Abstract
Many viruses are known to cause influenza‐like illness (ILI); however, in nearly 50% of patients, the etiologic agent remains unknown. The distribution of viruses in patients with ILI was investigated during the 2009 A/H1N1 influenza pandemic (A/H1N1p). From June 2009 to January 2010, 660 patients with suspected influenza were questioned and examined, and nasal swabs were collected. All patient samples were tested for influenza virus, and 286 negative nasal swabs were tested further for 18 other respiratory viruses using real‐time RT‐PCR. Two waves of ILI were observed in the epidemic curve (weeks 35–42 and 42–49). At least eight viruses co‐circulated during this period: human rhinovirus (HRV) (58), parainfluenza 1–4 viruses (PIV) (9), human Coronavirus (hCoV) OC43 (9), enterovirus (5), adenovirus (AdV) (4), and human metapneumovirus (hMPV) (2); however, 204 samples remained negative for all viruses tested. ILI symptoms, according to the Centers for Disease Control and Prevention criteria for ILI definition, were reported in 75% of cases. These patients had positive swabs for A/H1N1p, HRV, hCoV‐OC43, PIV, AdV, and hMPV without significant difference with non‐ILI patients. This study found that many respiratory viruses circulated during this period and that the A/H1N1p did not impact on the kinetics of other respiratory viruses. The proportion of non‐documented cases remains high. ILI could not distinguish A/H1N1p infection from that due to other respiratory viruses. However, in multivariate anlaysis, cough, chills, hyperemia, and dyspnea were associated significantly with influenza virus versus other respiratory viruses. J. Med. Virol. 84: 1071–1079, 2012. © 2012 Wiley Periodicals, Inc.
Keywords: respiratory viruses; influenza A virus, A/H1N1p subtype; rhinovirus; influenza‐like‐illness; acute respiratory tract infection
INTRODUCTION
Acute respiratory infection is one of the leading causes of child and adult morbidity and mortality throughout the world [Williams et al., 2002]. Determining the etiological diagnoses of patients who have respiratory symptoms remains a challenge both in the clinic and laboratory. Differentiating infections caused by influenza viruses from those caused by other respiratory viruses is essential for case management, as illustrated during the 2009 A/H1N1 influenza pandemic (A/H1N1p). Many definitions of influenza‐like illness (ILI) have been used worldwide in influenza surveillance; however, the sensitivity and positive predictive value of such definitions significantly vary depending on the co‐circulation of other respiratory viruses in the community [Boivin et al., 2000; Lee et al., 2011; Thursky et al., 2003]. The identification of the respiratory viruses that are responsible for influenza‐like illness has been reported in many countries, and the percentage of positive swabs for at least one virus ranges from 32% to 65% [Bellei et al., 2008; Laguna‐Torres et al., 2009; Ren et al., 2009; Buecher et al., 2010; Renois et al., 2010; Razanajatovo et al., 2011]. Influenza‐like illness can be attributed to a wide range of respiratory viruses, including influenza viruses, adenoviruses (AdV), respiratory syncytial virus (RSV), enteroviruses (EVs), human rhinovirus (HRV), and parainfluenza viruses (PiVs) [Bellei et al., 2008; Laguna‐Torres et al., 2009; Ren et al., 2009; Buecher et al., 2010; Renois et al., 2010; Razanajatovo et al., 2011]. Recently, several viruses have been associated with respiratory infections, including human metapneumovirus (hMPV) [van den Hoogen et al., 2001], human coronavirus NL63 (HCoV‐NL63) [van der Hoek et al., 2004], human coronavirus HKU1 (HCoVHKU1) [Woo et al., 2005], as well as human bocavirus (HBoV) [Allander et al., 2005]. Three novel polyomaviruses, KIPyV, WUPyV, and MCPyV, have been detected recently in the respiratory tracts of humans; however, their pathogenicity remains controversial [Norja et al., 2007; Babakir‐Mina et al., 2011].
The 2009 A/H1N1p influenza pandemic provided a unique opportunity to investigate the distribution of different viruses in patients with influenza‐like illness in a large sample of the general population. Few studies have described epidemiological and clinical data for different respiratory viruses that were identified to be circulating during the A/H1N1p pandemic [Hombrouck et al., 2011; Lee et al., 2011; Raboni et al., 2011; Smit et al., 2011a, b]. This study describes the prevalence of 19 viruses in patients suspected with influenza by the general practitioner and then sent to a referral center for nasal swab sampling and subsequent laboratory testing during the A/H1N1p outbreak.
MATERIALS AND METHODS
Respiratory Specimens
From June 2009 to January 2010, patients presenting with influenza‐like illness or suspected influenza were either referred to by their general practitioner, or consulted directly the doctor's group set up specifically for the management of suspect patients during the 2009 influenza pandemic at the Infectious Disease and Tropical Medicine Department of the North Hospital, Marseille, France.
Upon admission, patients were questioned and examined, and nasal swabs were collected and tested at the point‐of‐care (POC) laboratory by a rapid influenza diagnostic test (RIDT) and real‐time RT‐PCR (rRT‐PCR) [Ninove et al., 2010; Nougairede et al., 2010]. After obtaining oral consent, epidemiological and clinical questionnaires were completed while the patients waited for the RIDT result to be sent back by the POC lab. Patients with negative RIDT results returned home with isolation measures recommended until the result of the rRT‐PCR assay was obtained. When the rRT‐PCR results were obtained 12 hr later, patients with positive samples were contacted by telephone, and those with co‐morbidity risk factors were proposed for hospitalization, oseltamivir therapy, and isolation measures. For those without co‐morbidity risk factors, only symptomatic treatment was recommended.
Detection of Respiratory Viruses
RIDT was performed using the Directigen EZ influenza A + B test (BD EZ Flu A + B, Becton, Dickinson) according to the manufacturer's instructions.
RNA extraction: 200 µl of the respiratory sample prepared for RIDT were spiked with 10 µl of in‐house MS2/T4 phages internal control [Ninove et al., 2011]. RNA was extracted and eluted in 90 µl using the BioRobot EZ1 Workstation and the EZ1 Virus Mini Kit v2.0 (Qiagen, Courtaboeuf, France).
Reverse transcription was performed with the Taqman Reverse Transcription kit (Applied Biosystems, Branchburg, NJ) with 20 µl of RNA, 22 µl of MgCl2, 10 µl of 10× buffer, 20 µl of 10 mM dNTPs, 5 µl of hexamers (at 1/10 dilution), 2.5 µl of Multiscribe, and 2 µl of RNase inhibitor in a 100 µl final volume. The cycling program was 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min. For each sample, two reverse transcriptions in a 100 µl final volume were done, resulting in a 200 µl volume of cDNA to be used in PCR tests.
PCR assays were performed using the qPCR Mastermix‐No Rox kit (Eurogentec, Angers, France) with 10 µl of cDNA, 25 µl of Mastermix, 1 µl of each primer (10 mM), and 0.4 µl of probe (10 mM) in combination with a Stratagene MX3005P QPCR system (Agilent Technologies, La Jolla, CA). The primers and probes that were used in this study are listed in Table I. The cycling program was conducted at 50°C for 2 min, 95°C for 10 min, 45 cycles at 95°C for 15 sec, and 60°C for 60 sec.
Table I.
References for the PCR That Were Used for the Detection of Respiratory Viruses
| Viral etiology | Sequence of primers and probes | Protocol reference | |
|---|---|---|---|
| Influenza virus A virus A/H1N1psw 2009 | GRswH1‐349F | GAGCTAAGAGAGCAATTGA |
Duchamp et al. [2010 ] |
| GRswH1‐601R | GTAGATGGATGGTGAATG | ||
| GRswH1‐538Pr | FAM‐TTGCTGAGCTTTGGGTATGA–TAMRA | ||
| Influenza virus A virus H3N2 | INFA‐23 F | CATYCTGTTGTATATGAGGCCCAT |
van Elden et al. [2001 ] |
| INFA 1 R | GGACTGCAGCGTAGACGCTT | ||
| INFA Pr | FAM‐CTCAGTTATTCTGCTGGTGCACTTGCCA‐TAMRA | ||
| Rhinovirus | RHI2 F | C56 GCC 7GC GTG GC |
Lu et al. [2008 ] |
| RHI2 R | GAA ACA CGG ACA CCC AAA GTA | ||
| RHI2 Pr | FAM‐TCC TCC GGC CCC TGA ATG YGG C‐TAMRA | ||
| Metapneumovirus | NLN‐F | CATATAAGCATGCTATATTAAAAGAGTCTC |
Mackay et al. [2003 ] |
| NLN‐R | CCTATTTCTGCAGCATATTTGTAATCAG | ||
| NLN‐Pr | TGY AAT GAT GAG GGT GTC ACT GCG GTT G | ||
| Respiratory syncytial virus A | RSA 2bis F | GCA CAT CAT AAT TAG GAG TAT CAA T |
van Elden et al. [2003 ] |
| RSA 1 R | AGA TCA ACT TCT GTC ATC CAG CAA | ||
| RSA Pr | FAM CAC CAT CCA ACG GAG CAC AGG AGA T TAMRA | ||
| Respiratory syncytial virus B | RSB 2bis F | TGATATCCAGCATCTTTAAGTATCTTTATAGTG |
van Elden et al. [2003 ] |
| RSB 1 R | AAG ATG CAA ATC ATA AAT TCA CAG GA | ||
| RSB Pr | VIC AGG TAT GTT ATA TGC TAT GTC CAG GTT AGG AAG GGA A TAMRA | ||
| Human coronavirus 229E | COR 229 E2 F | AAA GGG CTA TAA AGA GAA TAA GGT ATT CT |
van Elden et al. [2004 ] |
| COR 229 E1 R | CAG TCA AAT GGG CTG ATG CA | ||
| COR 229 E Pr | CCC TGA CGA CCA CGT TGT GGT TCA | ||
| Human coronavirus OC43 | COR OC 43 1 F | CGA TGA GGC TAT TCC GAC TAG GT |
van Elden et al. [2004 ] |
| COR OC 43 2 R | CCT TCC TGA GCC TTC AAT ATA GTA ACC | ||
| COR OC 43 Pr | TCC GCC TGG CAC GGT ACT CCC T | ||
| Human coronavirus NL63 | hCoV‐NL63 F | CAG GGC TGA CAA GCC TTC TCA |
Tiveljung‐Lindell et al. [2009 ] |
| hCoV‐NL63 | R GCA TCA ACA CCA TTC TGA ACA AGA | ||
| hCoV‐NL63 Pr | FAM‐CGT TGG AAG CGT GTT CCT ACC AGA GAG G‐TAMRA | ||
| Human coronavirus KU1 | hCoV HKU‐1 F | CAC TTC TAT TCC CTC CGA TGT TTC |
Tiveljung‐Lindell et al. [2009 ] |
| hCoV‐HKU‐1 R | TTA GAA GCA GAC CTT CCT GAG CC | ||
| hCoV‐HKU‐1 Pr | FAM‐CGC CTG GTA CGA TTT TGC CTC AAG GCT‐TAMRA | ||
| Enterovirus | EV 1 F | CCC CTG AAT GCG GCT AAT CC |
Watkins‐Riedel et al. [2002 ] |
| EV 1 R | ATT GTC ACC ATA AGC AGC CA | ||
| Ent TM 1 Pr | FAM CAN GGA CAC CCA AAG TAG TCG GTT CC TAMRA | ||
| Parechovirus | AN345 F | GTA ACA SWW GCC TCT GGG SCC AAA AG |
Benschop et al. [2008 ] |
| AN344 R | GCC CCC WGR TCA GAT CCA YAG T | ||
| AN257 Pr | CCT RYG GGT ACC TYC WGG GCA TCC TTC | ||
| Polyomavirus KI/KU | PyV2263F | TTGGATGAAAATGGCATTGG |
Lindau et al. [2009 ] |
| PyV2404R | TAACCCTTCTTTGTCTAAARTGTAGCC | ||
| KIPyV Pr | FAM‐ACATTACTTGTGCAGATATGCTTGGAACAGC‐TAMRA | ||
| WU PyV Pr | FAM‐CATAACTTGTGCTGACCTTTTGGGAGTTAAC‐TAMRA | ||
| Parainfluenza virus 1/2/3/4 | PIV1 F | ACA GAT GAA ATT TTC AAG TGC TAC TTT AGT |
Tong et al. [2008 ] |
| PIV1 R | GCC TCT TTT AAT GCC ATA TTA TCA TTA GA | ||
| PIV1 Pr | FAM‐ATG GTA ATA AAT CGA CTC GCT‐TAMRA | ||
| PIV2 F | TGC ATG TTT TAT AAC TAC TGA TCT TGC TAA | ||
| PIV2 R | GTT CGA GCA AAA TGG ATT ATG GT | ||
| PIV2 Pr | FAM‐ACT GTC TTC AAT GGA GAT AT‐TAMRA | ||
| PIV3 F | TGC TGT TCG ATG CCA ACA A | ||
| PIV3 R | ATT TTA TGC TCC TAT CTA GTG GAA GAC A | ||
| PIV3 Pr | FAM‐TTG CTC TTG CTC CTC A‐TAMRA | ||
| PIV4 F | TGG CAA ATC GGC AAT TAA ACA | ||
| PIV4 R | GGC TCT GGC AGC AAT CAT AAG | ||
| PIV4 Pr | FAM‐TTC TGC ATT GAT GTG GCC TGT AAG GA‐TAMRA | ||
| Bocavirus | Boca NP1 F | AGA GGC TCG GGC TCA TAT CA |
Allander et al. [2007 ] |
| Boca NP1 R | CAC TTG GTC TGA GGT CTT CGA A | ||
| Boca NP1 Pr | 6FAM AGG AAC ACC CAA TCA RCC ACC TAT CGT CT TAMRA | ||
| Adenovirus | AQ1 F | GCC ACG GTG GGG TTT CTA AAC TT |
Heim et al. [2003 ] |
| AQ2 R | GCC CCA GTG GTC TTA CAT GCA CAT C | ||
| AP Pr | FAM‐TGC ACC AGA CCC GGG CTC AGG TAC TCC GA‐TAMRA | ||
Internal and External Controls
All steps (extraction, RT, PCR) were monitored with our universal internal control assay based on the use of DNA and RNA bacteriophages as described previously [Ninove et al., 2011]. PCR detection of T4 and MS2 bacteriophages was performed in parallel with other PCR using the same cycling program in a 15‐µl final volume with 3 µl of cDNA, 7.5 µl of Mastermix, 0.3 µl of each primer (10 mM), and 0.15 µl of probe (10 mM). For each sample, the run was validated by the results that were obtained for T4 and MS2 [Ninove et al., 2011].
Statistical Analyses
The questionnaire was entered anonymously into the database with Epidata 3.1 (Centers for Disease Control and Prevention criteria for influenza‐like illness, CDC, Atlanta, GA), and data were analyzed with SPSS, version 19.0 (SPSS Inc., Chicago, IL). To identify the clinical characteristics of each group of patients, all potential variables were first assessed individually in a univariate model, and P values were measured for qualitative variables using Pearson's chi‐square test or Fisher's exact test and for continuous variables using the Mann–Whitney non‐parametric test. Correlations were assessed using the Spearman non‐parametric test. Variables with P values <0.20 were retained and entered into a multivariate logistic regression analysis.
RESULTS
During the 29 weeks of the outbreak, 660 patients were seen at the outpatient clinic. Two peaks of influenza‐like illness were detected, the first peak (181 patients) from week 36 to 41 (wave 1) and the second peak (256 patients) from week 43 to 48 (wave 2) (Fig. 1). Among the 660 patients, 59.8% were female, more than half (53.9%) were between the ages of 20 and 40 and 6 (0.9%) were older than 65. The clinical characteristics and risk factors for serious illness are listed in Table II. The study was authorized by the ethics committee board of the university, number 10‐0010.
Figure 1.
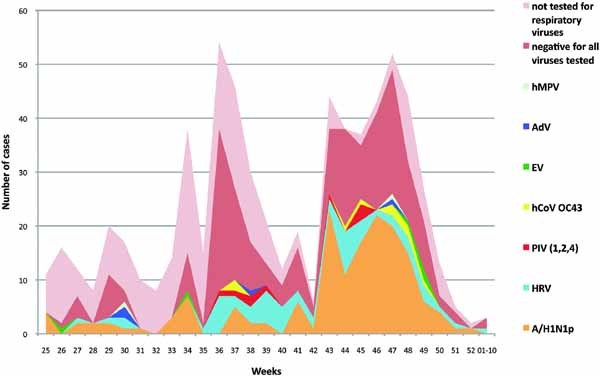
Weekly distribution of viruses causing respiratory infections from June 2009 (week 25) to January 2010 (week 01–10) in Marseille, France. ADV, adenovirus; hCoV OC43, human coronavirus OC43; EV, enterovirus; hMPV, human metapneumovirus; HRV, rhinovirus; A/H1N1p, 2009 pandemic A/H1N1 influenza virus; PIV, parainfluenza virus. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/jmv]
Table II.
Comparative Clinical and Epidemiological Characteristics of all of the Patients Stratified by A/H1N1p and HRV Infection
| Characteristics | N (%) total | N (%) A/H1N1p | N (%) HRV | A/H1N1p vs. non‐A/H1N1p | HRV vs. non‐HRV | A/H1N1p vs. HRV | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| P‐value | Odds ratio (95% CI) | P‐value | Odds ratio (95% CI) | P‐value | Odds ratio (95% CI) | |||||
| Sex | Female | 395 (59.8) | 79 (50) | 19 (32.8) | <0.01 | 1.7 (1.19; 2.44) | 0.6 | <0.05 | 0.5 (0.26; 0.92) | |
| Male | 265 (40.2) | 79 (50) | 39 (67.2) | |||||||
| Age | Mean (SD) | 31 (14.27) | 26.6 (12.12) | 32.6 (12.7) | <0.0001 | 0.8 | <0.01 | |||
| ≤10 | 40 (6.1) | 10 (6.4) | 1 (1.7) | <0.0001 | 1.0 | <0.05 | ||||
| 11–20 | 110 (16.7) | 40 (25.5) | 10 (17.2) | |||||||
| 21–30 | 195 (29.5) | 56 (35.7) | 17 (29.3) | |||||||
| 31–40 | 160 (24.2) | 35 (22.3) | 14 (24.1) | |||||||
| 41–50 | 78 (11.8) | 7 (4.5) | 9 (15.5) | |||||||
| 51+ | 76 (11.5) | 9 (5.7) | 7 (12.1) | |||||||
| Clinical characteristics | Tp° mean (SD) | 38.5 (0.90) | 38.5 (0.95) | 38.4 (0.79) | 0.75 | 0.9 | 0.5 | |||
| Influenza‐like illness | 358 (75.2) | 141 (89.2) | 43 (74.1) | <0.001 | 2.8 (1.64; 4.85) | 0.5 | <0.01 | 0.4 (0.16; 0.75) | ||
| Fever | 556 (84.9) | 143 (90.5) | 46 (79.3) | <0.05 | 1.9 (1.08; 3.47) | 0.5 | <0.05 | 0.4 (0.18; 0.92) | ||
| Cough | 545 (83.2) | 153 (96.8) | 50 (86.2) | <0.0001 | 8.2 (3.28; 20.49) | 0.5 | <0.01 | 0.2 (0.06; 0.65) | ||
| Sore throat | 424 (64.7) | 103 (65.2) | 40 (69) | 0.9 | 0.4 | 0.6 | ||||
| Asthenia | 611 (93.3) | 151 (95.6) | 51 (87.9) | 0.2 | 0.3 | 0.06 | 0.3 (0.11; 1.01) | |||
| Myalgia | 521 (79.7) | 126 (80.3) | 43 (74.1) | 0.8 | 0.5 | 0.3 | ||||
| Rhinorrhea | 414 (63.2) | 117 (74.1) | 47 (81) | <0.01 | 1.9 (1.29; 2.86) | <0.001 | 3.3 (1.61; 6.62) | 0.3 | ||
| Headache | 507 (77.4) | 123 (77.8) | 40 (69) | 0.8 | 0.07 | 0.6 (0.29; 1.06) | 0.2 | |||
| Chills | 427 (65.2) | 117 (74.1) | 30 (51.7) | <0.01 | 1.7 (1.16; 2.57) | <0.05 | 0.6 (0.31; 0.99) | <0.01 | 0.4 (0.20; 0.70) | |
| Arthralgia | 259 (39.7) | 65 (41.4) | 18 (31) | 0.6 | 0.3 | 0.2 | ||||
| Diarrhea | 109 (16.6) | 29 (18.4) | 7 (12.1) | 0.5 | 0.5 | 0.3 | ||||
| Nausea | 230 (35.2) | 62 (39.2) | 13 (22.8) | 0.2 | <0.05 | 0.5 (0.24; 0.93) | <0.05 | 0.5 (0.23; 0.92) | ||
| Vomiting | 104 (15.9) | 35 (22.2) | 8 (14) | <0.05 | 1.8 (1.12; 2.77) | 0.9 | 0.2 | |||
| Conjunctive hyperemia | 66 (11.1) | 23 (17.8) | 6 (12) | <0.01 | 2.1 (1.24; 3.71) | 1.0 | 0.4 | |||
| Dyspnea | 160 (24.5) | 54 (34.2) | 15 (26.3) | <0.01 | 1.9 (1.28; 2.82) | 0.7 | 0.3 | |||
| Medical history/risk factor | Recent travel | 106 (16.2) | 18 (11.4) | 8 (13.8) | 0.06 | 0.6 (0.35; 1.03) | 0.6 | 0.6 | ||
| Live locally | 658 (99.7) | 158 (100) | 57 (98.3) | 1.0 | 0.4 | 0.3 | ||||
| Pregnant (2nd trimester) | 36 (5.6) | 6 (4) | 7 (12.3) | 0.3 | 0.06 | 2.7 (1.01; 7.41) | <0.05 | 3.4 (1.08; 10.5) | ||
| Chronic bronchopathy | 92 (14.4) | 21 (14.1) | 13 (22.8) | 0.9 | 0.1 | 0.1 | ||||
| Cardiopathy | 11 (1.7) | 3 (2) | 1 (1.8) | 0.7 | 1.0 | 1.0 | ||||
| Neurologic | 10 (1.6) | 5 (3.4) | 1 (1.8) | 0.05 | 3.4 (0.97; 11.88) | 0.4 | 1.0 | |||
| Hematologic | 7 (1.1) | 2 (1.3) | 0 (0) | 0.7 | 0.6 | 1.0 | ||||
| Metabolic | 38 (5.9) | 12 (8.1) | 2 (3.5) | 0.2 | 0.7 | 0.4 | ||||
| Immunodeficiency | 11 (1.7) | 2 (1.3) | 2 (3.5) | 1.0 | 0.3 | 0.3 | ||||
| Obesity | 9 (1.4) | 0 (0) | 0 (0) | 0.1 | 0.6 | |||||
| Alcohol/hepatopathy | 9 (1.4) | 3 (2) | 0 (0) | 0.4 | 0.6 | 0.6 | ||||
| >65 Years old | 6 (0.9) | 1 (0.7) | 0 (0) | 1.0 | 0.6 | 1.0 | ||||
| Risk factor (one or more) | 189 (28.6) | 44 (27.8) | 21 (36.2) | 0.8 | 0.6 | 0.2 | ||||
| Hospitalization | 22 (3.3) | 5 (3.2) | 2 (3.4) | 0.9 | 0.7 | 1.0 | ||||
| Total | 660 | 186 | 58 | |||||||
HRV, rhinovirus; A/H1N1p, 2009 pandemic A/H1N1 influenza virus.
Virus Detection
Among the 660 patients, 158 were positive for A/H1N1p. Among the 502 patients with negative rRT‐PCR results for A/H1N1p, 286 samples (randomly chosen from the samples still available, 104 patients were seen during the wave 1, 123 patients were seen during the wave 2, and the remaining 59 patients during the other periods) were tested for 18 other respiratory viruses: 82 were positive for at least one virus (58 were positive for HRV, nine for HCoV OC43, five for EV, five for PIV1, one for PIV2, three for PIV4, four for AdV, and two for hMPV). The remaining 204 samples were found negative for all viruses tested in the study (Fig. 2).
Figure 2.
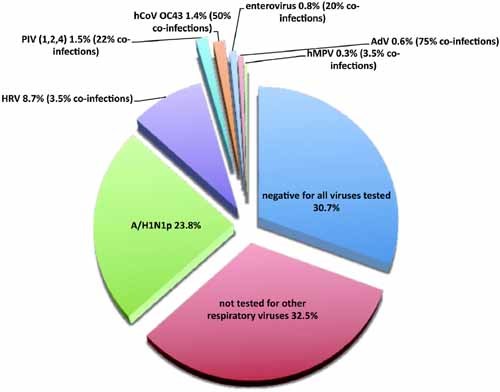
Distribution of viruses that were diagnosed in patients who presented at the hospital influenza group and were tested for all 19 agents. ADV, adenovirus; hCoV OC43, human coronavirus OC43; EV, enterovirus; hMPV, human metapneumovirus; HRV, rhinovirus; A/H1N1p, 2009 pandemic A/H1N1 influenza virus; PIV, parainfluenza virus. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/jmv]
During the first wave, 15 (8.3%) patients had positive swabs for A/H1N1p and among the 104 patients who were tested for other viruses, 25 (24%) had positive swabs for HRV and 72 (69%) had negative swabs. During the second wave, the percentage of patients who tested positive for A/H1N1p was significantly higher than that observed during the first wave (42.2%, P < 0.001, OR = 8, and 95% CI: 4.5; 14.5), whereas, among the 123 patients who were tested for other viruses, HRV had a consistent prevalence (16.3%, P = 0.143). Among the 286 samples tested for other viruses, co‐infection was identified in five patients: one HRV with ADV, one HRV with PIV4, one EV with hCoV OC43, one hMpV with ADV, and one PIV with ADV. No patients with co‐infections were hospitalized.
Clinical Features
Patient characteristics were stratified by infection status (Table II). In 75% of the cases, patients presented with an influenza‐like illness according to the definition proposed by the CDC with the following symptoms: a temperature >37.8°C and either a cough or sore throat [Babcock et al., 2006; CDC, 2010]. Only three patients were asymptomatic. Among the influenza‐like illness patients, 100 (27.9%) were positive for A/H1N1p and 27 (18.1%), 6 (4%), 5 (3.4%), 3 (2.1%), and 1 (0.7%) of the tested patients, were positive for HRV, PIV (1, 2, or 4), hCoV OC43, ADV, and hMpV, respectively (Table III). Of all of the patients, 22 were hospitalized later, 5 (22.7%) had confirmed A/H1N1p infection, and 2 (9%) tested positive for HRV.
Table III.
Etiologic Agent of Viral Respiratory Infection in Patients With or Without Influenza‐like Illness (According to the CDC's Definition) From June 2009 to January 2010 in Marseille, France
| Virus | Clinical presentation | P‐value | |
|---|---|---|---|
| Influenza‐like illness | Non‐influenza‐like illness | ||
| A/H1N1p | 100 (27.9) | 30 (25.4) | 0.596 |
| EV | 1 (0.3) | 1 (0.8) | 0.406 |
| HRV | 27 (7.5) | 7 (5.9) | 0.537 |
| hMPV | 1 (0.3) | 0 | 0.565 |
| hCoV OC43 | 5 (1.4) | 1 (0.8) | 0.641 |
| PIV 1 | 4 (1.1) | 1 (0.8) | 0.803 |
| PIV 2 | 0 | 0 | |
| PIV 4 | 2 (0.6) | 1 (0.8) | 0.728 |
| AdV | 3 (0.8) | 0 | 0.315 |
| Negative for all viruses tested | 108 (30.2) | 39 (33.1) | 0.311 |
| Not tested for respiratory viruses | 109 (30.4) | 39 (33.1) | 0.596 |
| Total | 358 | 118 | |
ADV, adenovirus; hCoV OC43, human coronavirus OC43; EV, enterovirus; hMPV, human metapneumovirus; HRV, rhinovirus; A/H1N1p, 2009 pandemic A/H1N1 influenza virus; PIV, parainfluenza virus.
Patients who tested positive for A/H1N1p were significantly younger (26.61 years with 95% CI: 24.70–28.52) than those who tested negative for A/H1N1p (32.28 years with 95% CI: 30.91–33.67; P = 0.0001). When analyzed by univariate analysis, cough (96.8%), self‐reported fever (90.5%), rhinorrhea (74%), chills (74%), vomiting (22.1%), conjunctive hyperemia (17.8%), and dyspnea (34.1%) were significantly more prevalent in A/H1N1p‐positive patients with odds ratios (OR) ranging from 1.72 to 8.20 (Table II). The presence of diarrhea was not associated significantly with A/H1N1p‐negative patients but was associated with traveling abroad, myalgia, nausea, and fever. Multivariate analyses demonstrated that cough (OR = 6.89 and 95% CI: 2.71–17.51; P < 0.001), chills (OR = 1.63 and 95% CI: 1.03–2.57; P = 0.035), conjunctive hyperemia (OR 1.93 and 95% CI: 1.07–3.48; P = 0.028), and dyspnea (OR = 1.58 and 95% CI: 1.01–2.47; P = 0.044) were associated independently with A/H1N1p infection when compared with A/H1N1p‐negative patients.
Since HRV was the second virus detected most frequently in swab samples, A/H1N1p‐positive patients were compared also with those who were positive for HRV, using univariate analyses. Fever (P = 0.027), cough (P = 0.007), chills (P = 0.002), and nausea (P = 0.026) were more frequent in patients with A/H1N1p‐positive swabs. Patients with HRV‐positive swabs were older (32.6 vs. 26.6, P = 0.002) (Table II). Multivariate analyses identified that cough (OR = 5 and 95% CI: 1.32–19.01; P = 0.018) and chills (OR = 3.18 and 95% CI: 1.57–6.47; P = 0.001) were associated independently with A/H1N1p infection.
DISCUSSION
In this prospective cohort, two waves of influenza‐like illness were observed in the epidemic curve. The first wave occurred in mid‐September, with a low prevalence of A/H1N1p (8.3%); in contrast with previously published data [Casalegno et al., 2009a, b], HRV (24%) could not explain completely this wave, and the majority of the etiologic agents were not identified. The second wave occurred in mid‐November, corresponding to the highest prevalence of A/H1N1p (42.2%) while the other viruses (as HRV or PIVs) was observed with the same prevalence.
After A/H1N1p, HRV was detected most commonly in patients (20.3% of tested patients). These results are consistent with previous data, which suggest that HRV is one of the most frequent causes of acute respiratory infection in adults and children, with a prevalence ranging from 6% to more than 40% [Bellei et al., 2008; Ren et al., 2009; Buecher et al., 2010; Renois et al., 2010; Razanajatovo et al., 2011; Tokarz et al., 2011].
Among the different hCoVs that were tested, hCoV OC 43 was the only strain found in 9 of the 286 samples that were tested (3.1%). This prevalence is similar to other studies that report hCoV OC43 in nearly 2% of patient samples [Bellei et al., 2008; Ren et al., 2009; Renois et al., 2010].
Nine patients were positive for at least one of the PiVs. Although PIV1, PIV2, and PIV3 are considered to be the most frequently identified PIVs [Henrickson, 2003], PIV1, and PIV4 were detected primarily, which is similar to the findings of Renois et al. [2010]. The lack of PIV3 detection may be due to the virus being commonly identified only during the first year of life [Renois et al., 2010].
The real‐time PCR method used for the diagnosis of EV was designed most specifically for diagnostics of central nervous system infections; however, recent studies have used a generic pan‐EV/rhinovirus real‐time PCR and have identified a novel respiratory EV that could not be detected by the systems designed for meningitis diagnostics [Watkins‐Riedel et al., 2002; Tapparel et al., 2009]. Although it is possible that some EVs remained undetected, five samples were tested positive for EVs (1.7% of tested patients), which is consistent with previous studies in patients with acute respiratory infection or influenza‐like illness [Bellei et al., 2008; Laguna‐Torres et al., 2009; Ren et al., 2009].
In this study, four samples (1.4%) were positive for AdV and two samples (0.7%) were positive for hMpV. These prevalences are comparable with previous reports [Bellei et al., 2008; Laguna‐Torres et al., 2009; Ren et al., 2009].
The absence of RSV infection could be explained by the mean age (31 year old) of the tested population, and by the delayed epidemic of RSV infection in France during the 2009–2010 winter season [Casalegno et al., 2009a, b].
The clinical significance of co‐infections is unclear [Jartti et al., 2004]. In this study, only 5 of the 286 tested swabs (1.7%) were positive for more than one respiratory virus, which was lower than previously reported [Esper et al., 2011; Hombrouck et al., 2011; Raboni et al., 2011; Tokarz et al., 2011], whereas this result is biased by the fact that only the patients who were negative for A/H1N1p were tested for the other viruses.
During the A/H1N1p pandemic, circulation of influenza B and A/H3N2 was null and very limited, respectively [Renois et al., 2010; Nakamura et al., 2011; Tokarz et al., 2011]. Therefore, patients who had negative RIDT results were not tested for influenza B.
There are multiple clinical definitions of influenza‐like illness. None are satisfactorily sensitive and specific for defining influenza virus infection [Thursky et al., 2003; CDC, 2010]. Although 75% of the clinical presentations were defined as influenza‐like illness according to the CDC definition [CDC, 2010], only 28% were confirmed influenza by laboratory documentation. Moreover, the percentage of patients with A/H1N1p‐positive swabs did not differ significantly between patients with symptoms of influenza‐like illness and those who did not present with these symptoms. Patients with influenza‐like illness also had swabs that tested positive for HRV, EV, hMpV, ADV, PIVS, or hCoV OC43. The clinical characteristics of patients with A/H1N1p infection have been reported in several countries [Crum‐Cianflone et al., 2009; Ong et al., 2009; Kim et al., 2010; Hombrouck et al., 2011; Lee et al., 2011; Smit et al., 2011a, b]. Of these eight studies, the clinical features that were associated mostly with A/H1N1p were the cough and the fever.
Several studies have reported the results from respiratory virus testing using respiratory samples that were obtained from patients with influenza‐like illness or acute respiratory infection throughout the world and during different times. Although the target populations, inclusion criteria, seasonality, climate, environment, diagnostic methods, and numbers of viruses or bacteria that were tested differed, the proportion of non‐documented cases remained relatively high [Bellei et al., 2008; Laguna‐Torres et al., 2009; Renois et al., 2010].
In conclusion, this study found (i) that many respiratory viruses circulated during the A/H1N1p pandemic in France, (ii) that A/H1N1p virus circulation did not impact on the kinetics of other respiratory viruses, (iii) that the percentage of non‐documented cases remains high and therefore justify to pursue technical development and to enlarge the variety of microorganisms in detection panels, (iv) that CDC definition of influenza‐like illness symptoms are not capable to distinguish A/H1N1p virus from other respiratory viruses, and finally, and (v) that the most specific criteria in favor of A/H1N1p infection was cough.
Systematic testing for respiratory viruses is necessary to improve the targeting of appropriate antiviral treatments. Specific studies exploring (i) the prevalence of co‐infections and their clinical characteristics, (ii) socio‐economic consequences of the different microorganisms involves in respiratory infections not only at the hospital level but more broadly inside and outside of the hospital are necessary for a better management of cases.
Acknowledgements
We thank Audrey Cortes for excellent technical assistance.
Conflicts of interest: None.
REFERENCES
- Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung‐Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA 102: 12891–12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allander T, Jartti T, Gupta S, Niesters HG, Lehtinen P, Osterback R, Vuorinen T, Waris M, Bjerkner A, Tiveljung‐Lindell A, van den Hoogen BG, Hyypia T, Ruuskanen O. 2007. Human bocavirus and acute wheezing in children. Clin Infect Dis 44: 904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babakir‐Mina M, Ciccozzi M, Perno CF, Ciotti M. 2011. The novel KI, WU, MC polyomaviruses: Possible human pathogens? New Microbiol 34: 1–8. [PubMed] [Google Scholar]
- Babcock HM, Merz LR, Fraser VJ. 2006. Is influenza an influenza‐like illness? Clinical presentation of influenza in hospitalized patients. Infect Control Hosp Epidemiol 27: 266–270. [DOI] [PubMed] [Google Scholar]
- Bellei N, Carraro E, Perosa A, Watanabe A, Arruda E, Granato C. 2008. Acute respiratory infection and influenza‐like illness viral etiologies in Brazilian adults. J Med Virol 80: 1824–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop K, Molenkamp R, van der Ham A, Wolthers K, Beld M. 2008. Rapid detection of human parechoviruses in clinical samples by real‐time PCR. J Clin Virol 41: 69–74. [DOI] [PubMed] [Google Scholar]
- Boivin G, Hardy I, Tellier G, Maziade J. 2000. Predicting influenza infections during epidemics with use of a clinical case definition. Clin Infect Dis 31: 1166–1169. [DOI] [PubMed] [Google Scholar]
- Buecher C, Mardy S, Wang W, Duong V, Vong S, Naughtin M, Vabret A, Freymuth F, Deubel V, Buchy P. 2010. Use of a multiplex PCR/RT‐PCR approach to assess the viral causes of influenza‐like illnesses in Cambodia during three consecutive dry seasons. J Med Virol 82: 1762–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalegno JS, Bouscambert‐Duchamp M, Morfin F, Lina B, Escuret V. 2009. Rhinoviruses, A(H1N1)v, RVS: The race for hivernal pandemics, France 2009–2010. Euro Surveill 14: pii:19390. [PubMed] [Google Scholar]
- Casalegno JS, Ottmann M, Bouscambert‐Duchamp M, Valette M, Morfin F, Lina B. Impact of the 2009 influenza A(H1N1) pandemic wave on the pattern of hibernal respiratory virus epidemics, France, 2009. Euro Surveill 15. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention . 2010. Overview of Influenza Surveillance in the United States. http://www.cdc.gov/flu/weekly/overview.htm.
- Crum‐Cianflone NF, Blair PJ, Faix D, Arnold J, Echols S, Sherman SS, Tueller JE, Warkentien T, Sanguineti G, Bavaro M, Hale BR. 2009. Clinical and epidemiologic characteristics of an outbreak of novel H1N1 (swine origin) influenza A virus among United States military beneficiaries. Clin Infect Dis 49: 1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchamp MB, Casalegno JS, Gillet Y, Frobert E, Bernard E, Escuret V, Billaud G, Valette M, Javouhey E, Lina B, Floret D, Morfin F. 2010. Pandemic A(H1N1)2009 influenza virus detection by real time RT‐PCR: Is viral quantification useful? Clin Microbiol Infect 16: 317–321. [DOI] [PubMed] [Google Scholar]
- Esper FP, Spahlinger T, Zhou L. 2011. Rate and influence of respiratory virus co‐infection on pandemic (H1N1) influenza disease. J Infect 63: 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim A, Ebnet C, Harste G, Pring‐Akerblom P. 2003. Rapid and quantitative detection of human adenovirus DNA by real‐time PCR. J Med Virol 70: 228–239. [DOI] [PubMed] [Google Scholar]
- Henrickson KJ. 2003. Parainfluenza viruses. Clin Microbiol Rev 16: 242–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombrouck A, Sabbe M, Van Casteren V, Wuillaume F, Hue D, Reynders M, Gerard C, Brochier B, Van Eldere J, Van Ranst M, Thomas I. 2011. Viral aetiology of influenza‐like illness in Belgium during the influenza A(H1N1)2009 pandemic. Eur J Clin Microbiol Infect Dis [Epub ahead of print]. DOI: 10.1007/s10096-011-1398-4 [DOI] [PubMed] [Google Scholar]
- Jartti T, Lehtinen P, Vuorinen T, Koskenvuo M, Ruuskanen O. 2004. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J Med Virol 72: 695–699. [DOI] [PubMed] [Google Scholar]
- Kim CO, Nam CM, Lee DC, Han SH, Lee JW. 2010. Clinical predictors of novel influenza A (H1N1)infection in Korea. Yonsei Med J 51: 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna‐Torres VA, Gomez J, Ocana V, Aguilar P, Saldarriaga T, Chavez E, Perez J, Zamalloa H, Forshey B, Paz I, Gomez E, Ore R, Chauca G, Ortiz E, Villaran M, Vilcarromero S, Rocha C, Chincha O, Jimenez G, Villanueva M, Pozo E, Aspajo J, Kochel T. 2009. Influenza‐like illness sentinel surveillance in Peru. PLoS One 4: e6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VJ, Yap J, Cook AR, Tan CH, Loh JP, Koh WH, Lim EA, Liaw JC, Chew JS, Hossain I, Chan KW, Ting PJ, Ng SH, Gao Q, Kelly PM, Chen MI, Tambyah PA, Tan BH. 2011. A clinical diagnostic model for predicting influenza among young adult military personnel with febrile respiratory illness in Singapore. PLoS One 6: e17468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindau C, Tiveljung‐Lindell A, Goh S, Ramqvist T, Allander T. 2009. A single‐tube, real‐time PCR assay for detection of the two newly characterized human KI and WU polyomaviruses. J Clin Virol 44: 24–26. [DOI] [PubMed] [Google Scholar]
- Lu X, Holloway B, Dare RK, Kuypers J, Yagi S, Williams JV, Hall CB, Erdman DD. 2008. Real‐time reverse transcription‐PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol 46: 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IM, Jacob KC, Woolhouse D, Waller K, Syrmis MW, Whiley DM, Siebert DJ, Nissen M, Sloots TP. 2003. Molecular assays for detection of human metapneumovirus. J Clin Microbiol 41: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Taira K, Tsukagoshi H, Itokazu K, Nidaira M, Okano S, Kudaka J, Noda M, Takeda M, Kimura H. 2011. Detection of various respiratory viruses in patients with influenza‐like illness before and after emergence of the 2009 pandemic H1N1 influenza virus in Okinawa. Jpn J Infect Dis 64: 87–89. [PubMed] [Google Scholar]
- Ninove L, Gazin C, Gould EA, Nougairede A, Flahault A, Charrel RN, Zandotti C, de Lamballerie X. 2010. A simple method for molecular detection of Swine‐origin and human‐origin influenza a virus. Vector Borne Zoonotic Dis 10: 237–240. [DOI] [PubMed] [Google Scholar]
- Ninove L, Nougairede A, Gazin C, Thirion L, Delogu I, Zandotti C, Charrel RN, De Lamballerie X. 2011. RNA and DNA bacteriophages as molecular diagnosis controls in clinical virology: A comprehensive study of more than 45,000 routine PCR tests. PLoS One 6: e16142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norja P, Ubillos I, Templeton K, Simmonds P. 2007. No evidence for an association between infections with WU and KI polyomaviruses and respiratory disease. J Clin Virol 40: 307–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougairede A, Ninove L, Zandotti C, de Lamballerie X, Gazin C, Drancourt M, La Scola B, Raoult D, Charrel RN. 2010. Point of care strategy for rapid diagnosis of novel A/H1N1 influenza virus. PLoS One 5: e9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong AK, Chen MI, Lin L, Tan AS, Nwe NW, Barkham T, Tay SY, Leo YS. 2009. Improving the clinical diagnosis of influenza—A comparative analysis of new influenza A (H1N1) cases. PLoS One 4: e8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboni SM, Stella V, Cruz CR, Franca JB, Moreira S, Goncalves L, Nogueira MB, Vidal LR, Almeida SM, Debur MC, Carraro H Jr, dos Santos CN. 2011. Laboratory diagnosis, epidemiology, and clinical outcomes of pandemic influenza A and community respiratory viral infections in southern Brazil. J Clin Microbiol 49: 1287–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razanajatovo NH, Richard V, Hoffmann J, Reynes JM, Razafitrimo GM, Randremanana RV, Heraud JM. 2011. Viral etiology of influenza‐like illnesses in Antananarivo, Madagascar, July 2008 to June 2009. PLoS One 6: e17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Gonzalez R, Wang Z, Xiang Z, Wang Y, Zhou H, Li J, Xiao Y, Yang Q, Zhang J, Chen L, Wang W, Li Y, Li T, Meng X, Zhang Y, Vernet G, Paranhos‐Baccala G, Chen J, Jin Q, Wang J. 2009. Prevalence of human respiratory viruses in adults with acute respiratory tract infections in Beijing, 2005–2007. Clin Microbiol Infect 15: 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renois F, Talmud D, Huguenin A, Moutte L, Strady C, Cousson J, Leveque N, Andreoletti L. 2010. Rapid detection of respiratory tract viral infections and coinfections in patients with influenza‐like illnesses by use of reverse transcription‐PCR DNA microarray systems. J Clin Microbiol 48: 3836–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit PM, Bongers KM, Kuiper RJ, von Rosenstiel IA, Smits PH, Brandjes DP. 2011a. Characterization of 2009 H1N1 pandemic influenza in a population of Dutch children with influenza‐like signs and symptoms. Acta Paediatr 101: 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit PM, Limper M, van Gorp EC, Smits PH, Beijnen JH, Brandjes DP, Mulder JW. 2011b. Adult outpatient experience of the 2009 H1N1 pandemic: Clinical course, pathogens, and evaluation of case definitions. J Infect 62: 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapparel C, Junier T, Gerlach D, Van‐Belle S, Turin L, Cordey S, Muhlemann K, Regamey N, Aubert JD, Soccal PM, Eigenmann P, Zdobnov E, Kaiser L. 2009. New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses. Emerg Infect Dis 15: 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thursky K, Cordova SP, Smith D, Kelly H. 2003. Working towards a simple case definition for influenza surveillance. J Clin Virol 27: 170–179. [DOI] [PubMed] [Google Scholar]
- Tiveljung‐Lindell A, Rotzen‐Ostlund M, Gupta S, Ullstrand R, Grillner L, Zweygberg‐Wirgart B, Allander T. 2009. Development and implementation of a molecular diagnostic platform for daily rapid detection of 15 respiratory viruses. J Med Virol 81: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarz R, Kapoor V, Wu W, Lurio J, Jain K, Mostashari F, Briese T, Lipkin WI. 2011. Longitudinal molecular microbial analysis of influenza‐like illness in New York City May 2009 through May 2010. Virol J 8: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Chern SW, Li Y, Pallansch MA, Anderson LJ. 2008. Sensitive and broadly reactive reverse transcription‐PCR assays to detect novel paramyxoviruses. J Clin Microbiol 46: 2652–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7: 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L, Pyrc K, Jebbink MF, Vermeulen‐Oost W, Berkhout RJ, Wolthers KC, Wertheim‐van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. 2004. Identification of a new human coronavirus. Nat Med 10: 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elden LJ, Nijhuis M, Schipper P, Schuurman R, van Loon AM. 2001. Simultaneous detection of influenza viruses A and B using real‐time quantitative PCR. J Clin Microbiol 39: 196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elden LJ, van Loon AM, van der Beek A, Hendriksen KA, Hoepelman AI, van Kraaij MG, Schipper P, Nijhuis M. 2003. Applicability of a real‐time quantitative PCR assay for diagnosis of respiratory syncytial virus infection in immunocompromised adults. J Clin Microbiol 41: 4378–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elden LJ, van Loon AM, van Alphen F, Hendriksen KA, Hoepelman AI, van Kraaij MG, Oosterheert JJ, Schipper P, Schuurman R, Nijhuis M. 2004. Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real‐time reverse‐transcriptase polymerase chain reaction. J Infect Dis 189: 652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins‐Riedel T, Woegerbauer M, Hollemann D, Hufnagl P. 2002. Rapid diagnosis of enterovirus infections by real‐time PCR on the LightCycler using the TaqMan format. Diagn Microbiol Infect Dis 42: 99–105. [DOI] [PubMed] [Google Scholar]
- Williams BG, Gouws E, Boschi‐Pinto C, Bryce J, Dye C. 2002. Estimates of world‐wide distribution of child deaths from acute respiratory infections. Lancet Infect Dis 2: 25–32. [DOI] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, Luk WK, Poon LL, Wong SS, Guan Y, Peiris JS, Yuen KY. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 79: 884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]


