale, M. (2005). Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG‐I. Journal of Virology, 79(5), 2689–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, O. , & Akira, S. (2009). Innate immunity to virus infection. Immunological Reviews, 227(1), 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornbrough, J. M. , Jha, B. K. , Yount, B. , Goldstein, S. A. , Li, Y. , Elliott, R. , … Weiss, S. R. (2016). Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio, 7(2), e00258–00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura, A. L. , & Baric, R. S. (2012). SARS coronavirus pathogenesis: Host innate immune responses and viral antagonism of interferon. Current Opinion in Virology, 2(3), 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veer, M. J. , Holko, M. , Frevel, M. , Walker, E. , Der, S. , Paranjape, J. M. , … Williams, B. R. (2001). Functional classification of interferon‐stimulated genes identified using microarrays. Journal of Leukocyte Biology, 69(6), 912–920. [PubMed] [Google Scholar]
- Vossen, M. , Westerhout, E. , Söderberg‐Nauclér, C. , & Wiertz, E. (2002). Viral immune evasion: A masterpiece of evolution. Immunogenetics, 54(8), 527–542. [DOI] [PubMed] [Google Scholar]
- Wong, L.‐Y. R. , Lui, P.‐Y. , & Jin, D.‐Y. (2016). A molecular arms race between host innate antiviral response and emerging human coronaviruses. Virologica Sinica, 31(1), 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Chen, X. , Bian, G. , Tu, J. , Xing, Y. , Wang, Y. , & Chen, Z. (2014). Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain‐like protease. Journal of General Virology, 95(3), 614–626. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Zhang, L. , Geng, H. , Deng, Y. , Huang, B. , Guo, Y. , … Tan, W. (2013). The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS‐CoV) are potent interferon antagonists. Protein & Cell, 4(12), 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying, T. , Li, W. , & Dimitrov, D. S. (2016). Discovery of T‐cell infection and apoptosis by Middle East respiratory syndrome coronavirus. The Journal of Infectious Diseases, 213(6), 877–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama, M. , Kikuchi, M. , Natsukawa, T. , Shinobu, N. , Imaizumi, T. , Miyagishi, M. , … Fujita, T. (2004). The RNA helicase RIG‐I has an essential function in double‐stranded RNA‐induced innate antiviral responses. Nature Immunology, 5(7), 730–737. [DOI] [PubMed] [Google Scholar]
- Zhang, N. , Jiang, S. , & Du, L. (2014). Current advancements and potential strategies in the development of MERS‐CoV vaccines. Expert Review of Vaccines, 13(6), 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Li, K. , Wohlford‐Lenane, C. , Agnihothram, S. S. , Fett, C. , Zhao, J. , … Perlman, S. (2014). Rapid generation of a mouse model for Middle East respiratory syndrome. Proceedings of the National Academy of Sciences of the United States of America, 111(13), 4970–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L. , Jha, B. K. , Wu, A. , Elliott, R. , Ziebuhr, J. , Gorbalenya, A. E. , … Weiss, S. R. (2012). Antagonism of the interferon‐induced OAS‐RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host and Microbe, 11(6), 607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Chu, H. , Chan, J. F.‐W. , & Yuen, K.‐Y. (2015). Middle East respiratory syndrome coronavirus infection: Virus‐host cell interactions and implications on pathogenesis. Virology Journal, 12(1), 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Chu, H. , Li, C. , Wong, B. H.‐Y. , Cheng, Z.‐S. , Poon, V. K.‐M. , … Yuen, K.‐Y. (2014). Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: Implications for pathogenesis. The Journal of Infectious Diseases, 209(9), 1331–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla, A. , Hui, D. S. , & Perlman, S. (2015). Middle East respiratory syndrome. The Lancet, 386(9997), 995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]


