Abstract
Human parechoviruses (HPeVs) infection is associated with a wide range of clinical syndromes such as respiratory, gastrointestinal, neurologic diseases, and neonatal sepsis‐like illness. The main objective of this study was to investigate the epidemiology of HPeVs infection in hospitalized patients in a period of 2 years. Respiratory samples from 3,525 patients with respiratory syndrome, cerebrospinal fluid (CSF) from 340 patients with neurologic syndrome as well as CSF and plasma samples from five neonatal patients with sepsis‐like illness collected from October 2008 to 2010 were tested retrospectively using HPeV‐specific real‐time RT‐PCR. Phylogenetic analysis of VP3/VP1 region was performed on the positive samples. Fourteen out of 3,525 (0.4%) patients with respiratory syndrome and five out of five patients with sepsis‐like illness were positive for HPeV. In 3/5 patients with sepsis‐like illness multiple samples (e.g., stool, plasma, CSF, or respiratory samples) were available, and HPeV was found in all specimens. In contrast, no positive CSF was detected among the 340 patients with neurologic syndromes. Eleven patients (57.9%) were infected with HPeV1 strain, 7 (36.8%) with HPeV3, and 1 (5.3%) with HPeV6 strains. Ten of the 14 HPeV patients with respiratory syndrome were co‐infected with other respiratory viruses (eight with rhinovirus and two with coronavirus OC43). All five patients with sepsis‐like illness were less than 1 month of age and were infected with HPeV3. Although not circulating at high frequency and unlikely to cause respiratory syndrome, HPeV was associated with severe clinical syndromes in a minority of newborns. J. Med. Virol. 84:686–690, 2012. © 2011 Wiley Periodicals, Inc.
Keywords: human parechovirus, respiratory syndromes, sepsis‐like illness, newborns
INTRODUCTION
Human parechoviruses (HPeVs) belong to the Picornaviridae family and were previously classified as echovirus 22 and echovirus 23 [Hyypiä et al., 1992; Oberste et al., 1999]. Fourteen HPeV genotypes have been described worldwide [Ito et al., 2004; Boivin et al., 2005; Benschop et al., 2006a, 2008; Al‐Sunaidi et al., 2007; Watanabe et al., 2007; Drexler et al., 2009; Li et al., 2009; Calvert et al., 2010; Kim et al., 2010]. Clinical manifestations of HPeV infection including gastrointestinal, respiratory, and neurologic syndrome. HPeV1 and 2 have been associated with mild respiratory and gastrointestinal infections [Abed and Boivin, 2006; Harvala et al., 2008; Harvala and Simmonds, 2009] and, in rare cases, with necrotizing enterocolitis [Birenbaum et al., 1997]. HPeV3 has been associated with sepsis‐like illness, severe neurologic manifestations [Ito et al., 2004; Boivin et al., 2005], and infant death [Levorson et al., 2009; Sedmak et al., 2010]. Although HPeV genotypes 4–14 were identified in a number of different countries, their clinical role remains to be clarified [Benschop et al., 2006a, 2008; Al‐Sunaidi et al., 2007; Watanabe et al., 2007; Drexler et al., 2009; Li et al., 2009; Calvert et al., 2010; Kim et al., 2010]. On the other hand, HPeVs are not included usually in diagnostic panels, thus, the HPeV infection rate might be underestimated.
The aims of present study were to investigate the prevalence of HPeV infections in hospitalized patients in the Northern Italy, analyze the clinical characteristics of HPeV infections, and characterize the circulating strains by phylogenetic analysis.
MATERIALS AND METHODS
In the period October 2008–2010 were tested retrospectively by HPeV‐specific real‐time RT‐PCR [Nix et al., 2008]: (i) 5,283 respiratory samples (nasal swab, nasopharyngeal aspirates, or bronchoalveolar lavage) from 3,525 patients (1,938 pediatric and 1,587 adult) with respiratory syndromes; (ii) 353 cerebrospinal fluid (CSF) from 340 patients with neurologic syndromes (80 pediatric and 260 adult); and (iii) multiple specimens (three CSF, three plasma, five respiratory, and three stool sample) from five neonates with sepsis‐like illness. Patients were stratified with respiratory syndrome according to Baumgarte et al. [2008] comparing patients <2 years of age with respect to patients >2 years of age.
All respiratory samples were tested prospectively with a diagnostic panel of 17 respiratory viruses [Piralla et al., 2009]. CSF samples were tested prospectively for herpes simplex, varicella zoster, cytomegalovirus, polyomavirus, and enterovirus. Virologic and clinical data for HPeV‐positive patients were retrieved from medical records.
Genotyping was performed by sequencing amplification products of the VP3/VP1 region [Harvala et al., 2008] using the BigDye Terminator cycle sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA) and the ABI Prism 3100 DNA sequencer (Applied Biosystems). Sequences were assembled with Sequencer software (4.6 version, Gene Code Corp., Ann Arbor, MI) and multiple sequence alignment was conducted using MEGA version 5.0 [Tamura et al., 2011]. Nucleotide sequences from the VP3/VP1 region have been submitted to GenBank under accession numbers JN112318‐JN112338.
RESULTS
Fourteen out of 3,525 (0.4%) patients with respiratory syndromes and five out of five patients with sepsis‐like illness were positive for HPeV (Table I). None of the patients with neurologic syndromes was positive for HPeV. Of the 14 HPeV‐positive patients with respiratory syndromes 6 (42.9%) had lower respiratory tract infections, while 8 (57.1%) had upper respiratory tract infections. In two premature patients (#11, and #13, born at 25 and 29 weeks, respectively) with sequential respiratory samples HPeV‐RNA persisted for up to 25 days (Table I). The number of patients <2 years of age (n = 831) was comparable with that of patients >2 years of age (n = 1,107). Considering the detection rate of respiratory syndrome caused by HPeV only in patients <2 years the rate increased to 1.6% (13/831) with respect to 0.4% of total rate.
Table I.
Clinical Manifestation of HPeV Infections
| Patient no. | Sex/age (years) | Date sample collection (month/year) | Positive clinical samples | Days follow‐up | Symptoms | Others signs or symptoms | Underlying disease | HPeV typing | Coinfecting virus |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M/14 mos | 10/2008 | NPA | Pharyngitis, cough, pneumonia | Leukocytosis | HPeV1 | HRV | ||
| 2 | M/11 mos | 11/2008 | NPA | Fever, cough, dyspnea | CRP: 1.7 mg/dl | HPeV1 | HRV | ||
| 3 | M/1 | 11/2008 | NPA | Rhinorrhea, cough | Leukocytosis | HPeV1 | hCoV‐OC43 | ||
| 4 | F/3 | 11/2008 | NPA | Fever, pharyngitis | Leucosytosis, CRP: 7.99 mg/dl otalgy, fever convulsion | HPeV6 | hCoV‐OC43 | ||
| 5 | M/1 | 11/2008 | NPA | Rhinorrhea, cough | Otalgy, CRP: 2,75 mg/dl, previous athmatic episode | HPeV1 | |||
| 6 | M/30 | 12/2008 | NPA | Rhinorrhea | Lung transplant (2005) | HPeV3 | HRV | ||
| 7 | F/29 days | 07/2009 | CSF | Fever, sepsis‐like illness | HPeV3 | ||||
| 8 | M/1 | 08/2009 | NPA | Fever, cough, wheezing, pneumonia | Leukocytosis | HPeV3 | |||
| 9 | M/6 mos | 11/2009 | NS | Rhinorrhea | HPeV1 | ||||
| 10 | F/13 mos | 11/2009 | NS | Rhinorrhea | HPeV1 | ||||
| 11 | F/2 mos | 12/2009 | NS | 0 | Cough | Premature (25 weeks), CPAP | HPeV1 | HRV | |
| NS | 11 | Cough | |||||||
| NS | 19 | Cough | |||||||
| NS | 26 | ||||||||
| 12 | F/50 days | 12/2009 | NPA | Rhinorrhea, cough, diarrhea | HPeV1 | HRV | |||
| 13 | F/30 days | 12/2009 | NPA | 0 | Rhinorrhea, cough, pneumonia | Premature (29 weeks) | HPeV1 | HRV | |
| NPA | 16 | Bronchopolmunary displasy | |||||||
| NPA | 29 | ||||||||
| 14 | F/17 mos | 04/2010 | NS | Fever, wheezing, dyspnea | Leukocytosis, saturation O2: 90%. CRP: 5.35 mg/dl, previous asthmatic episodes | HPeV1 | HRV | ||
| 15 | F/1 mos | 07/2010 | CSF, NS, Plasma | Fever, sepsis‐like illness | Diarrhea, otitis | HPeV3 | |||
| 16 | M/2 | 09/2010 | NS | Fever, cough, cough, dyspnea | Saturation O2: 96%, previous asthmatic episodes | HPeV1 | HRV | ||
| 17 | M/14 days | 09/2010 | Plasma | Sepsis‐like illness, encephalitis | HPeV3 | ||||
| 18 | F/11 days | 10/2010 | CSF, NPA, stool | Sepsis‐like illness | HPeV3 | ||||
| 19 | M/17 days | 10/2010 | Plasma, PS | Sepsis‐like illness | HPeV3 |
NPA, nasopharyngeal aspirate; CSF, cerebrospinal fluid; NS, nasal swab; PS, pharyngeal swab; CRP, C reactive protein; HPeV, human parechovirus; HRV, human rhinovirus; hCoV, human coronavirus; mo, month; NA, not available; CPAP, continuous positive airway pressure.
The patients with sepsis‐like illness were younger (median age, 17 days) with respect to the patients with respiratory infections (median age, 365 days; P < 0.01). Among the patients with sepsis‐like illness, HPeV was detected in different clinical specimens in three patients (#15, #18, and #19, Table I). Despite neurologic involvement, in two other patients (#17 and #19) CSF samples were not available and HPeV was detected in plasma samples. HPeV3 infection was community acquired in four patients with sepsis‐like illness (#7, #15, #17, and #19), while in one patient (#18) nosocomial infection was observed. All patients recovered from the infection.
HPeV‐positive patients were identified during the entire study period, 6 (31.6%) in 2008, 7 (36.8%) in 2009, and 6 (31.6%) in 2010. HPeV infections peaked during the fall season with 14/19 (73.7%) positive patients, followed by 5/19 (26.3%) in summer and 1/19 (5.3%) in spring. Ten of 14 (71.4%) HPeV‐positive patients with respiratory syndromes were co‐infected with other respiratory viruses (eight with rhinovirus and two with coronavirus OC43). In five patients with sepsis‐like illness no coinfecting viruses were found.
Eleven of 19 (57.9%) patients were infected with an HPeV1 virus strain, 7/19 (36.8%) patients were infected with an HPeV3 virus strain, and the remaining patient was infected with an HPeV6 virus strain (Fig. 1).
Figure 1.
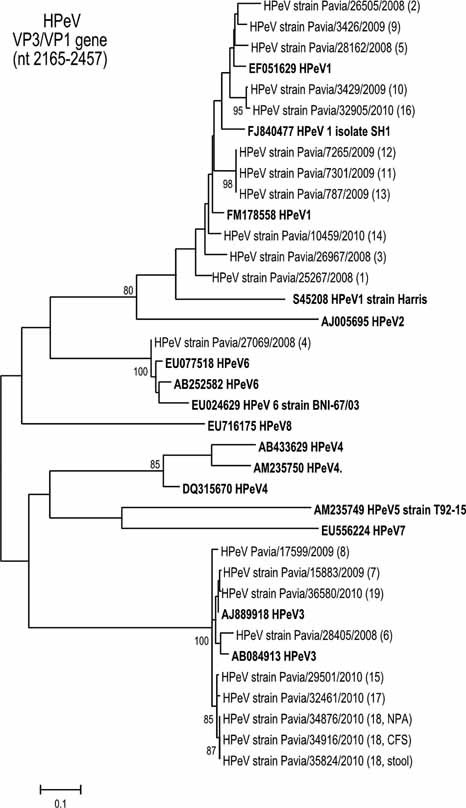
Phylogenetic tree constructed with VP3/VP1 (nt 2,165–2,457 with respect to HPeV1 strain Harris; accession number S45208) nucleotide sequences from reference strains (in bold) and Italian HPeV strains (n = 21) from 19 patients. Phylogenetic analysis was inferred by using the maximum likelihood method based on the Tamura 3‐parameter model as an evolutionary model. A discrete gamma distribution was used to model evolutionary rate differences among sites. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site.
Nucleotide identity among HPeV1 sequence strains ranged from 84.3% to 100%, and in HPeV3 sequences from 92.9% to 100%. HPeV1 sequence strains from patients #11, #12, and #13 were closely related (100% identity) and were indentified in patients hospitalized in the same pediatric unit (Table I). The HPeV3 sequences amplified from different clinical samples of patient #18 were identical to each other. HPeV3 sequence from patient #6 was amplified from a NPA sample collected in 2008 and was divergent from sequences circulating in 2009 and 2010.
DISCUSSION
In agreement with previous data a low prevalence of HPeV in respiratory samples was observed [Harvala et al., 2008] and detection was restricted to the patients <2 years of age. The frequent association of HPeV infections with other respiratory viruses may indicate a less pathogenic role for HPeV compared to the other viruses infecting the respiratory tract. The low number of single infections in our series did not allow to draw major conclusions on the role of HPeV respiratory syndromes. Few data have been published on the duration of virus shedding. In our series, HPeV infection persisted for 25 days in two neonates.
HPeV3 has been shown recently to be an important cause of severe infections in very young children including sepsis‐like illness and encephalitis [Ito et al., 2004; Boivin et al., 2005; Levorson et al., 2009]. In our series, five patients <1 month of age with sepsis‐like illness were infected with HPeV3. In three of these patients, multiple samples (plasma, stool, CSF, or respiratory sample) were HPeV‐positive. These findings support the hypothesis of a disseminated infection caused by HPeV3 [Piňeiro et al., 2010; Sedmak et al., 2010]. The detection of HPeV in different biologic specimens from patients with sepsis‐like illness may help to clarify the extent of the infection. On the other hand, none of patients with neurologic syndromes in the absence of sepsis‐like illness was positive for HPeV, thus suggesting that HPeV neurotropism might be limited to specific clinical conditions. In keeping with other reports [Benschop et al., 2006b, 2008; van der Sanden et al., 2008], the majority of HPeV infections in our study were observed in the fall season.
Despite the limitations of this retrospective study and the need for prospective studies to better define the clinical impact of HPeV infection, some conclusions can be drawn: (i) HPeV does not circulate with high frequency, (ii) the role of HPeV in the respiratory syndrome indicated a bystander phenomena; (iii) HPeV may be associated with severe clinical syndromes in a minority of newborns, and (iv) detection of HPeVs in different clinical samples should always be considered in newborns with sepsis‐like illness.
Acknowledgements
We thank Daniela Sartori for manuscript editing and Laurene Kelly for English revision.
REFERENCES
- Abed Y, Boivin G. 2006. Human parechovirus types 1, 2 and 3 infections in Canada. Emerg Infect Dis 12: 969– 975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Sunaidi M, Williams CH, Hughes PJ, Schnurr DP, Stanway G. 2007. Analysis of a new human parechovirus allows the definition of parechovirus types and the identification of RNA structural domains. J Virol 81: 1013– 1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarte S, de Souza Luna LK, Grywna K, Panning M, Drexler JF, Karsten C, Huppertz HI, Drosten C. 2008. Prevalence, types, and RNA concentrations of human parechoviruses, including a sixth parechovirus type, in stool samples from patients with acute enteritis. J Clin Microbiol 46: 242– 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop KS, Schinkel J, Luken ME, van den Broek PJ, Beersma MF, Menelik N, van Eijk HW, Zaaijer HL, VandenBroucke‐Grauls CM, Beld MG, Wolthers KC. 2006a. Fourth human parechovirus serotype. Emerg Infect Dis 12: 1572– 1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop KS, Schinkel J, Minnaar RP, Pajkrt D, Spanjerberg L, Kraakman HC, Berkhout B, Zaaijer HL, Beld MG, Wolthers KC. 2006b. Human parechovirus infections in Dutch children and the association between serotype and disease severity. Clin Infect Dis 42: 204– 210. [DOI] [PubMed] [Google Scholar]
- Benschop KS, Thomas X, Serpenti C, Molenkamp R, Wolthers K. 2008. High prevalence of human Parechovirus (HPeV) genotypes in the Amsterdam region and identification of specific HPeV variants by direct genotyping of stool samples. J Clin Microbiol 46: 3965– 3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birenbaum E, Handsher R, Kuint J, Dagan R, Raichman B, Mendelson E, Linder N. 1997. Echovirus type 22 outbreak associated with gastro‐intestinal disease in a neonatal intensive care unit. Am J Perinatol 14: 469– 473. [DOI] [PubMed] [Google Scholar]
- Boivin G, Abed Y, Boucher FD. 2005. Human parechovirus 3 and neonatal infections. Emerg Infect Dis 11: 103– 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert J, Chieochansin T, Benschop KS, McWilliam Leitch EC, Drexler JF, Grywna K, da Costa Ribeiro H Jr, Drosten C, Harvala H, Poovorawan Y, Wolthers KC, Simmonds P. 2010. Recombination dynamics of human parechoviruses: Investigation of type‐specific differences in frequency and epidemiological correlates. J Gen Virol 91: 1229– 1238. [DOI] [PubMed] [Google Scholar]
- Drexler JF, Grywna K, Stöcker A, Almeida PS, Medrado‐Ribeiro TC, Eschbach‐Bludau M, Petersen N, da Costa‐Ribeiro‐Jr H, Drosten C. 2009. Novel human parechovirus from Brazil. Emerg Infect Dis 15: 310– 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvala H, Simmonds P. 2009. Human parechoviruses: Biology, epidemiology and clinical significance. J Clin Virol 45: 1– 9. [DOI] [PubMed] [Google Scholar]
- Harvala H, Robertson I, McWilliam Leitch EC, Benschop K, Wolthers KC, Templeton K, Simmonds P. 2008. Epidemiology and clinical associations of human parechovirus respiratory infections. J Clin Microbiol 46: 3446– 3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyypiä T, Horsnell C, Maaronen M, Khan M, Kalkkinen N, Auvinen P, Kinnunen L, Stanway G. 1992. A distinct picornavirus group identified by sequence analysis. Proc Natl Acad Sci 89: 8847– 8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Yamashita T, Tsuzuki H, Takeda N, Sakae K. 2004. Isolation and identification of a novel human parechovirus. J Gen Virol 85: 391– 398. [DOI] [PubMed] [Google Scholar]
- Kim Pham NT, Trinh QD, Takanashi S, Abeysekera C, Abeygunawardene A, Shimizu H, Khamrin P, Okitsu S, Mizuguchi M, Ushijima H. 2010. Novel human parechovirus, Sri Lanka. Emerg Infect Dis 16: 130– 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levorson RE, Jantausch BA, Wiedermann BL, Spiegel HM, Campos JM. 2009. Human parechovirus‐3 infection: Emerging pathogen in neonatal sepsis. Pediatr Infect Dis J 28: 545– 547. [DOI] [PubMed] [Google Scholar]
- Li L, Victoria J, Kapoor A, Naeem A, Shaukat S, Sharif S, Alam MM, Angez M, Zaidi SZ, Delwart E. 2009. Genomic characterization of novel human parechovirus type. Emerg Infect Dis 15: 288– 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nix AW, Maher K, Johansson ES, Niklasson B, Lindberg AM, Pallansch MA, Oberste MS. 2008. Detection of all known parechoviruses by real‐time PCR. J Clin Microbiol 46: 2519– 2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberste MS, Maher K, Pallansch MA. 1999. Specific detection of echoviruses 22 and 23 in cell culture supernatants by RT‐PCR. J Med Virol 58: 178– 181. [DOI] [PubMed] [Google Scholar]
- Piňeiro L, Vicente D, Montes M, Hernández‐Dorronsoro U, Cilla G. 2010. Human Parechovirus in infants with systemic infection. J Med Virol 82: 1790– 1796. [DOI] [PubMed] [Google Scholar]
- Piralla A, Rovida F, Campanini G, Rognoni V, Marchi A, Locatelli F, Gerna G. 2009. Clinical severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J Clin Virol 45: 311– 317. [DOI] [PubMed] [Google Scholar]
- Sedmak G, Nix WA, Jentzen J, Haupt TE, Davis JP, Bhattacharyya S, Pallansch MA, Oberste MS. 2010. Infant deaths associated with human parechovirus infection in Wisconsin. Clin Infect Dis 50: 357– 361. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731– 2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sanden S, de Bruin E, Vennema H, Swanink C, Koopmans M, van der Avoort H. 2008. Prevalence of human parechovirus in The Netherlands in 2000 to 2007. J Clin Microbiol 46: 2884– 2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Oie M, Higuchi M, Nishikawa M, Fujii M. 2007. Isolation and characterization of novel human parechovirus from clinical samples. Emerg Infect Dis 13: 889– 895. [DOI] [PMC free article] [PubMed] [Google Scholar]


