Abstract
Influenza‐like illness can be caused by a wide range of respiratory viruses. In order to investigate the epidemiology of viral pathogens related to influenza‐like illness in children of Wuhan, the largest city in central China, throat swab samples were collected from 1,472 young patients, from July 2008 to June 2010, before and after the occurrence of the 2009 pandemic influenza A (H1N1) virus (pH1N1). It was found that 923 patients (62.7%) were positive for at least 1 virus and 90 patients (9.8%) were detected for multiple (≥2) respiratory viruses by real‐time PCR detection of 16 viruses. Seasonal influenza A virus was the predominant pathogen among all the 16 viruses with a positive rate of 13.3% (196/1,472), which was followed by pH1N1 (159/1,472). It was also noted that the viral distribution pattern in Wuhan changed upon the introduction of the pH1N1 virus. J. Med. Virol. 84:672–678, 2012. © 2011 Wiley Periodicals, Inc.
Keywords: influenza‐like illness, respiratory virus, children, China, epidemiology
INTRODUCTION
The concept of influenza‐like illness has widely been used in influenza surveillance globally. Influenza‐like illness can be attributed to a range of respiratory viruses, including seasonal influenza A and B viruses (FLUAV and FLUBV), respiratory syncytial viruses A and B (RSV A and RSV B), parainfluenza viruses 1–4 (PIVs 1–4), human rhinoviruses (HRV), human adenoviruses (HAdV), and human coronaviruses 229E and OC43 (HCoV‐229E and OC43) [Grondahl et al., 1999; Coiras et al., 2003; Druce et al., 2005; Pierangeli et al., 2007]. Recent advance in molecular detection techniques has facilitated the identification of several new respiratory viruses that may also cause influenza‐like illness symptoms, such as human metapneumovirus (HMPV), human bocavirus (HBoV) and novel strains of coronaviruses (HCoV‐NL63 and HKU1) [Drosten et al., 2003; Crowe, 2004; Allander et al., 2005; Fouchier et al., 2005; Vabret et al., 2005; Koetz et al., 2006; Lau et al., 2006]. Patients infected by these multiple viral pathogens may present with widely overlapping symptoms, which makes clinical diagnosis ambiguous and severely challenges the etiological studies [Eccles, 2005].
Since 2000, a nationwide surveillance network for influenza has been launched in the People's Republic of China. Sentinel hospitals of the network are required to upload their daily influenza‐like illness case numbers to the database of Chinese Centers for Disease Control and Prevention. The influenza‐like illness surveillance data are now used for rough assessment of influenza activity and considered as a rapid yet less specific data source. Unfortunately, the etiology and epidemiology of these influenza‐like illness cases are poorly understood, although the knowledge may play substantial roles in the clinical practice and decision‐making in policy of public health, such as vaccine selection.
Wuhan, the capital of Hubei Province, is the most populous city in central China. Wuhan has a population of approximately 9.8 million people (2010), with about 5.7 million residents in the urban area [Wuhan Bureau of Statistics, 2011]. Located at the heart of China, the city is recognized as a national transportation hub as well as the economic, financial, cultural, and educational center of central China. Wuhan is known for its oppressively humid summers. Years of influenza‐like illness surveillance in Wuhan has demonstrated a typical double peak pattern of southern China: a clear peak in the summer and a less pronounced peak in the winter [Shu et al., 2010].
In order to reveal the viral pathogens related to influenza‐like illness and their epidemiological characteristics, real‐time PCR assay was employed to screen 16 respiratory viruses including the 2009 pandemic influenza A(H1N1) virus (pH1N1) in throat swab samples from 1,472 young patients with influenza‐like illness in Wuhan city. Since the 2‐year surveillance work coincidentally covered the occurrence of pH1N1 in Wuhan, there has also been the opportunity to evaluate the impact of pH1N1 wave on the pattern of respiratory virus distribution.
MATERIALS AND METHODS
Patients and Clinical Specimens
The Children's Hospital of Wuhan, 1 of the 4 national influenza sentinel hospitals in Wuhan and a key pediatric hospital in China, was selected to be the study site from July 2008 to June 2010. The hospital, which has 920 beds, received over 1 million outpatient visits and admitted 30,000 inpatients in 2010, is a tertiary referral center providing both specialty care and primary care to young patients from Wuhan city as well as from other parts of the country. Influenza‐like illness patients visited the hospital were identified according to the WHO recommended definition: the sudden onset of fever >38°C and cough or sore throat in absence of other diagnoses [WHO, 1999]. Children who had fever for no longer than 3 days and were not treated with antiviral drugs were enrolled for this study (n = 1,472). Throat swabs were collected in 2.5 ml of viral transport medium and were delivered to the virology laboratory of the Wuhan Centers for Disease Prevention and Control (Wuhan CDC), the corresponding influenza laboratory in Wuhan. Demographic information of enrolled children was collected and oral informed consent was obtained from the parents of the study subjects. The protocol was approved by the Ethical Committees of the Children's Hospital of Wuhan and the Wuhan CDC.
Molecular Detection of Respiratory Viruses (See Supplementary Material 1 for Technical Details)
Nucleic acid was extracted from 200 µl of specimen using the MagNA Pure LC 2.0 system (Roche Diagnostics, Rotkreuz, Switzerland) following the manufacturer's instruction. Total nucleic acids were eluted in a final volume of 100 µl and tested for 16 respiratory viruses (FLUAV, FLUBV, pH1N1, RSV A, RSV B, PIVs 1–3, HRV, HAdV, HMPV, HBoV, HCoV‐229E, OC43, NL63, and HKU1) using real‐time PCR. The CDC protocol of real‐time RT‐PCR for pH1N1 was applied to detect pH1N1 virus [WHO, 2009]. Assays for the other 15 viruses were conducted as previously described [Tiveljung‐Lindell et al., 2009] (see Supplementary Table S1 for sequences of primers and probes).
Both cultured viral isolates and high‐load patient samples were used as positive controls in the study. Clinical isolates of FLUAV, FLUBV, and pH1N1 were provided by the virus laboratory of Wuhan CDC after confirmation by the WHO Collaborative Center in China (Chinese National Influenza Centre). Patient samples of the other 13 viruses with high viral titer were determined by previously described PCR assays [Esper et al., 2005; Sloots et al., 2006] or the Seeplex RV assay (S‐RV; Seegene, Inc., Seoul, Korea), a multiplex RT‐PCR method that has been shown to run 12 primer sets simultaneously for the detection of 12 respiratory viruses. All patient specimens used as positive controls were confirmed by DNA sequencing [Papadopoulos et al., 1999; Coiras et al., 2004; Huck et al., 2006; Lu and Erdman, 2006; Bharaj et al., 2009] (see Supplementary Table S2 for sequencing primers).
Statistical Analysis
Statistical analyses were conducted using SPSS 11.5. Descriptive statistics were used to characterize the median age and the infection rates. The chi‐squared test was used to compare the infection rates for respiratory viruses among different age groups. P‐value <0.05 was considered to be statistically significant.
RESULTS
General Findings
Clinical samples were collected from children with influenza‐like illness from July 2008 to June 2010. In the first year of the study, 5–15 throat swabs were collected each week. However, the weekly specimen number increased to 20–30 from August 2009, due to the extensive surveillance subsequent to the pandemic of pH1N1. Among the 1,472 enrolled patients, 875 (59.4%) were male and 597 (40.6%) were female. Their age ranged from ≤1 to 14 years with a median age of 3 years and a mean age of 3.71 years (SD = 3.29 years).
According to the results of real‐time PCR screening, 923 (62.7%) patients were found to be positive for at least 1 virus. The monthly positive rates of respiratory viruses ranged from 40% to 95.7% (Fig. 1). Almost all viruses were detected throughout the study years except PIV 2, which was detected only once in 2008. Expectedly, the infection of influenza viruses (include FLUAV 13.3%, FLUBV 6.8%, and pH1N1 10.8%) was dominant in this study population, detected in 455 (30.9%) patients. The detection rate of RSV was 8.3%, followed by HRV 7.6% and HMPV 6.7% (Table I).
Figure 1.
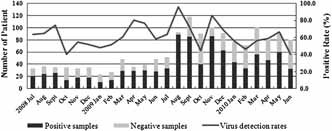
Patients' monthly distribution and virus detection rate from July 2008 to June 2010.
Table I.
The Frequency of Virus Infection in ILI in Different Age Groups
| Patients' no. | ≤1 Year, n = 344 | 1–3 Years, n = 522 | >3 Years, n = 606 | Total, n = 1,472 | P‐value |
|---|---|---|---|---|---|
| FLUAV | 55 (16.0)a | 69 (13.2) | 72 (11.9) | 196 (13.3) | 0.176 |
| pH1N1 | 9 (2.6) | 17 (3.3) | 133 (21.9) | 159 (10.8) | <0.001 |
| FLUBV | 3 (0.9) | 26 (5.0) | 71 (11.7) | 100 (6.8) | <0.001 |
| RSV A | 6 (1.7) | 5 (1.0) | 3 (0.5) | 14 (1.0) | 0.132 |
| RSV B | 43 (12.5) | 53 (10.2) | 12 (2.0) | 108 (7.3) | <0.001 |
| HRV | 22 (6.4) | 52 (10.0) | 38 (6.3) | 112 (7.6) | 0.105 |
| HMPV | 19 (5.5) | 50 (9.6) | 29 (4.8) | 98 (6.7) | 0.016 |
| HAdV | 14 (4.1) | 41 (7.9) | 31 (5.1) | 86 (5.8) | 0.221 |
| PIV 1 | 1 (0.3) | 9 (1.7) | 9 (1.5) | 19 (1.3) | 0.580 |
| PIV 2 | 0 | 1 (0.2) | 0 | 1 (0.07) | N/A |
| PIV 3 | 24 (7.0) | 28 (5.4) | 5 (0.8) | 57 (3.9) | <0.001 |
| HCoV | 20 (5.8) | 20 (3.8) | 11 (1.8) | 51 (3.5) | 0.040 |
| HBoV | 9 (2.6) | 11 (2.1) | 1 (0.2) | 21 (1.4) | 0.001 |
| Frequency of viral etiology | 55.80% | 64.90% | 64.70% |
FLUAV, seasonal influenza A virus; FLUBV, influenza B virus; pH1N1, the 2009 pandemic influenza A(H1N1) virus; RSV, respiratory syncytial virus; HRV, human rhinovirus; HMPV, human metapneumovirus; HAdV, human adenovirus; PIV, parainfluenza virus; HCoV, human coronavirus; HBoV, human bocavirus.
Numbers in parentheses are percentages.
Viral Distribution Across Age Groups and Genders
Viral frequency was analyzed with relation to age and three groups were studied: Group 1: ≤1 year (n = 344); Group 2: 1–3 years (n = 522), and Group 3: >3 years (n = 606). Among these age groups, Group 1 stood for the infants, Group 2 included toddlers who had not yet entered nursery and usually stayed at home, and Group 3 included children in the nursery and school who had more contacts with their peers and the outer environment. The total frequency of viral positives in the three age groups was 55.8%, 64.9%, 64.7%, respectively. The frequencies of FLUBV, pH1N1, RSV B, PIV 3, HCoVs, HBoV, and HMPV among three groups were significantly different, while FLUAV, PIV 1, RSV A, HAdV, and HRV did not show significant association with age. RSV B, HMPV, PIV 3, HCoVs, and HBoV associated influenza‐like illness predominated in children younger than 3 years while FLUBV and pH1N1 predominated in children older than 3 years (Table I). There was no significant difference in viral infection between male and female (data not shown).
Seasonal Prevalence of Respiratory Virus Infections
The rates of virus detection were not distributed equally during the study period. For FLUAV, the virus detected at the highest frequency, three major peaks were observed and two of them were in the summer (Fig. 2a). On the other hand, FLUBV was less frequent during summer, but had two major peaks in the spring of 2009 and 2010 (Fig. 2a; see Fig. S1 for detailed information about the subtyping of seasonal influenza viruses). pH1N1 was only present during September–December 2009. However, it was the second most frequent viral agent and contributed 78 of the 86 viral positive samples (90.7%) in the outbreak peaked in November 2009.
Figure 2.
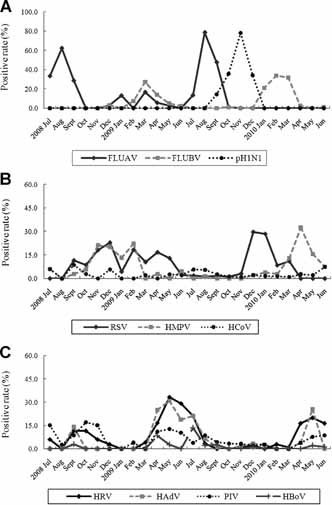
Monthly distribution of respiratory viruses detected by real‐time PCR. A: FLUAV, seasonal influenza A virus; FLUBV, influenza B virus; pH1N1, the 2009 pandemic influenza A(H1N1) virus; B: RSV, respiratory syncytial virus; HMPV, human metapneumovirus; HCoV, human coronavirus; C: HRV, human rhinovirus; HAdV, human adenovirus; PIV, parainfluenza virus; HBoV, human bocavirus.
It was noted that a peak of HMPV infection was coinciding with the RSV‐associated peak from October 2008 to March 2009 (Fig. 2b), but the temporal distribution of RSV and HMPV infections in the following season showed that the episodes shifted 1 and 4 months forward, respectively, where the rise in detection of HMPV occurred independently from the rate of RSV infection. HAdV and PIV showed three peaks of infection coinciding with the HRV‐associated peaks (Fig. 2c). It seemed that the temporal prevalence patterns of these three viruses were opposite to the circulation pattern of influenza viruses. HCoVs appeared sporadically during the study years (Fig. 2b) and HBoV, the least detected viral pathogen in the panel, was observed to be more prevalent in the summer (Fig. 2c).
Viral Etiology Before and After the Appearance of the pH1N1 Virus
Four time periods were assessed in the analysis: July–December, 2008 (1st period), January–June, 2009 (2nd period), July–December, 2009 (3rd period), and January–June, 2010 (4th period). These periods correspond to the time intervals before and soon after the first community‐acquired case of pH1N1 confirmed in Wuhan (August 26, 2009).
During the 1st period, 133 positive results were obtained from 121 positive patients (12 patients with co‐infections). Seasonal influenza A virus was predominant comprising 33% of the confirmed cases, followed by RSV (16%), PIV (15%), HMPV (14%), and HRV (10%), which accounted for 85% of the total viral agents. From July to December 2009 (3rd period), the pH1N1 virus was the most prevalent (37%) followed by FLUAV (31%), RSV (8%), HCoV (6%), HAdV (5%), and PIV (5%). Comparing the 2nd period with the 4th period, the predominant viruses from January to June, 2009 were HRV, HAdV, RSV, FLUBV, and FLUAV (in descending order of frequency), accounting for 71% of all positive results. Nevertheless, during January–June 2010 (4th period), FLUBV, HMPV, HRV, RSV, and HAdV accounted for most of the etiologies of influenza‐like illness (88%; Fig. 3).
Figure 3.
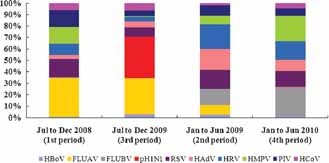
Viral etiology before and after the appearance of the novel pH1N1 virus in Wuhan. HBoV, human bocavirus; FLUAV, seasonal influenza A virus; FLUBV, influenza B virus; pH1N1, the 2009 pandemic influenza A(H1N1) virus; RSV, respiratory syncytial virus; HAdV, human adenovirus; Rhino, human rhinovirus; HMPV, human metapneumovirus; PIV, parainfluenza virus; HCoV, human coronavirus.
Co‐Detection of Respiratory Viruses
Multiple (≥2) respiratory viruses were detected in 90 positive cases. Dual infections were found in 82 of these patients, while triple infections were identified in 7 patients and only one case had tetramerous infections (Table II). Adenovirus, detected in 31 cases, was the most frequently found viral agent in co‐infections. Seasonal influenza A virus was co‐detected in 25 cases followed by RSV B in 20 cases and HRV in 19 cases. Co‐infection rates were significantly higher in the two younger groups as compared to the children over 3‐year old (<1 year, P < 0.001; 1–3 years, P = 0.002).
Table II.
Co‐Detection of Respiratory Viruses in Children With ILI
| Virus co‐detection status | Positive cases among age groups | Total cases, n = 1,472 | ||
|---|---|---|---|---|
| <1 Year, n = 344 | 1–3 Years, n = 522 | >3 Years, n = 606 | ||
| HAdV + HRV | 4 | 1 | 2 | 7 |
| FLUAV + HCoV‐NL63 | 2 | 4 | 1 | 7 |
| HAdV + HBoV | 2 | 3 | 0 | 5 |
| FLUAV + PIV3 | 2 | 2 | 1 | 5 |
| FLUAV + HCoV‐OC43 | 1 | 4 | 0 | 5 |
| RSV B + pH1N1 | 2 | 2 | 0 | 4 |
| HAdV + RSV B | 2 | 2 | 0 | 4 |
| HAdV + FLUAV | 0 | 2 | 1 | 3 |
| RSV B + HMPV | 3 | 0 | 0 | 3 |
| HAdV + PIV3 | 1 | 1 | 0 | 2 |
| HRV + PIV3 | 1 | 1 | 0 | 2 |
| PIV3 + HCoV‐OC43 + HMPV | 1 | 1 | 0 | 2 |
| RSV B + HCoV‐OC43 | 1 | 0 | 1 | 2 |
| HAdV + HMPV | 0 | 1 | 1 | 2 |
| FLUBV + HMPV | 0 | 1 | 1 | 2 |
| FLUAV + HMPV | 0 | 2 | 0 | 2 |
| HRV + HMPV | 0 | 2 | 0 | 2 |
| HRV + HBoV | 0 | 2 | 0 | 2 |
| HRV + HCoV‐HKU1 | 0 | 0 | 2 | 2 |
| HCoV‐OC43 + pH1N1 | 0 | 0 | 2 | 2 |
| FLUAV + HBoV | 1 | 0 | 0 | 1 |
| RSV B + HBoV | 1 | 0 | 0 | 1 |
| RSV A + PIV3 | 1 | 0 | 0 | 1 |
| RSV B + PIV3 | 1 | 0 | 0 | 1 |
| RSV B + HCoV‐HKU1 | 1 | 0 | 0 | 1 |
| RSV B + PIV3 + HBoV | 1 | 0 | 0 | 1 |
| HAdV + PIV3 + RSV B + HBoV | 1 | 0 | 0 | 1 |
| HRV + FLUBV | 0 | 1 | 0 | 1 |
| HRV + pH1N1 | 0 | 1 | 0 | 1 |
| HCoV‐OC43 + HCoV‐NL63 | 0 | 1 | 0 | 1 |
| FLUAV + RSV A | 0 | 1 | 0 | 1 |
| HBoV + PIV3 | 0 | 1 | 0 | 1 |
| PIV1 + HCoV‐229E | 0 | 1 | 0 | 1 |
| HAdV + HCoV‐NL63 | 0 | 1 | 0 | 1 |
| HAdV + PIV3 + HRV | 0 | 1 | 0 | 1 |
| FLUBV + HCoV‐NL63 | 0 | 0 | 1 | 1 |
| FLUBV + HCoV‐229E | 0 | 0 | 1 | 1 |
| HRV + PIV1 | 0 | 0 | 1 | 1 |
| pH1N1 + PIV1 | 0 | 0 | 1 | 1 |
| HAdV + PIV1 | 0 | 0 | 1 | 1 |
| HAdV + RSV A | 0 | 0 | 1 | 1 |
| HAdV + HCoV‐OC43 | 0 | 0 | 1 | 1 |
| HAdV + PIV3 + RSV A | 0 | 0 | 1 | 1 |
| RSV B + RSV A + HBoV | 0 | 0 | 1 | 1 |
| Total | 29 (8.4)a | 40 (7.7) | 21 (3.5) | 90 (6.1) |
Numbers in parentheses are percentages.
DISCUSSION
From July 2008 to June 2010, 1,472 outpatients aged from 1 month to 14 years were enrolled in this surveillance program of viral etiology in influenza‐like illness. To the best of authors' knowledge, this is the first study to investigate the etiologic agents associated with influenza‐like illness in Chinese children. Sixteen different respiratory viruses were detected in 923 samples (62.7% of the total), which was higher than the results (36–61.8%) of previous studies [Lina et al., 1996; Vabret et al., 2003; van Gageldonk‐Lafeber et al., 2005; Bellei et al., 2008]. Our data indicated that respiratory viruses might be the major pathogens responsible for influenza‐like illness in children in Wuhan, China. The most common viral pathogens detected in this study included seasonal influenza A virus, pH1N1, HRV, RSV B, influenza B virus, HMPV, and HAdV. Infections with multiple viruses were observed in 9.8% of positive cases, suggesting that co‐infection is not uncommon among children with influenza‐like illness.
Of interest, the change of viral distribution pattern was observed in this study upon the introduction of the pH1N1 virus. It was observed that the distribution pattern of influenza‐like illness related viruses in Wuhan during May–September 2009, was similar to that observed in 2008. However, because of occurrence of the pH1N1, the circulation pattern of several viruses changed in the following months: the activities of PIV and HRV were greatly reduced compared to the non‐pandemic period and the circulation of HMPV in 2009–2010 was delayed for over 3 months. Previous serologic studies have demonstrated that HMPV mainly circulated during the winter and early spring [Maggi et al., 2003; Peiris et al., 2003], which supports our observation in 2008–2009. However, in 2009–2010, the HMPV epidemic season was delayed to March–June 2010. It is unclear which factor(s) have contributed to this delay, yet several factors might be accounted for the pattern‐changing: weather conditions, increased hygiene measures implemented following the pandemic plan and viral interference. Nevertheless, it is worth noting that it was a 2‐year study and the possibility of the wider pattern in temporal dynamics which might not be reflected in the data from this timeframe could not be ruled out. Future studies in longer timeframe are necessary in order to better understand the etiologic pattern.
Intriguingly, the epidemiology for pH1N1 and HRV in Wuhan in September, 2009 was opposite to the pattern observed in European countries during the first part of autumn 2009 [Anestad and Nordbo, 2009; Casalegno et al., 2009, 2010; Linde et al., 2009]. Linde et al. reported that the pH1N1 circulation in Sweden increased after the end of the summer holiday, but after 4 weeks of increasing activity the spread suddenly declined. On the other hand, the number of samples sent for influenza testing was increasing until week 36 and HRV was responsible for the apparent increase of influenza‐like illness reported in early September, which may have delayed the onset of the pH1N1 pandemic [Linde et al., 2009]. A similar pattern was also observed in France [Casalegno et al., 2010]. However, result of this study was in good agreement with an earlier report from Peru [Laguna‐Torres et al., 2010], which tended to support higher transmission potential of pH1N1 than non‐pH1N1 influenza A viruses, as also reported elsewhere [Fraser et al., 2009; Munayco et al., 2009; Nishiura et al., 2009]. This phenomenon could be partially explained by higher population susceptibility to pH1N1 than non‐pH1N1 virus to which the population might have acquired partial immunity in previous years via prior natural exposure with antigenically related strains and annual immunization campaigns of fraction of the high‐risk population, with live attenuated vaccines [Frank and Taber, 1983; Belshe, 2004; Chowell et al., 2008]. Theoretical epidemiology suggests that in populations challenged by multiple infectious agents, the one with the highest reproduction number (fitness) will dominate the transmission dynamics [Anderson and May, 1991]. The reproduction number implicitly accounts for the intrinsic virus transmissibility and the background population immunity.
The multiple respiratory virus detection platform applied in this study may be used to monitor the circulation of existing viruses in the population, and current results could serve as the baseline for the surveillance of unusual prevalence of a specific virus and the detection of further changes of viral distribution patterns. In addition, results of this study also suggest the broad diagnostic testing could be a valuable tool in understanding the epidemic situation of newly emerging viruses like pH1N1 and helping public health policy makers choose proper disease preventive measures. The first pH1N1 case in Wuhan was an imported case diagnosed on May 31, 2009, while the first domestic case was not reported from sentinel surveillance until August. As the study discovered, during the period from June to August 2009, several respiratory viruses (FLUAV, HAdV, HRV, and HCoVs) were circulating simultaneously with the early spread of pH1N1, as reported elsewhere [Casalegno et al., 2009; Munayco et al., 2009]. These observations indicated that some rigorous, expensive preventive measures, such as the antiviral treatment of local influenza‐like illness patient, the quarantine and preventive dosing of their close contacts and the closure of schools having suspected pH1N1 patients [Ministry of Education and Ministry of Health, 2009], had no need to be implemented immediately after the report of the first imported case, since the relatively high influenza‐like illness rate was actually caused by pathogens other than the novel virus pH1N1.
One shortcoming of the sentinel surveillance program is the potential sampling bias. As this study was limited to the Children's Hospital of Wuhan, its results may not be representative of the entire children population. However, one advantage provided by sentinel surveillance systems is the ability to identify increasing trends in the number of patients seeking medical attention due to influenza‐like illness symptoms, and identifying the viruses related to such increases requires fewer resources than a population‐based study. Future studies will be warranted to determine the etiologic agents for the over 30% of influenza‐like illness cases that remain undiagnosed. Bacterial pathogens are likely responsible for some of those undiagnosed cases and will need to be further determined. Such analyses are necessary to better understand the influenza‐like illness‐associated pathogens in circulation in this region. The potential occurrence of false‐negative results due to the variable sensitivity of the laboratory techniques and the type of biological samples cannot be completely ruled out.
In conclusion, this is the first study in China to characterize 16 common respiratory viruses in pediatric influenza‐like illness during a 2‐year consecutive period in a city. Using real‐time PCR, it was able to identify a wide variety of viruses and differentiate highly pathogenic viruses from less virulent seasonal respiratory viruses. Results of this investigation provide the information on the etiology and epidemiology of influenza‐like illness in Chinese children, which is useful for the guidance of pediatric clinical practice as well as public health policy.
Supporting information
Additional Supporting Information may be found in the online version of this article.
Supplementary Material 1
Supplementary Material 2
Supplementary Tables
Acknowledgements
We thank the Chinese National Influenza Centre for the technical guidance in our study.
REFERENCES
- Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung‐Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA 102: 12891– 12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, May RM. 1991. Infectious diseases of humans: Dynamic and control. New York: Oxford University Press. [Google Scholar]
- Anestad G, Nordbo S. 2009. Interference between outbreaks of respiratory viruses. Euro Surveill 14: pii:19367. [PubMed] [Google Scholar]
- Bellei N, Carraro E, Perosa A, Watanabe A, Arruda E, Granato C. 2008. Acute respiratory infection and influenza‐like illness viral etiologies in Brazilian adults. J Med Virol 80: 1824– 1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshe RB. 2004. Current status of live attenuated influenza virus vaccine in the US. Virus Res 103: 177– 185. [DOI] [PubMed] [Google Scholar]
- Bharaj P, Sullender WM, Kabra SK, Mani K, Cherian J, Tyagi V, Chahar HS, Kaushik S, Dar L, Broor S. 2009. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007 Virol J 6: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalegno JS, Bouscambert‐Duchamp M, Morfin F, Lina B, Escuret V. 2009. Rhinoviruses, A(H1N1)v, RVS: The race for hivernal pandemics, France 2009–2010. Euro Surveill 14: pii:19390. [PubMed] [Google Scholar]
- Casalegno JS, Ottmann M, Duchamp MB, Escuret V, Billaud G, Frobert E, Morfin F, Lina B. 2010. Rhinoviruses delayed the circulation of the pandemic influenza A (H1N1) 2009 virus in France. Clin Microbiol Infect 16: 326– 329. [DOI] [PubMed] [Google Scholar]
- Chowell G, Miller MA, Viboud C. 2008. Seasonal influenza in the United States, France, and Australia: Transmission and prospects for control. Epidemiol Infect 136: 852– 864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coiras MT, Perez‐Brena P, Garcia ML, Casas I. 2003. Simultaneous detection of influenza A, B, and C viruses, respiratory syncytial virus, and adenoviruses in clinical samples by multiplex reverse transcription nested‐PCR assay. J Med Virol 69: 132– 144. [DOI] [PubMed] [Google Scholar]
- Coiras MT, Aguilar JC, Garcia ML, Casas I, Perez‐Brena P. 2004. Simultaneous detection of fourteen respiratory viruses in clinical specimens by two multiplex reverse transcription nested‐PCR assays. J Med Virol 72: 484– 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JE, Jr. 2004. Human metapneumovirus as a major cause of human respiratory tract disease. Pediatr Infect Dis J 23: S215– S221. [DOI] [PubMed] [Google Scholar]
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348: 1967– 1976. [DOI] [PubMed] [Google Scholar]
- Druce J, Tran T, Kelly H, Kaye M, Chibo D, Kostecki R, Amiri A, Catton M, Birch C. 2005. Laboratory diagnosis and surveillance of human respiratory viruses by PCR in Victoria, Australia, 2002–2003. J Med Virol 75: 122– 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles R. 2005. Understanding the symptoms of the common cold and influenza. Lancet Infect Dis 5: 718– 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper F, Weibel C, Ferguson D, Landry ML, Kahn JS. 2005. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J Infect Dis 191: 492– 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Rimmelzwaan GF, Kuiken T, Osterhaus AD. 2005. Newer respiratory virus infections: Human metapneumovirus, avian influenza virus, and human coronaviruses. Curr Opin Infect Dis 18: 141– 146. [DOI] [PubMed] [Google Scholar]
- Frank AL, Taber LH. 1983. Variation in frequency of natural reinfection with influenza A viruses. J Med Virol 12: 17– 23. [DOI] [PubMed] [Google Scholar]
- Fraser C, Donnelly CA, Cauchemez S, Hanage WP, Van Kerkhove MD, Hollingsworth TD, Griffin J, Baggaley RF, Jenkins HE, Lyons EJ, Jombart T, Hinsley WR, Grassly NC, Balloux F, Ghani AC, Ferguson NM, Rambaut A, Pybus OG, Lopez‐Gatell H, Alpuche‐Aranda CM, Chapela IB, Zavala EP, Guevara DM, Checchi F, Garcia E, Hugonnet S, Roth C. 2009. Pandemic potential of a strain of influenza A (H1N1): Early findings. Science 324: 1557– 1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondahl B, Puppe W, Hoppe A, Kuhne I, Weigl JA, Schmitt HJ. 1999. Rapid identification of nine microorganisms causing acute respiratory tract infections by single‐tube multiplex reverse transcription‐PCR: Feasibility study. J Clin Microbiol 37: 1– 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck B, Scharf G, Neumann‐Haefelin D, Puppe W, Weigl J, Falcone V. 2006. Novel human metapneumovirus sublineage. Emerg Infect Dis 12: 147– 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koetz A, Nilsson P, Linden M, van der Hoek L, Ripa T. 2006. Detection of human coronavirus NL63, human metapneumovirus and respiratory syncytial virus in children with respiratory tract infections in south‐west Sweden. Clin Microbiol Infect 12: 1089– 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguna‐Torres VA, Gomez J, Aguilar PV, Ampuero JS, Munayco C, Ocana V, Perez J, Gamero ME, Arrasco JC, Paz I, Chavez E, Cruz R, Chavez J, Mendocilla S, Gomez E, Antigoni J, Gonzalez S, Tejada C, Chowell G, Kochel TJ. 2010. Changes in the viral distribution pattern after the appearance of the novel influenza A H1N1 (pH1N1) virus in influenza‐like illness patients in Peru. PLoS ONE 5: e11719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Yip CC, Tse H, Tsoi HW, Cheng VC, Lee P, Tang BS, Cheung CH, Lee RA, So LY, Lau YL, Chan KH, Yuen KY. 2006. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol 44: 2063– 2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lina B, Valette M, Foray S, Luciani J, Stagnara J, See DM, Aymard M. 1996. Surveillance of community‐acquired viral infections due to respiratory viruses in Rhone‐Alpes (France) during winter 1994 to 1995. J Clin Microbiol 34: 3007– 3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linde A, Rotzén‐Ostlund M, Zweygberg‐Wirgart B, Rubinova S, Brytting M. 2009. Does viral interference affect spread of influenza? Euro Surveill 14: pii:19354. [PubMed] [Google Scholar]
- Lu X, Erdman DD. 2006. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch Virol 151: 1587– 1602. [DOI] [PubMed] [Google Scholar]
- Maggi F, Pifferi M, Vatteroni M, Fornai C, Tempestini E, Anzilotti S, Lanini L, Andreoli E, Ragazzo V, Pistello M, Specter S, Bendinelli M. 2003. Human metapneumovirus associated with respiratory tract infections in a 3‐year study of nasal swabs from infants in Italy. J Clin Microbiol 41: 2987– 2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ministry of Education and Ministry of Health . 2009. Guidelines for the H1N1 influenza prevention and control in the school (Trial).
- Munayco CV, Gomez J, Laguna‐Torres VA, Arrasco J, Kochel TJ, Fiestas V, Garcia J, Perez J, Torres I, Condori F, Nishiura H, Chowell G. 2009. Epidemiological and transmissibility analysis of influenza A(H1N1)v in a southern hemisphere setting: Peru. Euro Surveill 14: pii:19299. [PubMed] [Google Scholar]
- Nishiura H, Castillo‐Chavez C, Safan M, Chowell G. 2009. Transmission potential of the new influenza A(H1N1) virus and its age‐specificity in Japan. Euro Surveill 14: pii:19227. [DOI] [PubMed] [Google Scholar]
- Papadopoulos NG, Hunter J, Sanderson G, Meyer J, Johnston SL. 1999. Rhinovirus identification by BglI digestion of picornavirus RT‐PCR amplicons. J Virol Methods 80: 179– 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JS, Tang WH, Chan KH, Khong PL, Guan Y, Lau YL, Chiu SS. 2003. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis 9: 628– 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierangeli A, Gentile M, Di Marco P, Pagnotti P, Scagnolari C, Trombetti S, Lo Russo L, Tromba V, Moretti C, Midulla F, Antonelli G. 2007. Detection and typing by molecular techniques of respiratory viruses in children hospitalized for acute respiratory infection in Rome, Italy. J Med Virol 79: 463– 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu YL, Fang LQ, de Vlas SJ, Gao Y, Richardus JH, Cao WC. 2010. Dual seasonal patterns for influenza, China. Emerg Infect Dis 16: 725– 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD, Mackay IM. 2006. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol 35: 99– 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiveljung‐Lindell A, Rotzen‐Ostlund M, Gupta S, Ullstrand R, Grillner L, Zweygberg‐Wirgart B, Allander T. 2009. Development and implementation of a molecular diagnostic platform for daily rapid detection of 15 respiratory viruses. J Med Virol 81: 167– 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A, Mourez T, Gouarin S, Petitjean J, Freymuth F. 2003. An outbreak of coronavirus OC43 respiratory infection in Normandy, France. Clin Infect Dis 36: 985– 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret A, Mourez T, Dina J, van der Hoek L, Gouarin S, Petitjean J, Brouard J, Freymuth F. 2005. Human coronavirus NL63, France. Emerg Infect Dis 11: 1225– 1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gageldonk‐Lafeber AB, Heijnen ML, Bartelds AI, Peters MF, van der Plas SM, Wilbrink B. 2005. A case–control study of acute respiratory tract infection in general practice patients in The Netherlands. Clin Infect Dis 41: 490– 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhan Bureau of Statistics . 2011. Communiqué on major data of the Sixth National Census in Wuhan.
- WHO . 1999. WHO recommended surveillance standards. Second edition. [Google Scholar]
- WHO . 2009. CDC protocol of realtime RTPCR for influenza A (H1N1).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article.
Supplementary Material 1
Supplementary Material 2
Supplementary Tables


